Abstract
As marine environments change, the greatest ecological shifts—including resource usage and species interactions—are likely to take place in or near regions of biogeographic and phylogeographic transition. However, our understanding of where these transitional regions exist depends on the defining criteria. Here we evaluate phylogeographic transitions using a bootstrapping procedure that allows us to focus on either the strongest genetic transitions between a pair of contiguous populations, versus evaluation of transitions inclusive of the entire overlap between two intraspecific genetic lineages. We compiled data for the Atlantic coast of the United States, and evaluate taxa with short- and long-dispersing larval phases separately. Our results are largely concordant with previous biogeographic and phylogeographic analyses, indicating strong biotic change associated with the regions near Cape Cod, the Delmarva Peninsula, and eastern Florida. However, inclusive analysis of the entire range of sympatry for intraspecific lineages suggests that broad regions—the Mid-Atlantic Bight and eastern Florida – already harbor divergent intraspecific lineages, suggesting the potential for ecological evaluation of resource use between these lineages. This study establishes baseline information for tracking how such patterns change as predicted environmental changes take place.
1. Introduction
To date, there has been intense exploration of the relationship between recognized biogeographic patterns and the concordance of phylogeographic transitions—changes in allele frequency within a taxon that reflect either the historical origins of diversity, or the ecological processes that maintain these patterns [1,2,3,4,5,6]. In the marine realm, these comparisons have proliferated because of the ease of comparing approximately linear coastlines [7], the intrigue of understanding how the biphasic life cycle of most marine species influences the distribution of diversity [8,9,10,11,12,13], and how oceanographic features structure the vast oceans [14,15,16,17,18,19].
However, we know that the oceans are changing, with the most obvious changes being increased surface temperatures, and increasing homogenization of communities through anthropogenic introduction [20]. Marine species are moving their ranges to closely track ocean temperatures [21] and other factors [22,23]. The distributional independence of some species shifts [24,25] highlights the potential for climate change to promote novel ecological interactions as well. Therefore, it is at the boundaries or transitions between biotic regions that the independent movement of ranges can lead to the greatest ecological change, including shifts of mean trait values for the local community [22,26,27,28].
Thus, identifying regions of concordant biogeographic and phylogeographic transition allows us to suggest the regions in which the greatest ecological change may be coming [22,25,29,30,31]. These boundaries, most notably defined for marine biota by Briggs [32] as the demarcation between areas of high endemic diversity [33], can themselves be elusive to draw on a map [34] because of the assortment of independent lineages into niches of varying breadths, as well as spatial and temporal sampling uncertainties [3].
As an example, Point Conception (near Santa Barbara, California, USA) is a coastal region notable for the large number of species range limits associated with a transition in water temperature and oceanographic features. When evaluated for the distribution of species range limits, Point Conception appears to be a very strong feature that in particular limits the distribution of marine species with broadly-dispersing planktonic larvae [4,35,36]. However, the divisions between phylogeographic lineages in this area, across many species, are distributed over hundreds of kilometers [4] and when the depth distribution of species is also considered near Point Conception, the latitudinal overlap may be quite broad [37].
It has been argued that to understand the distribution of diversity best, we need information both on the overlap of distributional ranges [22] as well as the pattern of distributional limits [38]; both approaches may be informative for where ecological interactions will be most volatile in the coming decades [39]. Here, we synthesize data from available population genetic and phylogeographic studies from the east coast of North America, expanding previous work by Wares [3], to understand the extent to which canonical biogeographic transitions are reflected in intraspecific genetic data, and how these patterns differ for species with pelagic larval dispersal versus those with limited dispersal. In doing this, we combine and compare the lineage range bootstrapping methods of Wares [3] and Pelc et al. [4] to explore how “transitions” between regions differ based on analysis of how two evolutionarily distinct lineages are fully distributed, versus analysis of where those lineages change the most in relative abundance (Figure 1). As much work has gone into understanding how species and (phylogeographic) lineage ranges interact with each other, and with the environment [30,40,41,42,43,44,45,46,47,48,49,50], identifying the potential breadth of these codistributions could be an insight both into our uncertainty about biogeographical and phylogeographical boundaries and species coexistence.
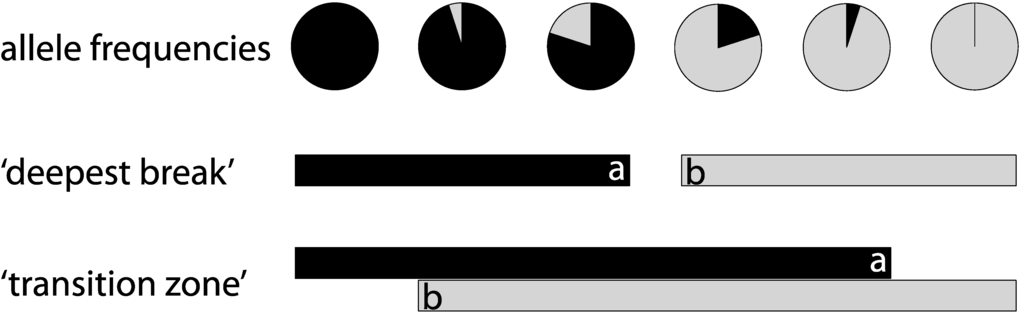
Figure 1.
Given the frequency of two primary genetic lineages (alleles, clades, etc.) at a series of locations, phylogeographic methods would tend to identify the change in frequency between the two central locations as being of the greatest magnitude. The “deepest break” analysis in this paper thus indicates that the environmental mechanism influencing this pattern is found between a and b along a transect. However, both lineages are found in four of the six locations represented, and the “transition zone” analysis in this paper again assumes that the environmental mechanism influencing this pattern is found between a and b, typically reflecting greater uncertainty in the specific location of interest.
2. Methods
2.1. Literature Search/Study Compilation
We identified marine/estuarine taxa along the east coast of North America for which population genetic information is available. A comprehensive list of taxa and studies were established through literature searches of phylogeographical and population genetic studies, published between 2002–2012, using the Web of Science academic citation index, Google Scholar, and other resources, as in Small and Wares [31]. In addition, previous phylogeographical studies published between 1986–2002 were included from two previous syntheses [3,4]. Published studies were considered even if no significant phylogeographical break (statistically significant spatial disjunction in allele groups) was identified. All studies included here represent species in their native ranges that included at least 3 sampling locations.
Of the studies that reported a phylogeographical break, we considered this break to be significant if genetic structure was statistically significant (p < 0.05) according to the author’s reported results and was not considered a signal of isolation by distance. Given the variety of methodologies, sampling efforts, molecular markers, and statistical analyses used across studies, we did not attempt to standardize these measures of differentiation across studies or reanalyze the data for standardized or absolute levels of differentiation; Weersing and Toonen [51] showed that all such factors contribute to variance among results for different studies.
Taxa were separated into two dispersal classes, short (no pelagic phase or pelagic larval dispersal <3 days) and long (planktonic, >3 days). This division is based on observations that the distribution of larval duration is not continuous, but bimodal [52,53,54] and strongly influenced by life history, the strong relationship between larval duration and dispersal distance [55], and theoretical values that separate the two classes in Lagrangian dispersal models [7,41]. The sensitivity of our results to this criterion is discussed below. To determine the significance of the association between long and short dispersers with and without apparent genetic structure, we applied Pearson’s Chi-square tests and Fisher’s exact tests on the total number of species and published studies found in each group (long/short, break/no break).
2.2. Analysis of Phylogenetic Concordance
Following the methodology of Pelc et al. [4], we used a bootstrapping technique to analyze the concordance of reported phylogenetic breaks for all taxa in which they occurred. This method tests for non-random distribution of potential phylogenetic breaks along the coast of interest [4], with each sample location in each study assigned a linearized distance along the east coast of North America. Including all sampled locations takes into account the variation in scale (number and distribution of sites) among studies. For each study, the location and span of the phylogenetic break, consisting of the entire length of the coastline between upper and lower boundaries, was defined in two ways: (1) using the two sampled locations within each study between which the greatest degree of genetic change was reported (based on p values), boundaries were defined as the “deepest break” as in [4]; (2) using the southernmost site occupied by individuals in the northern genetic lineage and the northernmost site occupied by individuals from the southern lineage, the break was defined as the “transition zone” as in Wares [3] (see Figure 1). This latter definition is a more inclusive descriptor of the break, allowing for overlap between clades and often representing a broader range. Although in some studies multiple significant breaks are described, we only focus on the strongest (greatest significance) separation for this analysis.
The summed number of apparent phylogenetic breaks was calculated at each sampling location across all studies. Using 10,000 bootstrap simulations in an R package modified from Pelc et al. [4], we determined whether the number of apparent phylogenetic breaks observed at any location along the coast was greater than expected by chance. That is, the null hypothesis was that the distribution of lineage breaks was randomly sampled from the distribution of the evaluated species. Each bootstrap replicate chose a random location (or a bounding pair of locations, as in [3]) to simulate a break (transition) for each study and the summed simulated breaks were calculated across all studies. We then determined which locations contained more apparent phylogenetic breaks than a proportion β of simulated phylogenetic breaks (with results showing β at 0.90, 0.95, and 0.99). A smaller β leads to a larger region where any break must be; for example the region for a β of 0 would be the region that contains all the potential breaks—the entire domain. We ran separate analyses for the two methods of identifying breaks as described above, for both short and long dispersal taxa. When multiple studies were published for the same taxon, the data from each study were used but individual studies were weighted proportionally so that each individual species is equally weighted in overall analysis. Studies with multiple markers (typically, a mitochondrial locus and one or more nuclear loci) were discarded if markers did not exhibit concordance for the primary break, as a number of distinct mechanisms may lead to such discordance [56]. All analyses were run using the R programming environment [57].
3. Results
We identified a total of 52 studies representing 50 estuarine and marine species for which phylogenetic analyses have been documented (Supplement 1). Two studies described phylogeographical data in algal species; the remaining studies focused on animals. The majority of the studies (38) described long dispersers. Within the long dispersers in the present analysis, 63% identified at least one phylogeographical break, while 86% of the short dispersers exhibited such a pattern. Chi-square and Fisher’s exact tests revealed that these trends were not significant when all species are included in the analyses (Pearson’s Χ2 = 3.0857, df = 1, p-value = 0.079; Fisher’s Exact Test p = 0.11); however, removing the algal species, we find that there is a significantly larger proportion of short dispersers with phylogeographical breaks (Pearson’s Χ2 = 6.9333, df = 1, p-value = 0.008; Fisher’s Exact Test p = 0.01). Power analyses reveal that we have sufficient sample size to detect high effect size differences whether we focus on the total species or studies (alpha = 0.05, power = 0.94 and 0.95, respectively). Study sites across all species and dispersal types were evenly dispersed along the east coast of North America (Supplement 2).
Results from bootstrap analysis revealed different regions of phylogeographical transition depending on larval dispersal type. Long dispersers on this coast show a large likely region for phylogeographical transition (allowing for overlap of types) ranging from 35.9–42.5°N latitude (e.g., from North Carolina to Massachusetts, Figure 2A). When breaks were defined using the area of deepest phylogenetic transition as in Pelc et al. [4], a somewhat different picture was revealed (Figure 2B). In this case, five discrete regions were identified to be more likely to contain phylogenetic breaks than expected by chance. These regions included Cape Cod, MA (41.7–42.6°N), the New Jersey/Delaware shore and Chesapeake Bay (38.64–39.3°N), Morehead City, NC (34.7°N), southern GA/northern Florida (30.0–31.2°N), and the coast of Florida from St. John’s River to Sarasota (28.8–27.3°N).
Short distance dispersers showed a different pattern in which regions of transition were more concentrated to the south. Using either of the two described definitions to define phylogeographical breaks, bootstrap analysis revealed a region of phylogeographical breaks beginning on the Georgia coast and extending into Florida, slightly past Cape Canaveral, FL (~31°–28.5°N) or farther south to the Florida Keys (24.4°N; Figure 2C,D). In addition, at the lowest confidence level (β of 90%), regions of phylogeographical transition were identified south of Cape Canaveral, FL and in Narragansett/Fisher’s Island Sound (41.4°–41.2°N) and Western Long Island Sound (41.3°N; Figure 4). With increased confidence levels, significant ranges of transition were identified north and south of Cape Canaveral (Figure 2C,D). When breaks were defined using the sites that encompass the deepest phylogenetic break, the Chesapeake Bay region was also identified as a significant transition region (Figure 2D).

Figure 2.
The Atlantic coast of North America, with coincidence of phylogeographic breaks indicated for regions that have more observed transitions than 90, 95, or 99% of location-bootstrapped “null” distributions. Results vary for “long” dispersing species (A, B; 22 species) depending on whether the overlap of lineages (A) or deepest phylogeographic break (B) is analyzed. Similarly, results differ for “short” dispersing species (C, D; 11 species) depending on whether the overlap of lineages (C) or deepest phylogeographic break (D) is analyzed. Sampling effort and taxa included in each plot are found in Supplemental Data.
4. Discussion
Contrasting the phylogeographic patterns that emerge from the strongest disjunctions in allele frequencies (“strongest breaks”), versus those based on the range of overlap of one distinct allelic group with another (see Figure 1), requires consideration of what information is summarized by each analysis. When the deepest phylogeographic break is recorded for each species, for example, many lesser breaks may not be analyzed, and the environmental or ecological difference between regions may not be identifiable as the location of such boundaries is not necessarily associated with environmental features [58]. Analysis of the transition zone, or overlap of ranges, captures better the full information on presence of lineages, but at the expense of not sharply defining phylogeographic break points [40,44,59]. Each way of looking at phylogeographic transition, however, intends to identify regions of the coast that could mechanistically be important in maintaining the transition.
Thus, it is useful to identify which regions are robust to analytical assumptions. In our results (Figure 2), we can focus on the regions near Cape Cod, the Delmarva Peninsula, and Cape Canaveral. As perhaps might be assumed, there are many phylogeographic transitions associated with each of these regions. For Cape Cod, there is clearly an excess of phylogeographic transition (relative to random localization of transitions) for long-dispersing taxa whether we focus on the “deepest break” or “transition region” (Figure 2A,B), but less support for the importance of Cape Cod for short-dispersing taxa (Figure 2C,D). The Delmarva peninsula is also certainly an important transition zone (Figure 2A,B,D) that is not easily distinguished in short-dispersing taxa if their entire range of overlap is considered (Figure 2C). Similarly, the east coast of Florida is clearly an important transition region (Figure 2B,C,D), but the signal for this is lost in long-dispersing taxa if the entire range of overlap for these species is considered. It is worth noting that our results imply a differential effect of how the data are analyzed combined with how taxa are split into “short” versus “long” dispersal; our three-day cutoff between these two classes is based on observations of the bimodality of larval strategy [52,53,54] as well as considerations based on oceanographic modeling [7] and enables greater statistical power for our analysis than if we further split based on life history variation, e.g. separating out the small number of taxa with intermediate pelagic phases or splitting out those species with no pelagic phase at all. As very few of our “long” dispersers would fall into an intermediate class (and some, such as Streblospio benedecti, are actually polymorphic in dispersal strategy [60]), the results shown illuminate the patterns of greatest generality.
These results suggest that, because greater analytical weight is put on adjoining pairs of locations with the strongest phylogeographic disjunction—for example, the highest or most significant Fst value—focus on the “deepest breaks” (as in [4]) is able to resolve narrower geographic regions where there is concordance in transition across lineages. Although the range of overlap of these divergent lineages may also be informative for individual taxa or life history types with regard to how much ecological divergence is associated with a given population genetic divergence, it is less clear how to incorporate this information, from varying types of data, sampling effort, and initial analysis, into a post hoc comparative phylogeographic analysis [61]. Our results indicate broadly similar patterns to coastal biogeography based on estuarine diversity [62] as well as benthic and pelagic diversity [63]. The presence of such spatial concordance suggests that similar environmental processes are maintaining these transitions, whether among species or among intraspecific lineages.
The early days of phylogeography, in which cladistically distinct groups were identified on either side of (typically) apparent barriers to dispersal (“Type I breaks” [64]), necessarily put a focus on strong patterns that heretofore had not been recognized. In the past decade, however, the field has become much more quantitative as subtler questions of temporal divergence, migration, and colonization are involved [65,66,67]. It has become clear that many strong phylogeographical breaks are associated with how populations are sampled. For example, Wares [68] identified a deep break between populations of the isopod Idotea balthica sampled in Virginia and Rhode Island. However, Bell [69] showed that with higher-resolution sampling of the same taxon, there is a broad range of overlap between the two lineages along the coast. Simply put, as with species range boundaries, a clear and spatially discrete phylogeographical disjunction is unlikely without a physical boundary involved.
Environmental gradients can be important mechanisms for maintaining clines and phylogeographical (as well as biogeographical) transitions. Endler [70] argued, through use of simulation, that historical events leading to transient population allopatry would be unlikely to generate the strong concordant rapid transitions seen in biogeographical and comparative phylogeographical analyses, implicating environmental transitions and natural selection. Similarly, Endler [71], and Pringle and Wares ([7] and references therein) indicated that while such transitions may originate through disparate and selectively neutral processes (e.g., transient allopatry), they will still be attracted, over time, to shared environmental gradients and disruptions. This suggests a hypothesis that is in agreement with the partial success phylogeographers have had in reconstructing biogeographical boundaries: population genetics and phylogeographical patterns may originate through many processes, but are also quite dynamic, as evidenced by recent work on changes in distribution of species and genetic lineages as climate change advances [24,72,73,74,75]. Thus, the concordance of these patterns necessitates that we focus on contemporary mechanisms, more than the origins of these divergent lineages.
Moving forward with Comparative Phylogeography
Despite the importance of contemporary mechanisms for maintenance of phylogeographic and biogeographic patterns, there may be great advantage to evaluating large, comparable data sets in which the temporal origins of such patterns are inferred, either through coalescent analysis [65] or the use of standardized divergence metrics [76]. Ideally, a comparative dataset either identifies and limits analysis to taxa that share diversification events [77], or can evaluate a range of events that may be associated with ecological or environmental diversification among those taxa. Further comparison of species patterns will be possible with improved and comparable sampling of populations and genomic markers across species [78,79].
Overall, what we show here is that there are certain associations between biogeographic and phylogeographic patterns on the Atlantic coast of North America, but that to an extent improved sophistication in exploring these discontinuities with greater spatial sampling, greater genomic sampling, and a greater ability to compare data sets through statistical models will be necessary [67]. Many of the taxa explored here have not been evaluated using genetic data in over ten years. Given the number of recent studies showing rapid temporal biogeographic responses to marine climate change [80,81] in temperate waters, this suggests a number of key update studies that, if coordinated appropriately, could lead to a much greater understanding of how nearshore currents, environmental transitions and changes, and species interactions contribute to diversity patterns on this coast [18,82]. More and more, fields of biogeography and community ecology are recognizing the quantitative nature of global patterns: we can draw lines between regions, but those lines often depend heavily on our starting assumptions and information.
5. Conclusions
Our results are spatially concordant with prior analyses of phylogeographic boundaries on this coast, indicating significant intraspecific transitions associated with the regions near Cape Cod, the Delmarva Peninsula, and eastern Florida. However, across broad regions of the Mid-Atlantic Bight and eastern Florida, many of these divergent intraspecific lineages coexist, suggesting the need for further evaluation of resource use and other patterns of ecological divergence within these taxa. Our study establishes important baseline information for tracking how these spatial patterns change in the coming decades.
Supplementary Materials
Supplementary materials can be accessed at: http://www.mdpi.com/2071-1050/5/2/263/s1.
Acknowledgments
The authors thank C. Ewers and K. Bockrath for substantive comments during the development of this paper, and Robin Pelc for the R code used in her similar analyses. Two anonymous reviewers improved the manuscript substantially. This work was financially supported by the National Science Foundation (NSF-OCE-0961830).
References
- Burton, R.S. Intraspecific phylogeography across the Point Conception biogeographic boundary. Evolution 1998, 52, 734–745. [Google Scholar] [CrossRef]
- Dawson, M.N. Phylogeography in coastal marine animals: a solution from California? J. Biogeogr. 2001, 28, 723–736. [Google Scholar] [CrossRef]
- Wares, J.P. Community Genetics in the Northwestern Atlantic Intertidal. Mol. Ecol. 2002, 11, 1131–1144. [Google Scholar] [CrossRef]
- Pelc, R.A.; Warner, R.R.; Gaines, S.D. Geographical patterns of genetic structure in marine species with contrasting life histories. J. Biogeogr. 2009, 36, 1881–1890. [Google Scholar] [CrossRef]
- Marko, P.B. “What’s larvae got to do with it?” Disparate patterns of post-glacial population structure in two benthic marine gastropods with identical dispersal potential. Mol. Ecol. 2004, 13, 597–611. [Google Scholar] [CrossRef]
- Puritz, J.B.; Toonen, R.J. Coastal pollution limits pelagic larval dispersal. Nature Comm. 2011. [Google Scholar] [CrossRef]
- Pringle, J.M.; Wares, J.P. The maintenance of alongshore variation in allele frequency in a coastal ocean. Mar. Ecol. Prog. Series 2007, 335, 69–84. [Google Scholar] [CrossRef]
- Eckman, J.E. Closing the larval loop: linking larval ecology to the population dynamics of marine benthic invertebrates. J. Exp. Mar. Biol. Ecol. 1996, 200, 207–237. [Google Scholar] [CrossRef]
- Emlet, R.B. Developmental mode and species geographic range in regular sea urchins (Echinodermata: Echinoidea). Evolution 1995, 49, 476–489. [Google Scholar] [CrossRef]
- Grantham, B.A.; Eckert, G.L.; Shanks, A.L. Dispersal potential of marine invertebrates in diverse habitats. Ecol. Appl. 2003, 13, 108–116. [Google Scholar] [CrossRef]
- Havenhand, J.N. Evolutionary ecology of larval types. In Ecology of Marine Invertebrate Larvae; McEdward, L., Ed.; CRC Press: New York, NY, USA, 1995; pp. 79–122. [Google Scholar]
- Levin, L.A.; Bridges, T.S. Pattern and Diversity in Reproduction and Development; McEdward, L., Ed.; CRC Press: New York, NY, USA, 1995; pp. 1–48. [Google Scholar]
- McEdward, L. Ecology of Marine Invertebrate Larvae; CRC Press: New York, NY, USA, 1995; p. 464. [Google Scholar]
- Baums, I.B.; Paris, C.B.; Chérubin, L.M. A bio-oceanographic filter to larval dispersal in a reef-building coral. Limn. Ocean. 2006, 51, 1969–1981. [Google Scholar] [CrossRef]
- Connolly, S.R.; Roughgarden, J. A latitudinal gradient in northeast Pacific intertidal community structure: evidence for an oceanographically based synthesis of marine community theory. Amer. Nat. 1998, 151, 311–326. [Google Scholar]
- Dawson, M.N. Phylogeography in coastal marine animals: a solution from California? J. Biogeogr. 2001, 28, 723–736. [Google Scholar] [CrossRef]
- Galindo, H.M.; Olson, D.B.; Palumbi, S.R. Seascape genetics: A coupled oceanographic-genetic model predicts population structure of Caribbean corals. Curr. Biol. 2006, 16, 1622–1626. [Google Scholar] [CrossRef]
- Selkoe, K.A.; Watson, J.R.; White, C.; Horin, T.B.; Iacchei, M.; Mitarai, S.; Siegel, D.A.; Gaines, S.D.; Toonen, R.J. Taking the chaos out of genetic patchiness: seascape genetics reveals ecological and oceanographic drivers of genetic patterns in three temperate reef species. Mol. Ecol. 2010, 19, 3708–3726. [Google Scholar] [CrossRef]
- Zakas, C.; Binford, J.; Navarrete, S.A.; Wares, J.P. Restricted gene flow in Chilean barnacles reflects an oceanographic and biogeographic transition zone. Mar. Ecol. Prog. Ser. 2009, 394, 165–177. [Google Scholar] [CrossRef]
- Carlton, J.T. Community assembly and historical biogeography in the Atlantic Ocean: the potential role of human-mediated dispersal vectors. Hydrobiologia 2003, 503, 1–8. [Google Scholar] [CrossRef]
- Sunday, J.M.; Bates, A.E.; Dulvy, N.K. Thermal tolerance and the global redistribution of animals. Nature Clim. Change. 2012, 2, 686–690. [Google Scholar]
- Wiens, J.J. The niche, biogeography and species interactions. Phil. Trans. R. Soc. Lond. B 2011, 366, 2336–2350. [Google Scholar] [CrossRef]
- Sexton, J.P.; McIntyre, P.J.; Angert, A.L.; Rice, K.J. Evolution and ecology of species range limits. Ann. Rev. Ecol. Evol. Syst. 2009, 40, 415–436. [Google Scholar] [CrossRef]
- Dawson, M.N.; Grosberg, R.K.; Stuart, Y.E.; Sanford, E. Population genetic analysis of a recent range expansion: mechanisms regulating the poleward range limit in the volcano barnacle Tetraclita rubescens. Mol. Ecol. 2010, 19, 1585–1605. [Google Scholar] [CrossRef]
- Zacherl, D.; Gaines, S.D. The limits to biogeographical distributions: Insights from the northward range extension of the marine snail, Kelletia kelletia. J. Biogeography 2003, 30, 913–924. [Google Scholar] [CrossRef]
- Sanford, E.; Roth, M.S.; Johns, G.C.; Wares, J.P.; Somero, G.N. Local selection and latitudinal variation in a marine predator-prey interaction. Science 2003, 300, 1135–1137. [Google Scholar] [CrossRef]
- Harley, C.D.G.; Pankey, M.S.; Wares, J.P.; Grosberg, R.K.; Wonham, M.J. The impacts of climate change in coastal systems. Ecol. Lett. 2006, 9, 228. [Google Scholar] [CrossRef]
- Norberg, J.; Urban, M.C.; Vellend, M.; Klausmeier, C.A.; Loeuille, N. Eco-evolutionary responses of biodiversity to climate change. Nature Clim. Change 2012, 2, 747–751. [Google Scholar] [CrossRef]
- Wethey, D.S. Biogeography, competition, and microclimate: the barnacle Chthamalus fragilis in New England. Int. Comp. Biol. 2002, 42, 872–880. [Google Scholar] [CrossRef]
- Sanford, E.; Holzman, S.B.; Haney, R.A.; Rand, D.M.; Bertness, M.D. Larval tolerance, gene flow, and the northern geographic range limit of fiddler crabs. Ecology 2006, 87, 2882–2894. [Google Scholar] [CrossRef]
- Small, S.T.; Wares, J.P. Phylogeography and marine retention. Journal of Biogeography 2010, 37, 781–784. [Google Scholar] [CrossRef]
- Briggs, J.C. Marine zoogeography; McGraw Hill: New York, NY, USA, 1974. [Google Scholar]
- Briggs, J.C.; Bowen, B.W. A realignment of marine biogeographic provinces with particular reference to fish distributions. J. Biogeogr. 2012, 39, 12–30. [Google Scholar] [CrossRef]
- Olivero, J.; Márquez, A.L.; Real, R. Integrating fuzzy logic and statistics to improve the reliable delimitation of biogeographic regions and transition zones. Syst. Biol. 2012, 26, 1–21. [Google Scholar]
- Wares, J.P.; Gaines, S.D.; Cunningham, C.W. A Comparative Study of Asymmetric Migration Events across a Marine Biogeographic Boundary. Evolution 2001, 55, 295–306. [Google Scholar]
- Blanchette, C.A.; Miner, C.M.; Raimondi, P.T.; Lohse, D.; Heady, K.E.K.; Broitman, B.R. Biogeographical patterns of rocky intertidal communities along the Pacific coast of North America. J. Biogeogr. 2008, 35, 1593–1607. [Google Scholar] [CrossRef]
- Newman, W.A.; Abbott, D.P.; Haderlie, E.C. Cirripedia; Morris, R.H., Ed.; Stanford University Press: Stanford, CA, USA, 1980; pp. 504–535. [Google Scholar]
- Fortin, M.J.; Keitt, T.H.; Maurer, B.A.; Taper, M.L.; Kaufman, D.M.; Blackburn, T.M. Species’ geographic ranges and distributional limits: pattern analysis and statistical issues. Oikos 2005, 108, 7–17. [Google Scholar] [CrossRef]
- Kelly, M.W.; Sanford, E.; Grosberg, R.K. Limited potential for adaptation to climate change in a broadly distributed marine crustacean. Proc. Roy. Soc. B 2012, 279, 349–356. [Google Scholar] [CrossRef]
- Blackburn, T.M.; Cassey, P.; Gaston, K.J. Variations on a theme: sources of heterogeneity in the form of the interspecific relationship between abundance and distribution. J. Anim. Ecol. 2006, 75, 1426–1439. [Google Scholar] [CrossRef]
- Byers, J.E.; Pringle, J.M. Going against the flow: retention, range limits and invasions in advective environments. Mar. Ecol. Prog. Ser. 2006, 313, 27–41. [Google Scholar] [CrossRef]
- Case, T.J.; Taper, M.L. Interspecific competition, environmental gradients, gene flow, and the coevolution of species’ borders. Amer. Nat. 2000, 155, 583–605. [Google Scholar] [CrossRef]
- Gaines, S.D.; Lester, S.; Eckert, G.; Kinlan, B.P.; Sagarin, R.D.; Gaylord, B. Dispersal and geographic ranges in the sea. In Marine Macroecology; Witman, J.D., Roy, K., Eds.; University of Chicago Press: Chicago, IL, USA, 2009; pp. 227–249. [Google Scholar]
- Gaston, K.J. The structure and dynamics of geographic ranges. In Oxford Series Ecology and Evolution; Oxford University Press: Oxford, UK, 2003; p. 266. [Google Scholar]
- Gaylord, B.; Gaines, S.D. Temperature or transport? Range limits in marine species mediated solely by flow. Amer. Nat. 2000, 155, 769–789. [Google Scholar] [CrossRef]
- Gilman, S.E. Experimental manipulations at the northern geographic range limit of an intertidal snail. Ph.D. Thesis, University of California, Davis, CA, USA, 2003. [Google Scholar]
- Helmuth, B.; Mieszkowska, N.; Moore, P.; Hawkins, S.J. Living on the edge of two changing worlds: Forecasting the responses of rocky intertidal ecosystems to climate change. Ann. Rev. Ecol. Evol. Syst. 2006, 37, 373–404. [Google Scholar] [CrossRef]
- Holt, R.D. On the evolutionary ecology of species’ ranges. Evolutionary Ecology Research 2003, 5, 159–178. [Google Scholar]
- Jablonski, D.; Valentine, J.W. From regional to total geographic ranges: testing the relationship in Recent bivalves. Paleobiology 1990, 16, 126–142. [Google Scholar]
- Kirkpatrick, M.; Barton, N.H. Evolution of a species’ range. The American Naturalist 1997, 150, 1–23. [Google Scholar]
- Weersing, K.; Toonen, R.J. Population genetics, larval dispersal, and connectivity in marine systems. Mar. Ecol. Prog. Ser. 2009, 393, 1–12. [Google Scholar] [CrossRef]
- Strathman, R.R. Feeding and nonfeeding larval development and life-history evolution in marine invertebrates. Ann. Rev. Ecol. Syst. 1985, 16, 339–361. [Google Scholar]
- Strathman, R.R. What controls the type of larval development. Bull. Mar. Sci. 1986, 39, 616–622. [Google Scholar]
- Levin, L.A. Recent progress in understanding larval dispersal: new directions and digressions. Integr. Comp. Biol. 2006, 46, 282–297. [Google Scholar] [CrossRef]
- Shanks, A.L. Pelagic larval duration and dispersal distance revisited. Biol. Bull. 2009, 216, 373–385. [Google Scholar]
- Toews, D.P.L.; Brelsford, A. The biogeography of mitochondrial and nuclear discordance in animals. Mol. Ecol. 2012, 21, 3907–3930. [Google Scholar] [CrossRef]
- R_Development_Core_Team, R: A Language and Environment for Statistical Computing, R Foundation for Statistical Computing: Vienna, Austria, 2009.
- Irwin, D.E. Phylogeographic breaks without geographic barriers to gene flow. Evolution 2002, 56, 2383–2394. [Google Scholar]
- Gilman, S.E. Life at the edge: An experimental study of a poleward range boundary. Oecologia 2006, 148, 270–279. [Google Scholar] [CrossRef]
- Zakas, C.; Wares, J.P. Consequences of a poecilogonous life history for genetic structure in coastal populations of the polychaete Streblospio benedicti. Mol. Ecol. 2012, 21, 5447–5460. [Google Scholar] [CrossRef]
- Williams, P.H. Mapping variations in the strength and breadth of biogeographic transition zones using species turnover. Proc. R. Soc. Lond. B 1996, 263, 579–588. [Google Scholar] [CrossRef]
- Engle, V.D.; Summers, J.K. Latitudinal gradients in benthic community composition in Western Atlantic estuaries. J. Biogeogr. 1999, 26, 1007–1023. [Google Scholar] [CrossRef]
- Spalding, M.D.; Fox, H.E.; Halpern, B.S.; McManus, M.A.; Molnar, J.; Allen, G.R.; Davidson, N.; Jorge, Z.A.; Lombana, A.L.; Lourie, S.A.; et al. Marine ecoregions of the world: A bioregionalization of coastal and shelf areas. BioScience 2007, 57, 573–583. [Google Scholar] [CrossRef]
- Avise, J.C. Phylogeography; Harvard University Press: Cambridge, MA, USA, 2000. [Google Scholar]
- Hickerson, M.J.; Meyer, C.P. Testing comparative phylogeographic models of marine vicariance and dispersal using a hierarchical Bayesian approach. BMC Evol. Biol. 2008, 8, 322. [Google Scholar] [CrossRef]
- Nielsen, R.; Beaumont, M.A. Statistical inferences in phylogeography. Mol. Ecol. 2009, 18, 1034–1047. [Google Scholar]
- Huang, W.; Takebayashi, N.; Qi, Y.; Hickerson, M.J. MTML-msBayes: Approximate Bayesian comparative phylogeographic inference from multiple taxa and multiple loci with rate heterogeneity. BMC Bioinformatics 2011, 12, 1. [Google Scholar]
- Wares, J.P. Intraspecific variation and geographic isolation in Idotea balthica (Isopoda: Valvifera). J. Crust. Biol. 2001, 21, 1007–1013. [Google Scholar] [CrossRef]
- Bell, T.M. The maintenance of genetic diversity in the Atlantic isopod, Idotea balthica. Ph.D. Thesis, University of Georgia, Athens, GA, USA, 2009. [Google Scholar]
- Endler, J.A. Problems in distinguishing historical from ecological factors in Biogeography. Amer. Zoolo. 1982, 22, 441–452. [Google Scholar]
- Endler, J.A. Geographic Variation, Speciation, and Clines; Princeton University Press: Princeton, NJ, USA, 1977; p. 246. [Google Scholar]
- Barry, J.P.; Baxter, C.H.; Sagarin, R.D.; Gilman, S.E. Climate-related, long-term faunal changes in a California rocky intertidal community. Science 1995, 267, 672–675. [Google Scholar] [CrossRef]
- Southward, A.J. Forty years of changes in species composition and population density of barnacles on a rocky shore near Plymouth, England, UK. J. Mar. Biol. Assoc. UK 1991, 71, 495–514. [Google Scholar] [CrossRef]
- Perry, A.L.; Low, P.J.; Ellis, J.R.; Reynolds, J.D. Climate change and distribution shifts in marine fishes. Science 2005, 308, 1912–1915. [Google Scholar] [CrossRef]
- Hoffmann, A.A.; Daborn, P.J. Towards genetic markers in animal populations as biomonitors for human-induced environmental change. Ecol. Lett. 2007, 10, 63–76. [Google Scholar] [CrossRef]
- Meirmans, P.G. Using the AMOVA framework to estimate a standardized genetic differentiation measure. Evolution 2006, 60, 2399–2402. [Google Scholar]
- Dawson, M.N. Parallel phylogeographic structure in ecologically similar sympatric sister taxa. Mol. Ecol. 2012, 21, 987–1004. [Google Scholar] [CrossRef]
- Kelly, R.P.; Palumbi, S.R. Genetic Structure Among 50 Species of the Northeastern Pacific Rocky Intertidal Community. PloS One 2010, 5, e8594. [Google Scholar]
- Ilves, K.; Huang, W.; Wares, J.P.; Hickerson, M.J. Colonization and/or mitochondrial selective sweeps across the Atlantic intertidal assemblage revealed by multi-taxa approximate Bayesian computation. Mol. Ecol. 2010, 19, 4505–4519. [Google Scholar] [CrossRef]
- Ling, S.D.; Johnson, C.R.; Ridgway, K.; Hobday, A.J.; Haddon, M. Climate-driven range extension of a sea urchin: inferring future trends by analysis of recent population dynamics. Global Change Biol. 2009, 15, 719–731. [Google Scholar] [CrossRef]
- Wernberg, T.; Russell, B.D.; Moore, P.J.; Ling, S.D.; Smale, D.A.; Campbell, A.; Coleman, M.A.; Steinberg, P.D.; Kendrick, G.A.; Connell, S.D. Impacts of climate change in a global hotspot for temperate marine biodiversity and ocean warming. J. exp. mar. Biol. Ecol. 2011, 400, 7–16. [Google Scholar] [CrossRef]
- Williams, J.W.; Kharouba, H.M.; Veloz, S.; Vellend, M.; McLachlan, J.; Liu, Z.Y.; Otto-Bliesner, B.; He, F. The ice age ecologist: testing methods for reserve prioritization during the last global warming. Global Ecol. Biogeogr. 2012, 22, 289–301. [Google Scholar]
© 2013 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).