In the original version of our article [1], insufficient acknowledgement was given for the source of some of the DNA samples used. We apologize for the original error. To correct this oversight, Yvonne Willi has been added as an author, a recent paper by Willi, Y.
et al. (2010) has been added, the acknowledgements have been altered to more appropriately recognize support and funding, and the sources of samples collected have been corrected in
Table 1.
The corrected author list, acknowledgements, references and Table are provided below:
Author List and Affiliations:
Andrew Tedder 1,2, Peter N. Hoebe 1,3, Yvonne Willi 4, Stephen W. Ansell 5 and Barbara K. Mable 1,*
1 Division of Ecology and Evolutionary Biology, University of Glasgow, Glasgow, G12 8QQ, UK; Email: a.tedder@botinst.uzh.ch (A.T.); peter.hoebe@sac.ac.uk (P.N.H.)
2 Institute of Plant Biology, University of Zürich, Zollikerstrasse 107, CH-8008 Zürich, Switzerland
3 Scottish Agricultural College, West Mains Road, EH9 3JG, Edinburgh, UK
4 Institute of Biology, Evolutionary Botany, University of Neuchâtel, CH-2000 Neuchâtel, Switzerland; Email: yvonne.willi@unine.ch
5 Department of Botany, The Natural History Museum, Cromwell road, London, SW7 5BD, UK; Email: s.ansell@nhm.ac.uk
* Author to whom correspondence should be addressed; Email: barbara.mable@glasgow.ac.uk; Tel.: +44-141-330-3532; Fax: +44-141-330-5971.
Acknowledgements
We thank Aileen Adam for technical assistance and Marc Stift for comments on the manuscript and fruitful discussions. Seeds from samples from North Carolina were generously provided by David Remington. Collection permits were granted by Parks Canada, Ontario Parks, U.S. National Park Service, Palisades Interstate Park Commission, the Nature Conservancy of Eastern New York, the New York State Office of Parks, the Commonwealth of Pennsylvania, the Illinois Department of Natural Resources, the Michigan Department of Natural Resources, the Ohio Department of Natural Resources, the Ohio Nature Conservancy, and John Haataja. AT was supported on a Natural Environment Research Council (NERC) PhD studentship tied to a grant awarded to BKM (NE/D013461/1), PH was supported on a University of Glasgow Institute of Biomedical and Life Sciences PhD studentship, and BKM on a NERC Advanced Research Fellowship (NE/B500094X/1). YW was supported by the Swiss National Science Foundation (31003A-116270 and PP00P3-123396⁄1), the Genetic Diversity Centre of ETH Zürich, and the Foundation Pierre Mercier pour la Science.
References:
1. Tedder, A.; Hoebe, P.N.; Ansell. S.W.; Mable, B.K. Using chloroplast trnF pseudogenes for phylogeography in Arabidopsis lyrata. Diversity 2010, 2, 653-678.
22. Willi, Y.; Määttänen, K. Evolutionary dynamics of mating system shifts in Arabidopsis lyrata. J. Evolutionary Biol. 2010, 23, 2123-2131.
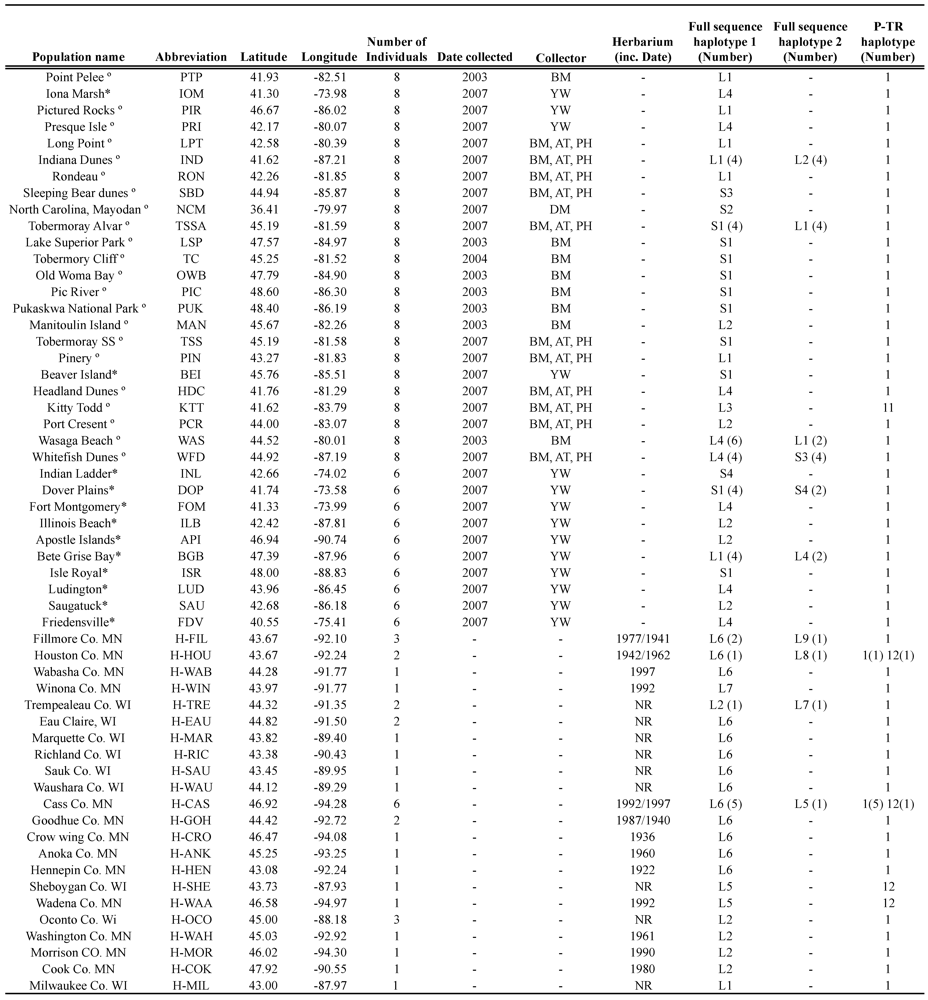
Table 1.
Sampled populations of Arabidopsis lyrata, geographic co-ordinates, date collected, collector and trnF haplotypes. Haplotype assignments are based on the P-TR assignment method and the full sequence haplotype method. º Data taken from Foxe et al. [21]. * New populations described in this paper and collected by Yvonne Willi [22].
Table 1.
Sampled populations of Arabidopsis lyrata, geographic co-ordinates, date collected, collector and trnF haplotypes. Haplotype assignments are based on the P-TR assignment method and the full sequence haplotype method. º Data taken from Foxe et al. [21]. * New populations described in this paper and collected by Yvonne Willi [22].
![Diversity 04 00161 i001]() |