1. Introduction
Plants are exposed to various stress factors at the same time or at different times of their lives. Therefore, plant plasticity is essential to overcome such stresses and to establish acclimation mechanisms that allow plants to respond to reiterated stresses [
1]. The ability of the plants to respond and adapt to recurring biotic and abiotic stresses is essential for their survival because of their sessile nature. The ability to survive is linked to population fitness and this is positively correlated to heterozygosity, population size, and quantitative genetic variation [
2]. The study of links between genetic diversity and phenotypic changes that result from habitat-mediated adaptation gives insight into how the mosaic of the natural landscape shapes the genomes of the organisms distributed across its vastness [
3].
Mangrove forests occur along tropical and subtropical coastlines that are subjected to tide, salinity and nutrient variations [
4,
5]. Plants from different taxonomic groups have developed mechanisms to survive exclusively in the mangrove ecosystem. The three most important adaptive traits found in mangrove plants are vivipary, salt exclusion, and aerial roots [
6]. However, each mangrove species has its own physiological mechanism of achieving these traits.
All mangrove plant species are well adapted to their ecosystem. However, the natural variation of salinity, nutrient, and tides in these habitats results in an environmental gradient usually causing morphological differences in mangrove plants. The salt marsh is a plain where brackish water enters with higher tides, accumulates, and then evaporates leading to high salt content in both the soil and interstitial water. The unfavorable conditions near a salt marsh, such as low levels of nutrients, high salt content, hypoxic soil are very restrictive for the growth of mangrove plants [
4,
7,
8,
9]. The most favorable areas are located at the riverside. Daily tides bring nutrients and propagules (seedlings) dispersed by water [
10]. The conditions at the riverside are favorable to plantlet establishment and plant survival, allowing for increased genetic diversity with the constant arrival of new genotypes with the tides.
Environmental heterogeneity may shape the distribution and abundance of genotypes by influencing gene flow and selection [
11]. Due to the stressful conditions near the salt marsh, only a few plants would be able to grow and survive producing viable propagules. In the long-term, the limited numbers of genotypes present in the stressful area and its reduced population size would result in the loss of genetic diversity near the salt marsh by either selection or drift. It is possible that genetic diversity is correlated to morphological divergence and survival of the plants. Consequently, in this stressful environmental condition, gene flow might not counteract genetic differentiation because of the effects of evolutionary forces such as selection or genetic drift.
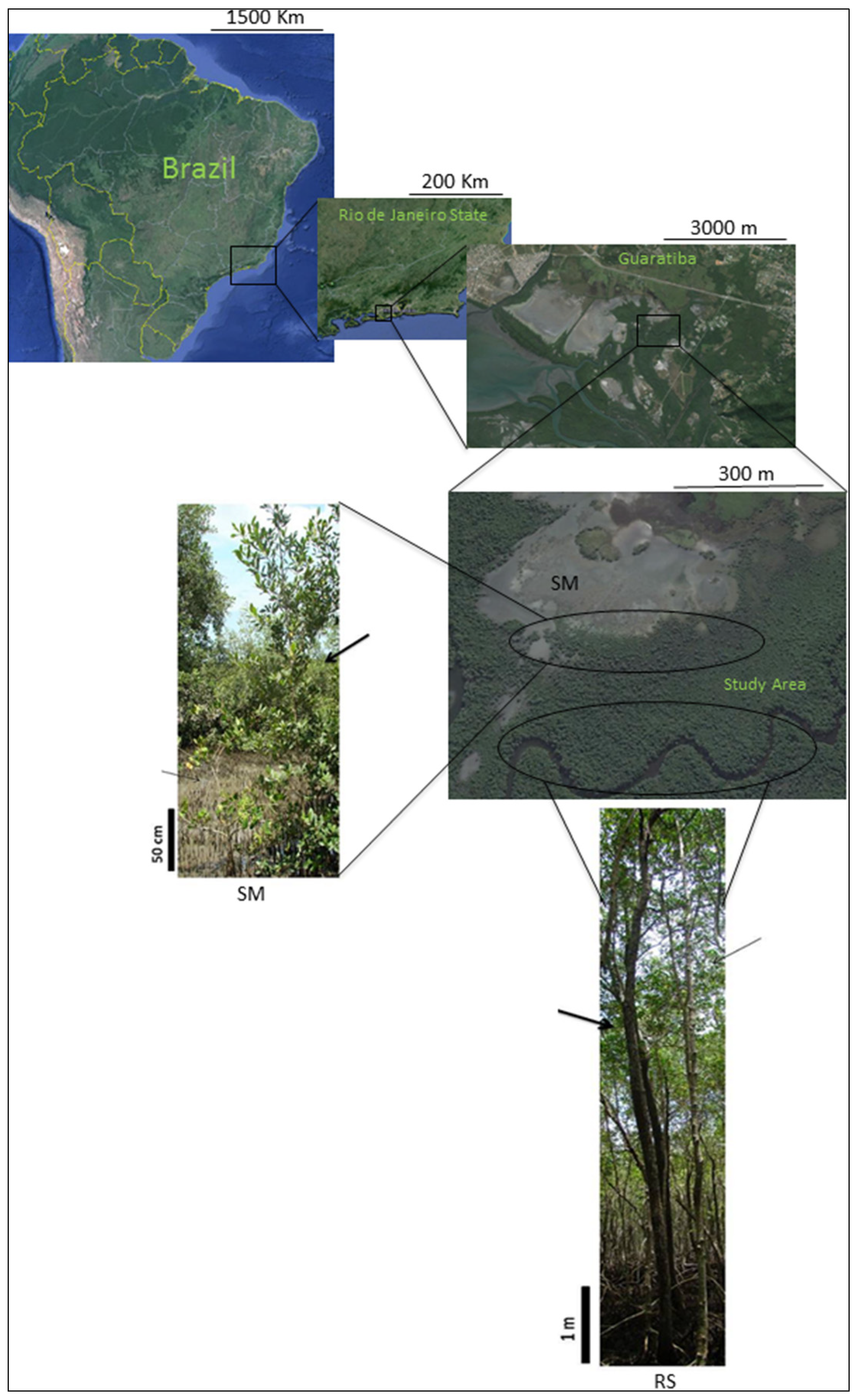
Inside the Sepetiba Bay’s mangrove habitat, salt marsh formations induce morphological alterations in the species
Avicennia schaueriana Stapf & Leechm. ex Moldenke (Acanthaceae) and
Laguncularia racemosa (L.) Gaertn (Combretaceae), such as reduced tree height, contorted, thin trunks, and an increase in the number of lower branches.
Avicennia schaueriana’s plants reach 3 meters in height and
Laguncularia racemosa plants reach only 1.5 meters. They both have contorted stems and many lower branches. By contrast, these species develop very well at the riverside where they reach 8 meters in height with a long, straight trunk and canopy branches (
Figure 1). Due to the proximity of those areas inside Sepetiba’s Bay mangrove, the divergent morphologies of the plants located in each area raise questions about the level of genetic differentiation between the areas. The morphological variation found inside these natural populations could be correlated to genetic variation of the plants from each area. Our aim was to assess genetic diversity of two mangrove species, occurring in these areas,
Avicennia schaueriana and
Laguncularia racemosa, which have morphological alterations.
Figure 1.
Location map of Guaratiba’s populations, with a picture illustrating the morphological alterations of Avicennia schaueriana and Laguncularia racemosa near the salt marsh (SM) and at the riverside (RS). The maps show Brazil, Rio de Janeiro State, Guaratiba and its urban pressures, and the study area. The plant pictures are from SM area, and RS area showing A. schaueriana (thick arrows) and L. racemosa (thin arrows) and their natural morphological alterations. Scales are shown on each map.
Figure 1.
Location map of Guaratiba’s populations, with a picture illustrating the morphological alterations of Avicennia schaueriana and Laguncularia racemosa near the salt marsh (SM) and at the riverside (RS). The maps show Brazil, Rio de Janeiro State, Guaratiba and its urban pressures, and the study area. The plant pictures are from SM area, and RS area showing A. schaueriana (thick arrows) and L. racemosa (thin arrows) and their natural morphological alterations. Scales are shown on each map.
Neutral molecular markers measure overall genetic diversity of a species or population, which is traditionally associated to long-term survival and potential of a species/population to survive in a changing environment [
11]. For this purpose, we used Amplified Fragment Length Polymorphism (AFLP) markers, which provide overall genetic diversity without the need of previous DNA sequence knowledge [
12]. Our results show that the two mangrove species have different levels of genetic differentiation and genetic diversity between and within the studied areas. Consequently these two mangrove species might have reached different levels of variation because of the different effect of stochastic evolutionary forces acting on this stressful environmental condition near the salt marsh.
2. Materials and Methods
The mangrove population of Sepetiba Bay is located in the Biological Reserve of Guaratiba. This reserve protects the last 3360 ha of mangrove in the metropolitan area of the city of Rio de Janeiro, the second largest mangrove population of Rio de Janeiro State. Our study area was limited by the south edge of a big salt marsh (ca. 30 ha) and by the Piracão riverbank. The study area was approximately 14 ha where adult plants were collected randomly, some near the salt marsh (SM) and some near the Piracão River bank (riverside-RS). Plants were at least 10 meters distant from each other. The greatest distance between the salt marsh edge and the riverbank was 300 meters but with a small transition area where only a third mangrove species, Rhizophora mangle, was present. The plants near the salt marsh had morphological alterations with poor development, nevertheless they were still able to flower and fructify.
We collected leaf material in silica-gel of 22 plants of
A. schaueriana (12 near the salt marsh) and 21 plants of
L. racemosa (9 near the salt marsh) inside the study area. DNA extraction followed the protocol of Lira-Medeiros
et al. [
13], based on Doyle and Doyle [
14]. A total of 50 mg of dried leaves were ground through the Retsch MM 301 Mixer for 1 minute at 30 Hz. We added 10 mg of PVP 40,000 to grinded samples and 700 μL of preheated buffer MATAB (0.1M Tris-HCl pH 8.0, 1.25 M NaCl, 0.02 M EDTA, Matab 4%) with ß-mercaptoethanol (0.1%). We used vortex and incubated samples at 65 °C for 1 hour. We then did two organic extractions with 600 μL of chloroform:isoamyl alcohol (24:1) mixed by inversion for 10 minutes and centrifugation at 12,000 rpm for 10 minutes. Subsequently, the first precipitation was made with 0.8 volumes of ice-cold isopropyl alcohol, followed by centrifugation 12,000 rpm for 15 minutes. After solubilization of the precipitate in 500 μL of sterile milliQ water, the second precipitation was performed with 0.1 volumes of 5M NaCl and 2 volumes of cold absolute ethanol. The samples were now incubated for 30 minutes in freezer (−20 °C). Ethanol was discarded and pellet was washed with 500 μL of 70% ethanol for 20 minutes. Ethanol was discarded again and pellet was dissolved in 100 μL of Tris-HCl 10 mM (pH 8.0). The quality and quantity of DNA was estimated on agarose gel 1% and Nanodrop 2000 (Thermo Electron North America LLC.: West Palm Beach, FL, USA).
AFLP markers were amplified with radioactive-labeled primer [
12,
15]. We used the AFLP
® Analysis System I (Life Technologies Inc.: São Paulo, Brazil) for the following procedure. We used 300 ng genomic DNA in a final volume of 15 μL for digestions with 10 U of EcoRI and 10 U of MseI. The digestions were incubated at 37 °C for 3 hours and the enzymes were denatured for 15 minutes at 70 °C. After confirmation of total digestion in 1% agarose gels, adapters were ligated using 6 μL of the solution of adapters and 3 U of T4 DNA ligase. This reaction was incubated at 20 °C for 2 hours. Pre-amplification reactions were performed using the primers complementary to EcoRI and MseI adapter sequences, but with a selective nucleotide. We used the ligated DNA diluted 5 to 10 times, depending on the intensity of digestion observed in the gel, with the aim of maintaining the samples with approximately the same concentration of DNA. The PCR program of pre-amplification consisted of 29 cycles of 30 s at 94 °C, 60 s at 56 °C and 60 s at 72 °C with a final extension of 2 minutes. The PCR products were checked in 1% agarose gel and proportionally diluted 10 to 50 times so that all the samples remain with similar DNA concentrations.
The final amplification reaction was performed in 20 μL final volume. The primer with sequence complementary to the EcoRI adapter was radioactively labeled with 1 μCi of 32P-ATP and 0.2 U of T4 Polynucleotide Kinase. They were incubated at 37 °C for 1 hour, then at 70 °C for 10 minutes and then another 10 minutes on ice. For the PCR reaction, we used 5 μL of pre-amplified and diluted DNA, primers complementary to the adapters EcoRI and MseI with three selective bases each in a final volume of 20 μL. The program of this reaction was a touchdown for the first 12 cycles with 30 s at 94 °C, 30 s of annealing temperature, starting at 65 °C decreasing 0.7 °C per cycle; and 1 minute at 72 °C. The program continued for another 25 cycles with annealing temperature of 56 °C. The primer combinations used were EcoRI-ACC/MseI-CAT, EcoRI-ACG/MseI-CAG and EcoRI-AAG/MseI-CAT.
The results were analyzed after separation by electrophoresis at 60W for 2 hours, in denaturing 6% polyacrylamide gel with 7 M urea and TBE (1X). The gel was exposed to a Kodak film XK1 one or two days. The revelation of the film was made using developer and fixer solutions of Kodak.
Data were recorded in a matrix of 1 and 0, where one is the presence of a DNA fragment and zero the absence of the fragment. DNA fragments of the same size (similar mobility) were considered to be from the same locus. Fragments between 70 and 500 bp were used in the analysis. The diversity index of Nei and Shannon, the differentiation index G
ST were calculated by POPGENE software version 1.32 [
16]. Nei’s diversity index is often used in population genetic studies, though it is based exclusively on allele frequency and relies on Hardy-Weinberg Equilibrium (HWE). On the other hand, Shannon index is calculated based on allele richness considering both the number and frequency of the alleles, consequently not assuming HWE [
17]. Both indexes were calculated for comparison with other studies. Although Shannon index would be more reliable for small sample sizes, Nei’s index has been used in mangrove population studies.
The G
ST index assumes that populations are in HWE. This is not ideal for dominant markers because it assumes there is no selfing in the population [
18]. To overcome this problem, Bayesian and multivariate analyzes were also carried out. The Bayesian method uses Markov Chain Monte Carlo (MCMC) to calculate deviation from HWE. The Bayesian differentiation index, θ
ST, was calculated with f-free model, 250,000 runs and 50,000 burn-ins by Hickory version 1.1 [
19]. This model excludes uncertainties about inbreeding values (f) that is more indicated for dominant marker’s datasets and species without previous information on f. The multivariate analysis is based on the presence and absence of fragments, ignoring HWE deviation. The multivariate differentiation index, β
ST, was computed by Between-Class Analysis of the Principal Component Analysis using ADE-4 [
20] package for R software [
21]. Permutation test was done for β
ST with 999 permutations also on ADE-4 package [
20].
3. Results
Dominant AFLP markers were highly polymorphic for both species,
Avicennia schaueriana and
Laguncularia racemosa, but with some differences in their patterns of polymorphism.
Avicennia schaueriana had 212 amplified loci, with 200 polymorphic (94.3%) among all samples (
Table 1). Among riverside (RS) samples 198 (93.4%) loci were polymorphic, and salt marsh (SM) samples had 98 (46.2%) polymorphic loci. The divergent number of polymorphic loci between RS and SM samples was also reflected on the number of rare polymorphic loci, amplified in only one or two samples (52 and 27 respectively). The diversity index of Nei was 0.307 (SD = 0.144) for RS plants and 0.11 (SD = 0.143), for SM plants. The diversity index of Shannon was 0.468 (SD = 0.186) and 0.183 (SD = 0.218), respectively, for RS and SM plants (
Table 1). Interestingly, some loci had no amplification and were called as absent loci. Although absent loci could be associated to low number of samples and/or rare loci, they were mostly found on SM area indicating a tendency inside this area. There were 26 absent loci on SM plants of
A. schaueriana, and only one in RS.
Table 1.
Genetic analysis of Avicennia schaueriana and Laguncularia racemosa from riverside (RS) and near salt marsh (SM) areas obtained with AFLP markers. Number of polymorphic loci both total and rare (present in 1 or 2 samples only), absent loci (no amplification) and the diversity index of Nei and Shannon, is shown. Standard deviations were calculated by Popgene.
Table 1.
Genetic analysis of Avicennia schaueriana and Laguncularia racemosa from riverside (RS) and near salt marsh (SM) areas obtained with AFLP markers. Number of polymorphic loci both total and rare (present in 1 or 2 samples only), absent loci (no amplification) and the diversity index of Nei and Shannon, is shown. Standard deviations were calculated by Popgene.
| Species | Area | Polymorphic Loci | Absent Loci | Diversity index of Nei | Diversity index of Shannon |
|---|
| Total | Rare |
|---|
| Avicennia schaueriana | RS | 198 (93.4%) | 52 | 1 | 0.307 (SD = 0.144) | 0.468 (SD = 0.186) |
| SM | 98 (46.2%) | 27 | 26 | 0.111 (SD = 0.143) | 0.183 (SD = 0.218) |
| All samples | 200 (94.3%) | 53 (25%) | - | 0.228 (SD = 0.128) | 0.372 (SD = 0.172) |
| Laguncularia racemosa | RS | 245 (96.5%) | 66 | 6 | 0.324 (SD = 0.138) | 0.492 (SD = 0.173) |
| SM | 224 (88.2%) | 51 | 21 | 0.357 (SD = 0.162) | 0.520 (SD = 0.219) |
| All samples | 252 (99.2%) | 72 (28.3%) | - | 0.357 (SD = 0.131) | 0.532 (SD = 0.158) |
Laguncularia racemosa samples were highly polymorphic. Of 254 amplified loci, only three (1.2%) were monomorphic among all samples (
Table 1). The number of polymorphic loci was similar in RS and SM plants, 245 (96.5%) and 224 (88.2%), respectively. The number of rare polymorphic loci was also similar, 66 and 51 for RS and SM, respectively. Similar levels of polymorphism were reflected in similar values of diversity indexes. The diversity index of Nei was 0.324 (SD = 0.138) for RS and 0.357 (SD = 0.162) for SM. The diversity index of Shannon was 0.492 (SD = 0.173) and 0.520 (SD = 0.219), respectively, for RS and SM (
Table 1). Both areas had diversity levels similar to the overall genetic diversity calculated for this species (Nei = 0.357 and Shannon = 0.532) and higher than
A. schaueriana indexes. The only divergence found between areas was the number of absent loci, six in RS and 21 in SM. Both species had higher number of absent loci among SM samples.
The genetic differentiation between RS and SM plants was moderate for
A. schaueriana, G
ST = 0.118, θ
ST = 0.065 (SD = 0.011), β
ST = 0.122 (
p < 0.005), but very low and not significant for
L. racemosa plants, G
ST = 0.053, θ
ST = 0.013 (SD = 0.004), β
ST = 0.052 (
p = 0.317;
Table 2).
Table 2.
Genetic differentiation index calculated between RS and SM plants by three statistical methods for both Avicennia schaueriana and Laguncularia racemosa. GST: frequency-based method assuming HWE allelic frequencies; θST: Bayesian analysis that calculates HWE deviation; βST: Multivariate analysis that bypasses HWE. SD: Standard deviation. Level of significance was calculated by permutation test.
Table 2.
Genetic differentiation index calculated between RS and SM plants by three statistical methods for both Avicennia schaueriana and Laguncularia racemosa. GST: frequency-based method assuming HWE allelic frequencies; θST: Bayesian analysis that calculates HWE deviation; βST: Multivariate analysis that bypasses HWE. SD: Standard deviation. Level of significance was calculated by permutation test.
| Species | GST | θST | βST |
|---|
| A. schaueriana | 0.118 | 0.065 (SD = 0.011) | 0.122* |
| L. racemosa | 0.053 | 0.013 (SD = 0.004) | 0.052† |
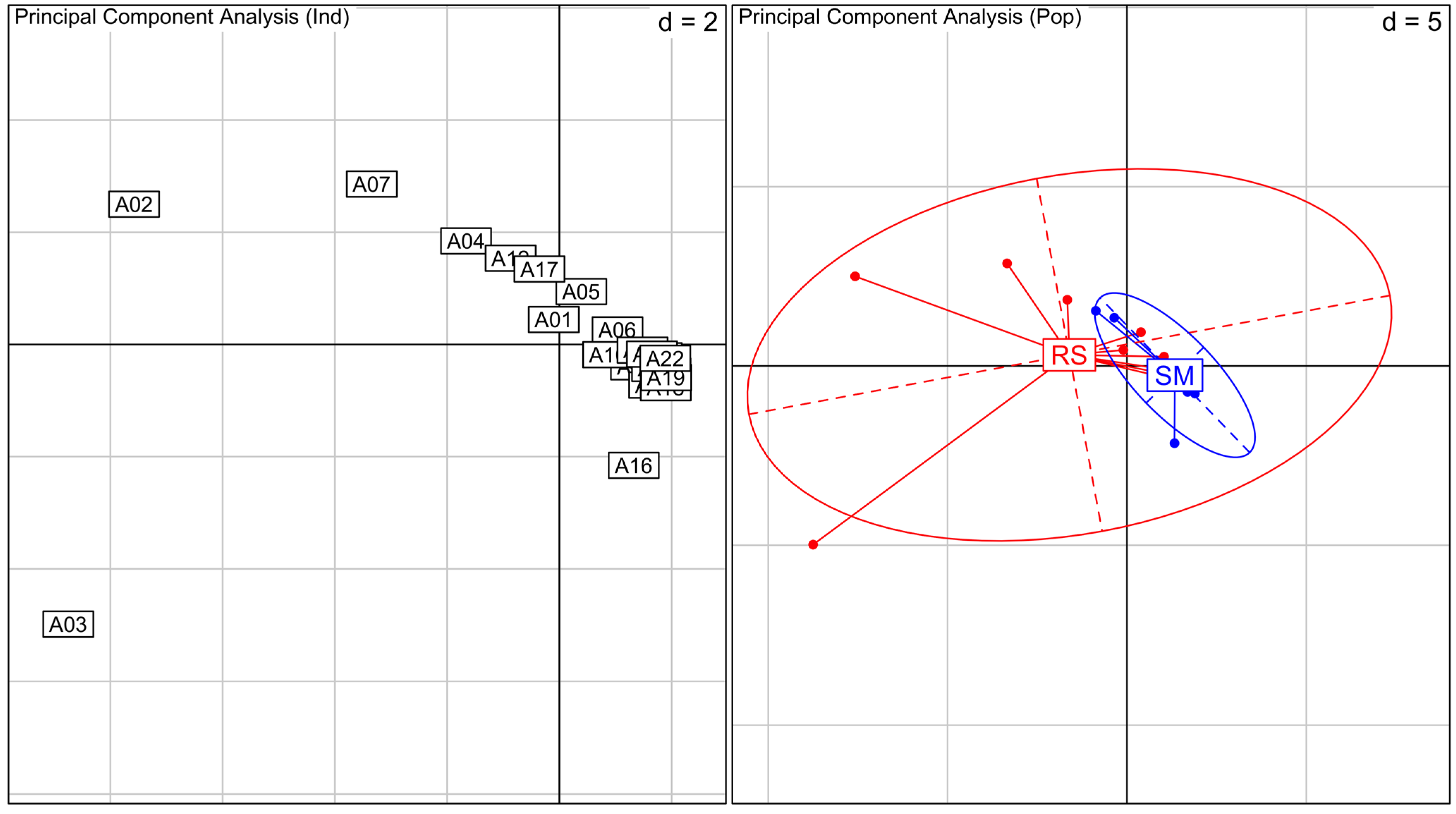
The PCA analyses show that most of
A. schaueriana’s genetic diversity is present in RS plants, which are broadly distributed on the graphic (
Figure 2). In contrast, SM plants are clustered together representing a small part of RS diversity. The PCA of
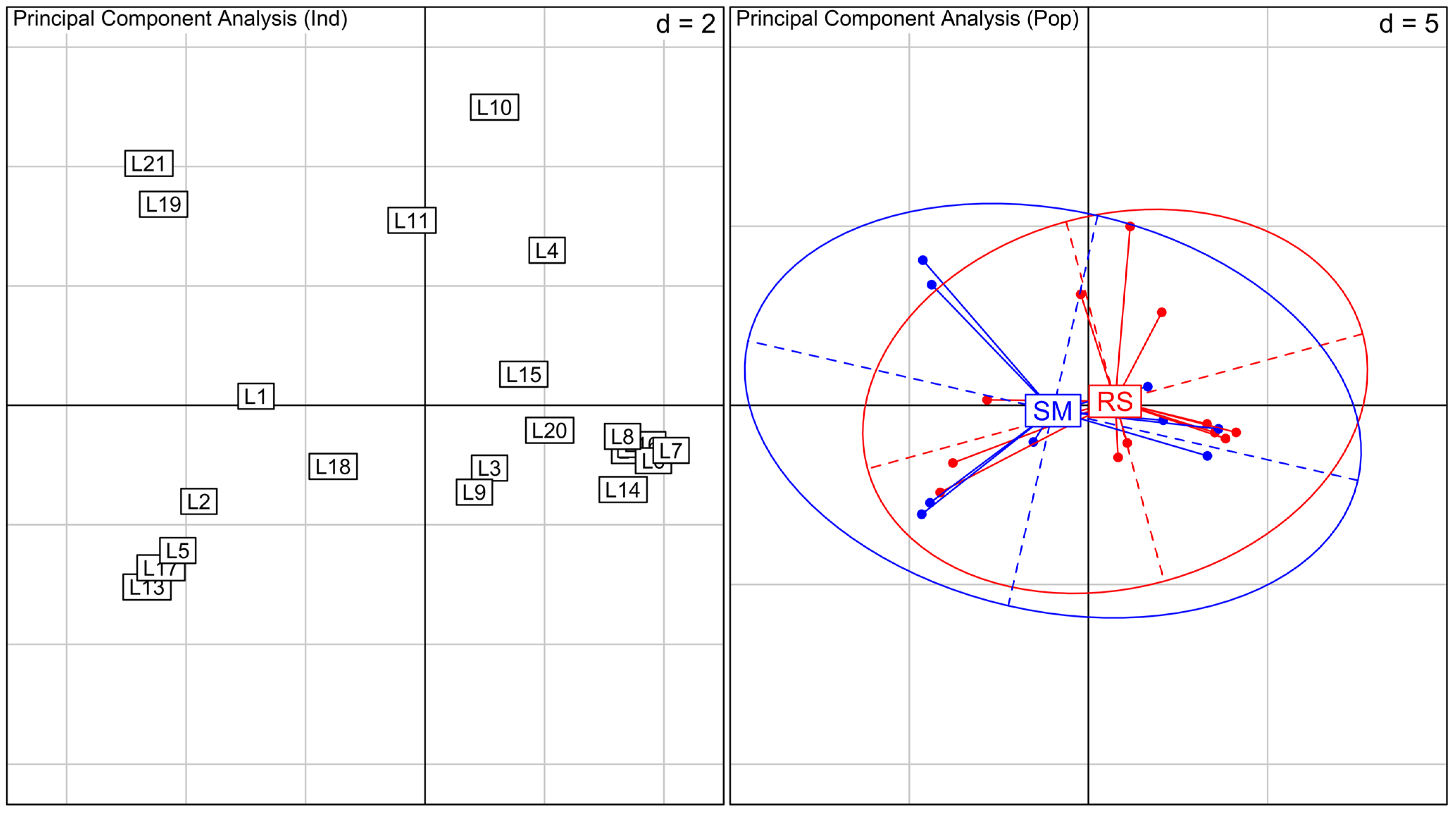
Laguncularia racemosa show an overlap of the variation observed for RS and SM areas, with no cluster formation (
Figure 3).
Figure 2.
Principal Component Analysis (PCA) for Avicennia schaueriana species. On the left, PCA between all individuals analyzed, numbers indicate samples. Samples A1 to A10 are from RS and A11 to A22 from SM. On the right, PCA between individuals grouped by their location (RS in red and SM in blue). Inertia ellipses represent 95% confidence interval of the variance in each area. Values represented as d in the graphs are related to grid size.
Figure 2.
Principal Component Analysis (PCA) for Avicennia schaueriana species. On the left, PCA between all individuals analyzed, numbers indicate samples. Samples A1 to A10 are from RS and A11 to A22 from SM. On the right, PCA between individuals grouped by their location (RS in red and SM in blue). Inertia ellipses represent 95% confidence interval of the variance in each area. Values represented as d in the graphs are related to grid size.
Figure 3.
Principal Component Analysis (PCA) for Laguncularia racemosa species. On the left, PCA between all individuals analyzed, numbers indicate samples. Samples L1 to L12 are from RS and L13 to L21 are from SM. On the right, PCA between individuals grouped by their location (RS in red and SM in blue). Inertia ellipses represent 95% confidence interval of the variance in each area. Values represented as d in the graphs are related to grid size.
Figure 3.
Principal Component Analysis (PCA) for Laguncularia racemosa species. On the left, PCA between all individuals analyzed, numbers indicate samples. Samples L1 to L12 are from RS and L13 to L21 are from SM. On the right, PCA between individuals grouped by their location (RS in red and SM in blue). Inertia ellipses represent 95% confidence interval of the variance in each area. Values represented as d in the graphs are related to grid size.
4. Discussion
We assessed the genetic diversity in a natural population of mangrove with extensive morphological alterations on
Avicennia schaueriana and
Laguncularia racemosa plants. Although morphological alterations in mangrove plants are well described [
4,
7,
22], little is known about its correlation to the genetic diversity of the plants.
The two species showed divergent patterns on their overall genetic diversity within and between the studied areas. The genetic diversity of Avicennia schaueriana was reduced near the salt marsh, with moderate genetic differentiation between the areas. However, Laguncularia racemosa plants showed non-significant genetic differentiation between RS and SM areas. The overall genetic diversity was similar between areas but Nei and Shannon indexes were significantly divergent being higher near the salt marsh. Further unraveling of factors leading to morphological differentiation between the two sites may be undertaken with a population genomics approach or, alternatively, candidate loci may be identified from functional studies. Significant genetic differentiation between A. schaueriana plants from the two studied areas was found, but Laguncularia racemosa had no significant or very little genetic differentiation between areas.
For
Avicennia schaueriana, the high genetic diversity found in RS plants (Nei = 0.307, Shannon = 0.468) is similar to the Rio de Janeiro State mean diversity, though slightly lower (Nei = 0.408, Shannon = 0.595, unpublished data [
23]). However SM diversity was depleted (0.111 and 0.183, for Nei and Shannon respectively). Overall genetic diversity of
A. schaueriana within this study (Nei = 0.228, Shannon = 0.372) was similar to its congeneric sympatric species,
A. germinans, within its natural range from the east Atlantic, west Atlantic and east Pacific (Nei = 0.212 using AFLP markers [
24]); but lower than within-Colombian populations of
A. germinans (Nei = 0.398 using SSR marker [
25]). A recent work on Brazilian mangroves found strong genetic structure of
A. schaueriana populations due to isolation by distance grouping them in two main areas, called North and South of the northeastern extremity of South America [
26]. Populations of the latter group had lower genetic diversity than the former, mean H
E = 0.192 and 0.310, respectively [
26], but still consistent with our findings, since Sepetiba’s Bay is located in the South group area. Although different molecular markers were used in the cited work and in ours, we believe that Sepetiba’s Bay mangrove is an important population to maintain the levels of genetic diversity within this species on the South group, especially the RS area that had higher levels of diversity.
Our results indicate that A. schaueriana’s genetic diversity from SM is likely a small part of the genetic diversity of RS plants, as shown on PCA. It is likely that near SM gene flow is limited resulting in loss of genetic diversity. Although neutral molecular markers do not investigate adaptive loci, we would not discard the possibility that selection is acting in favor of genotypes capable of surviving on the unfavorable conditions near the salt marsh (less tidal inundation, higher salinity, and lower nutrient input), limiting the genetic variation within SM area. Further investigations with codominant markers and on adaptive loci are needed to resolve gene flow levels and selection effects on SM plants.
The
Laguncularia racemosa plants of both areas, RS and SM, had similar genetic diversity levels and very little or no significant genetic differentiation between areas. The overall genetic diversity of
L. racemosa calculated in this work (0.357 and 0.532, Nei and Shannon respectively) was higher than the overall diversity found within Rio de Janeiro State (Nei = 0.216, Shannon = 0.344, unpublished data [
23]). Similar levels of genetic diversity were reported for two mangrove species of the Combretaceae family from the Indo-West Pacific Ocean,
Lumnitzera racemosa (Shannon = 0.403 using ISSR marker [
27]) and
Lumnitzera littorea (Shannon = 0.365 using ISSR marker [
28]). Interestingly, the mean genetic diversity within SM area was not significant different than RS, indicating no detrimental effect of evolutionary forces over plants located in the unfavorable area near the salt marsh. It is likely that an efficient gene flow is maintaining the genetic diversity within these areas counteracting genetic drift, although an analysis with co-dominant markers would be ideal to confirm it.
The PCA show no structure between RS and SM plants, with clear overlapping of the whole variation found within these areas. No differentiation between RS and SM plants indicates the absence of correlation between overall genetic diversity and morphological variation for L. racemosa. This result raises questions about the mechanisms behind the observed morphological divergence in the studied areas.
The expanding field of epigenetics shows that acquired traits can be passed to progeny without changing DNA sequences, and that such traits can be inherited [
29]. Several studies have suggested that epigenetic variation can be linked to natural phenotypic divergence under different environmental conditions [
30,
31,
32]. It is believed that selection can act directly upon epigenetic variation, if genetic and epigenetic components of the genome are decoupled, with the potential to change the adaptive landscape of genes, resulting in long-term effects on evolution, such as transgenerational phenotypic changes [
33]. For this to occur, epigenetic variation should be inherited from one generation to another and clear environmental-based epigenetic alterations should be seen. Although these two points are still unclear on
L. racemosa species, MSAP markers showed overall epigenetic differentiation between RS and SM plants [
13]. So it’s possible that morphological alterations found for this species at Sepetiba’s Bay, could be explained mostly by epigenetic variation instead of genetic variation.
The present work indicates that two mangrove species similarly adapted to adverse environmental conditions and showing comparable morphological alterations might have different genomic mechanisms responsible for these alterations. This difference is probably the consequence of different evolutionary forces acting on those species. Though we cannot discuss the possible effects of natural selection because we used neutral markers in this study, it is likely that A. schaueriana has suffered more intensely the negative effects of genetic drift near the salt marsh, whereas L. racemosa plants have counteracted this force probably by an efficient migration between areas. In this latter case, we believe that morphological alterations might be resulting from epigenetic mechanisms that are not dependent of DNA sequence variation.
The different genomic responses found for these two mangrove species occurring in the same location are very surprising. Although these species are from different taxonomic families, they are similarly adapted to the stressful environment of the mangroves, and their dispersal mechanisms of pollen and propagules by small insects and water respectively are very similar [
10,
22]. Clearly
Avicennia schaueriana is being affected by genetic drift, leading to depleted genetic diversity near the salt marsh. This might be the result of bottleneck events and/or high selfing rates near the salt marsh. However, this is not likely to affect
Laguncularia racemosa plants, which even with strikingly divergent morphologies, should be considered as one population with no differentiation or structure at the studied area.
A high selfing rate is associated with less efficient gene flow. Although
A. schaueriana inbreeding level was recently assessed on the Brazilian coast for the first time [
26], the population of Sepetiba’s Bay was not included. The lack of genetic population studies on Brazil, using different molecular markers, limits our knowledge on their selfing rates, migration and gene flow within and between mangroves. But the gene flow of
L. racemosa predicted as efficient is corroborated by its successful and broader geographical distribution than
A. schaueriana. The species
L. racemosa is widespread on Western mangroves, located in the neotropics and Western Africa, both Atlantic coasts, while
A. schaueriana is mostly located in Brazil (90% of the documented occurrences) and a few countries in the neotropics [
22]. Besides, the sympatric distribution in Brazil of
A. schaueriana with a congeneric widespread species,
A. germinans, might decrease the former’s ability to expand its populations or colonize new areas.