Abstract
Honey is produced by honeybees from nectar and from secretions of living plants. It reflects the honeybees’ diet and the local plant communities. Honey also shows different plant compositions in different geographical locations. We propose a new method for studying the plant diversity and the geographical origin of honey using a DNA barcoding approach that combines universal primers and massive parallel pyrosequencing. To test this method we use two commercial honeys, one from a regional origin and one composed of a worldwide mix of different honeys. We demonstrate that the method proposed here is fast, simple to implement, more robust than classical methods, and therefore suitable for analyzing plant diversity in honey.
1. Introduction
Honey is a natural sweet substance produced by honeybees from the nectar of plants, from secretions of living parts of plants, or from excretions of plant-sucking insects. Once collected, it is subsequently transformed by different substances produced by the honeybee, by dehydration and by maturation in honeycombs [1]. It is mainly composed of carbohydrates, which represent 95% of honey dry weight [2]. It also contains organic acids, proteins, amino acids, minerals, polyphenols, vitamins and aroma compounds.
Honey is an important natural sweetener that was used relatively early in human history, with the first written evidence found in a Sumerian tablet dating back to 2100–2000 B.C. [3]. It has been used for centuries for its medical proprieties for anti-microbial, anti-viral, anti-parasitary, anti-inflammatory, anti-oxidant, anti-mutagenic and anti-tumoral effects [4,5,6,7,8]. These medical proprieties principally depend on the plant composition. For example, it was suggested that a part of the antibacterial activity might be of plant origin [9]. Some groups of plants, such as Senecioneae, Eupatorieae (both tribes of Asteraceae), Boraginaceae, Apocynaceae, and the Crotalaria genus of Fabaceae, produce Pyrrilizidine alkaloids (PA); toxic compounds for humans [10]. Therefore, the exact knowledge of honey composition is very important for human health and it has been demonstrated that PA can be found in honey and pollen products [10,11].
Several methods have been proposed for the determination of botanical and geographical origins of honey. The traditional approach consists of the microscopic examination of pollen present in honey (melissopalynology) [12]. However this method is very tedious to implement, and requires a considerable amount of training. Several chemical methods have also been proposed, such as aroma compounds [13], free-amino acids [14] or minerals and trace elements [15]. Even if all these methods work well for identifying the geographic origins and for distinguishing honey with different botanical origins, they provide only limited information on the plant composition. A recent study has demonstrated that molecular genetics can be used for analyzing the composition and geographical origin of honey [16]: the authors developed real-time PCR systems for detecting the different plant species commonly found in Corsican honey (acacia, linden, citrus, clover, heather, olive, rape, sunflower and rosemary). However, this method requires an a priori knowledge of the plant species that can be found, and must be adjusted according to the potential geographical origin.
In this study, our main objective is to test a universal approach based on DNA barcoding i.e., taxon identification using a standardized DNA region [17]. We used the trnL approach [18] that combines the highly parallel sequencing systems [19] with the amplification of the P6 loop of the the intron of the chloroplast transfer RNA gene for Leucine, codon UAA (trnL (UAA) intron) using universal primers [20]. This technique has already been successfully used in different diet analyses of herbivorous animals [18,21]. To test this method we analyzed two commercial honeys with a potentially complex composition, one from a regional origin (Pyrenean honey) and one composed of a worldwide mix of different honeys (wild flower honey).
2. Results and Discussion
Both honeys were composed of a large variety of different plant taxa: 22 for the Pyrenean honey and 26 for the wild flower honey (Figure 1, Table 1 and Table 2). Results obtained for Pyrenean honey are consistent with expectations for a honey collected in a mountainous area. The high content of Castanea found in the Pyrenean honey was also confirmed by the dark amber colours of the samples typical of unifloral Castanea honey [22]. In the wild flower honey several plant taxa identified were not the result of nectar collection, such as Bryum sp., Athyrium vidalii and Pinus. Their presence is probably the result of the proximity of the hives to a forest and/or the high windiness of the honey collection region. The presence of Athyrium vidalii can give an indication of the honey’s geographical origin. This fern is distributed in China, Japan, Korea and Taiwan and it grows on broad-leaf forest edges or in the undergrowth of Pinus forests [23]. The product label does not give precise indication on the geographical origins; it just states that the product comes from a mix of honey from EU and non-EU countries. The wild flower honey might have been collected in China, the world’s major honey producer (http://faostat.fao.org/site/339/default.aspx). The results obtained for this sample are consistent with this hypothesis.
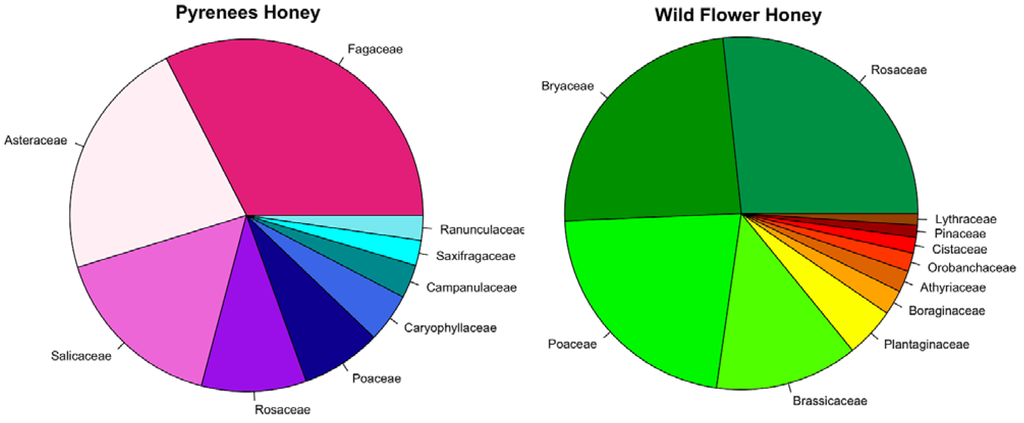
Figure 1.
Pie chart showing the proportion of plant families found in the two different honeys. Only families that compose more than 1% of the total composition were considered.

Table 1.
Plant taxa identified in the Pyrenean Honey.
| Family | Identified Plant | Identification Rank | Nb of Sequences |
|---|---|---|---|
| Asteraceae | Asteraceae | family | 785 |
| Fagaceae | Castanea | genus | 620 |
| Salicaceae | Salicea | tribe | 569 |
| Fagaceae | Fagaceae | family | 527 |
| Rosaceae | Alchemilla | genus | 264 |
| Poaceae | Poeae | tribe | 189 |
| Caryophyllaceae | Sagina | genus | 159 |
| Campanulaceae | Campanulaceae | family | 108 |
| Saxifragaceae | Saxifraga hirculus | species | 82 |
| Ranunculaceae | Ranunculaceae | family | 80 |
| Rosaceae | Rosoideae | subfamily | 75 |
| Poaceae | Avenula aetolica | species | 70 |
| Fabaceae | Robinia | genus | 34 |
| Convolvulaceae | Convolvulaceae | family | 23 |
| Ranunculaceae | Anemone amurensis | species | 18 |
| Gentianaceae | Gentiana | genus | 16 |
| Ophioglossaceae | Botrychium lunaria | species | 14 |
| Onagraceae | Ludwigia hyssopifolia | species | 11 |
| Oleaceae | Ligustrum | genus | 11 |
| Lycopodiaceae | Lycopodiaceae | family | 6 |
| Rosaceae | Torminalis clusii | species | 5 |
| Fabaceae | Trifolium | genus | 5 |
| Total | 3,671 |

Table 2.
Plant taxa identified in the Wild Flower Honey.
| Family | Identified Plant | Identification Rank | Nb of Sequences |
|---|---|---|---|
| Bryaceae | Bryum | genus | 497 |
| Rosaceae | Rosaceae | family | 472 |
| Brassicaceae | Brassicaceae | family | 271 |
| Poaceae | BEP clade | ---- | 241 |
| Poaceae | Poeae | tribe | 216 |
| Plantaginaceae | Plantago | genus | 95 |
| Rosaceae | Rosa | genus | 57 |
| Boraginaceae | Boraginaceae | family | 47 |
| Athyriaceae | Athyrium vidalii | species | 41 |
| Orobanchaceae | Orobanche | genus | 35 |
| Cistaceae | Cistus ladanifer | species | 30 |
| Rosaceae | Alchemilla | genus | 23 |
| Pinaceae | Pinus | genus | 23 |
| Lythraceae | Lythraceae | family | 22 |
| Rosaceae | Rosoideae | subfamily | 21 |
| Cucurbitaceae | Cucurbitaceae | family | 21 |
| Juncaceae | Juncus | genus | 13 |
| Campanulaceae | Campanulaceae | family | 13 |
| Lamiaceae | Nepetoideae | subfamily | 10 |
| Polytrichaceae | Polytrichaceae | family | 9 |
| Ranunculaceae | Aconitum violaceum | species | 7 |
| Asteraceae | Asteraceae | family | 7 |
| Brachytheciaceae | Brachytheciaceae | family | 6 |
| Caryophyllaceae | Sagina nivalis | species | 6 |
| Solanaceae | Solanaceae | family | 4 |
| Salicaceae | Populus | genus | 4 |
| Total | 2,191 |
One of the potential weaknesses of the trnL approach is the low resolution of the P6 loop at the species level. Many closely related species exhibit the same P6 loop sequence. Taberlet et al. have demonstrated that the resolution is better when using a trnL (UAA) intron database restricted to plant species available in the study area instead of the database at the world-wide level retrieved from the public database [20]. Furthermore, some genera exhibit a low variation in their P6 loop (e.g., Carex) or almost no variation (e.g., Salix, Pinus). In this case, the trnL approach can be coupled with one or several additional systems, specially designed for amplifying a short and variable region for these "problematic" genera, as suggested in Valentini et al. [18]. The high carbohydrate concentration in honey helps DNA preservation. Sugars stabilize nucleic acids [24] with a dehydrating and an antibiotic effect, and in a similar way to tree resin, honey provides an airtight seal that prevents oxygen from entering, thus preserving DNA from being degraded. Another strategy that would strongly increase the resolution would be to additionally amplify the whole trnL intron using the universal primers c and d designed by Taberlet et al. [25]. These primers amplify a longer fragment (254–767 bp) showing a higher resolution than the P6 loop fragment [20].
One of the advantages of the trnL approach is that no prior knowledge of honey composition is needed. It does not have any limitation in the number of plants species that can be detected, contrary to the method proposed by Laube and co-authors [16]. Thus, the trnL approach allows the detection of unexpected species in honey, as in the case of Bryum species in the wild flower honey. The exact knowledge of honey composition can be of prime importance for allergic or poisoning agent detection. Even if honey allergy seems uncommon [26], honey poisoning has been well documented in Turkey (mad honey [27]). The use of this approach can lead to more accurate labeling. The 2001/110/EC [28] states that except in the case of filtered or baker's honey, the product names may be supplemented by information referring to: (1) floral or vegetable origin, if the product comes wholly or mainly from the indicated source and possesses the organoleptic, physico-chemical and microscopic characteristics of the source, (2) regional, territorial or topographical origin, if the product comes entirely from the indicated source, (3) specific quality criteria. Another important advantage of the trnL approach is its robustness and simplicity. This approach is particularly well suited for large-scale and standardized analyses, with the possibility of analyzing hundreds of samples in a single sequencing experiment.
3. Experimental Section
3.1. DNA Extraction, Amplification and Sequencing
Total DNA was extracted from about 10 mg of sample with the DNeasy Tissue Kit (Qiagen GmbH), following the manufacturer’s instructions. The DNA extracts were recovered in a total volume of 200 μL. Mock extractions without samples were systematically performed to monitor possible contaminations. DNA amplifications were carried out in a final volume of 25 μL, using 2.5 μL of DNA extract as template. The amplification mixture contained 1 U of AmpliTaq R Gold DNA Polymerase (Applied Biosystems), 10 mM Tris-HCl, 50 mM KCl, 2 mM of MgCl2, 0.2 mM of each dNTP, 0.1 μM of each primer, and 0.005 mg of bovine serum albumin (BSA, Roche Diagnostic). The mixture was denatured at 95 °C for 10 min, followed by 35 cycles of 30 s at 95 °C, and 30 s at 55 °C; the elongation was removed in order to reduce the +A artefact [29,30]. Samples were amplified using the universal primers g and h described by Taberlet et al. [20]. The addition of a specific tag on the 5’ end allowed the assignment of sequences to the respective samples. After amplification all the samples were pooled for the pyrosequencing run. Each sample was recognized by a specific four base long tag for a better assignation of sequences to samples during bioinformatic segregation of sequences.
PCR products were purified using the MinElute PCR purification kit (Qiagen GmbH). DNA quantification was carried out using the NanoDrop ND-1000 UV-Vis Spectrophotometer (NanoDrop Technologies). Then, a mix was made taking into account these DNA concentrations in order to obtain roughly the same number of molecules per PCR product corresponding to the different samples. Large-scale pyrosequencing was carried out using the GS 20 sequencing system (Roche Diagnostic) following the manufacturer’s instructions.
The different output sequences from the pyrosequencing were sorted according to the tag present on the 5’ end of the primers. Thus, a new file was generated for each, containing all the sequences having the relevant tag. Then, these sequences were analyzed to determine the plant diversity. To limit the influence of sequence errors [31], only sequences that were present more than three times were considered in the subsequent analyses.
3.2. Data Analysis
The sequences were compared to a plant sequence database. The sequences were retrieved from the full plant division of GenBank downloaded from NCBI server on the October 16, 2009 (ftp://ftp.ncbi.nlm.nih.gov/genbank) using simulated PCR with the g-h primers (ecoPCR [18], http://www.grenoble.prabi.fr/trac/ecoPCR). The taxon was assigned to each sequence in the dataset by similarity assessment with a reference database using the FASTA [32] algorithm, implemented in ecoTag software (https://www.grenoble.prabi.fr/svn/OBISofts/OBITools/tags/OBITools-1.0.0/src/). An alignment was retrieved if there was at least 98% identity between the query and database sequences, 100% query coverage and 100% subject sequence coverage. If two or more taxa could be assigned the same score for a given sequence, we assigned this sequence to the higher taxonomic level that included both taxa. This method resulted in some sequenced taxa being assigned to the rank of genus or family
4. Conclusions
In this article we demonstrate that the trnL approach is a suitable method for the study of plant composition in honey. It does not need an a priori knowledge of the possible plant species composition. This approach is fast, simple to implement, does not need special skills for the analysis and is more robust than classical methods. It is particularly well adapted for analysing complex situations, when the honey is composed of many different species and it can be easily applied to large-scale studies. It opens new perspectives in the analysis of the honey diversity.
Acknowledgements
We thank two anonymous reviewers for constructive comments on the manuscript. We also thank Eric Coissac and Jessica Rocha for help with the data analysis, and with the English, respectively.
References and Notes
- FAO, Standard for Honey (CODEX STAN 12). In Codex Alimentarius: Sugars, Cocoa Products and Chocolate and Miscellaneous Products; FAO: Rome, Italy, 1981; Vol. 11.
- White, J. Composition of honey. In Honey: A Comprehensive Survey; Crane, E., Ed.; Heinemann: London, UK, 1975; pp. 175–206. [Google Scholar]
- Crane, E. The World History of Beekeeping and Honey Hunting; Gerald Duckworth & Co: London, UK, 1999. [Google Scholar]
- Viuda-Martos, M.; Ruiz-Navajas, Y.; Fernàndez-Lòpez, J.; Pérez-Alvarez, J.A. Functional properties of honey, propolis, and royal jelly. J. Food Sci. 2008, 73, R117–R124. [Google Scholar] [CrossRef]
- Ferreira, I.C.F.R.; Aires, E.; Barreira, J.C.M.; Estevinho, L.M. Antioxidant activity of Portuguese honey samples: Different contributions of the entire honey and phenolic extract. Food Chem. 2009, 114, 1438–1443. [Google Scholar] [CrossRef]
- Socha, R.; Juszczak, L.; Pietrzyk, S.; Fortuna, T. Antioxidant activity and phenolic composition of herbhoneys. Food Chem. 2009, 113, 568–574. [Google Scholar]
- Peterson, J.; Dwyer, J. Flavonoids: Dietary occurrence and biochemical activity. Nutr. Res. 1998, 18, 1995–2018. [Google Scholar] [CrossRef]
- Tsiapara, A.V.; Jaakkola, M.; Chinou, I.; Graikou, K.; Tolonen, T.; Virtanen, V.; Moutsatsou, P. Bioactivity of Greek honey extracts on breast cancer (MCF-7), prostate cancer (PC-3) and endometrial cancer (Ishikawa) cells: Profile analysis of extracts. Food Chem. 2009, 116, 702–708. [Google Scholar] [CrossRef]
- Bogdanov, S. Nature and origin of the antibacterial substances in honey. LWT-Food Sci. Technol. 1997, 30, 748–753. [Google Scholar] [CrossRef]
- Kempf, M.; Heil, S.; Hasslauer, I.; Schmidt, L.; von der Ohe, K.; Theuring, C.; Reinhard, A.; Schreier, P.; Beuerle, T. Pyrrolizidine alkaloids in pollen and pollen products. Mol. Nutr. Food Res. 2010, 54, 292–300. [Google Scholar] [CrossRef]
- Kempf, M.; Reinhard, A.; Beuerle, T. Pyrrolizidine alkaloids (PAs) in honey and pollen-legal regulation of PA levels in food and animal feed required. Mol. Nutr. Food Res. 2010, 54, 158–168. [Google Scholar] [CrossRef]
- Anklam, E. A review of the analytical methods to determine the geographical and botanical origin of honey. Food Chem. 1998, 63, 549–562. [Google Scholar] [CrossRef]
- Cuevas-Glory, L.F.; Pino, J.A.; Santiago, L.S.; Sauri-Duch, E. A review of volatile analytical methods for determining the botanical origin of honey. Food Chem. 2007, 103, 1032–1043. [Google Scholar] [CrossRef]
- Hermosin, I.; Chicon, R.M.; Cabezudo, M.D. Free amino acid composition and botanical origin of honey. Food Chem. 2003, 83, 263–268. [Google Scholar] [CrossRef]
- Fernandez-Torres, R.; Perez-Bernal, J.L.; Bello-Lopez, M.A.; Callejon-Mochon, M.; Jimenez-Sanchez, J.C.; Guiraum-Perez, A. Mineral content and botanical origin of Spanish honeys. Talanta 2005, 65, 686–691. [Google Scholar] [CrossRef]
- Laube, I.; Hird, H.; Brodmann, P.; Ullmann, S.; Schone-Michling, M.; Chisholm, J.; Broll, H. Development of primer and probe sets for the detection of plant species in honey. Food Chem. 2010, 118, 979–986. [Google Scholar] [CrossRef]
- Hebert, P.D.N.; Ratnasingham, S.; deWaard, J.R. Barcoding animal life: cytochrome c oxidase subunit 1 divergences among closely related species. P. Roy. Soc. B-Biol. Sci. 2003, 270, S96–S99. [Google Scholar] [CrossRef]
- Valentini, A.; Miquel, C.; Nawaz, M.A.; Bellemain, E.; Coissac, E.; Pompanon, F.; Gielly, L.; Cruaud, C.; Nascetti, G.; Wincker, P.; Swenson, J.E.; Taberlet, P. New perspectives in diet analysis based on DNA barcoding and parallel pyrosequencing: the trnL approach. Mol. Ecol. Res. 2009, 9, 51–60. [Google Scholar]
- Margulies, M.; Egholm, M.; Altman, W.E.; Attiya, S.; Bader, J.S.; Bemben, L.A.; Berka, J.; Braverman, M.S.; Chen, Y.-J.; Chen, Z.; Dewell, S.B.; Du, L.; Fierro, J.M.; Gomes, X.V.; Godwin, B.C.; He, W.; Helgesen, S.; Ho, C.H.; Irzyk, G.P.; Jando, S.C.; Alenquer, M.L.I.; Jarvie, T.P.; Jirage, K.B.; Kim, J.-B.; Knight, J.R.; Lanza, J.R.; Leamon, J.H.; Lefkowitz, S.M.; Lei, M.; Li, J.; Lohman, K.L.; Lu, H.; Makhijani, V.B.; McDade, K.E.; McKenna, M.P.; Myers, E.W.; Nickerson, E.; Nobile, J.R.; Plant, R.; Puc, B.P.; Ronan, M.T.; Roth, G.T.; Sarkis, G.J.; Simons, J.F.; Simpson, J.W.; Srinivasan, M.; Tartaro, K.R.; Tomasz, A.; Vogt, K.A.; Volkmer, G.A.; Wang, S.H.; Wang, Y.; Weiner, M.P.; Yu, P.; Begley, R.F.; Rothberg, J.M. Genome sequencing in microfabricated high-density picolitre reactors. Nature 2005, 437, 376. [Google Scholar]
- Taberlet, P.; Coissac, E.; Pompanon, F.; Gielly, L.; Miquel, C.; Valentini, A.; Vermat, T.; Corthier, G.; Brochmann, C.; Willerslev, E. Power and limitations of the chloroplast trnL (UAA) intron for plant DNA barcoding. NAR 2007, 35, e14. [Google Scholar] [CrossRef]
- Soininen, E.M.; Valentini, A.; Coissac, E.; Miquel, C.; Gielly, L.; Brochmann, C.; Brysting, A.K.; Sonstebo, J.H.; Ims, R.A.; Yoccoz, N.G.; Taberlet, P. Analysing diet of small herbivores: the efficiency of DNA barcoding coupled with high-throughput pyrosequencing for deciphering the composition of complex plant mixtures. Front. Zool. 2009, 6, 16. [Google Scholar] [CrossRef]
- Persano Oddo, L.; Piazza, M.G.; Sabatini, A.G.; Accorti, M. Characterization of unifloral honeys. Apidologie 1995, 26, 453–465. [Google Scholar] [CrossRef]
- Liu, Y.-C. A Taxonomic Study of Athyrium in Taiwan; National Sun Yat-sen University: Kaohsiung, Taiwan, 2008. [Google Scholar]
- Schill, R.O.; Mali, B.; Dandekar, T.; Schnölzer, M.; Reuter, D.; Frohme, M. Molecular mechanisms of tolerance in tardigrades: New perspectives for preservation and stabilization of biological material. Biotechnol. Adv. 2009, 27, 348–352. [Google Scholar] [CrossRef]
- Taberlet, P.; Gielly, L.; Pautou, G.; Bouvet, J. Universal primers for amplification of 3 noncoding regions of chloroplast DNA. Plant Mol. Biol. 1991, 17, 1105–1109. [Google Scholar] [CrossRef]
- Bogdanov, S.; Jurendic, T.; Sieber, R.; Gallmann, P. Honey for nutrition and health: a review. J. Am. Coll. Nutr. 2008, 27, 677–689. [Google Scholar] [CrossRef]
- Koca, I.; Koca, A.F. Poisoning by mad honey: A brief review. Food Chem. Toxicol. 2007, 45, 1315–1318. [Google Scholar] [CrossRef]
- Council Directive 2001/110/EC of 20 December 2001 realting honey. Offic. J. Eur. Communities, 2001; Vol. 2001/110/EC, 6.
- Brownstein, M.J.; Carpten, J.D.; Smith, J.R. Modulation of non-templated nucleotide addition by Taq DNA polymerase: primer modifications that facilitate genotyping. Biotechniques 1996, 20, 1004-1006, 1008-1010. [Google Scholar]
- Magnuson, V.L.; Ally, D.S.; Nylund, S.J.; Karanjawala, Z.E.; Rayman, J.B.; Knapp, J.I.; Lowe, A.L.; Ghosh, S.; Collins, F.S. Substrate nucleotide-determined non-templated addition of adenine by Taq DNA polymerase: implications for PCR-based genotyping and cloning. Biotechniques 1996, 21, 700–709. [Google Scholar]
- Huse, S.M.; Huber, J.A.; Morrison, H.G.; Sogin, M.L.; Welch, D.M. Accuracy and quality of massively parallel DNA pyrosequencing. Genome Biol. 2007, 8. [Google Scholar]
- Pearson, W.R.; Lipman, D.J. Improved tools for biological sequence comparison. Proc. Natl. Acad. Sci. USA 1988, 85, 2444–2448. [Google Scholar] [CrossRef]
© 2010 by the authors; licensee MDPI, Basel, Switzerland. This article is an open-access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).