Global Amphibian Declines, Loss of Genetic Diversity and Fitness: A Review
Abstract
:1. Introduction
1.1. Why Are Amphibians Particularly Vulnerable
1.1.1. Small effective population sizes and whole clutch mortality
1.1.2. Population declines
1.1.3. Habitat fragmentation
1.1.4. Low dispersal rates
2. Genetic Diversity and Fitness in Amphibians
2.1. Genetic-Fitness Correlations
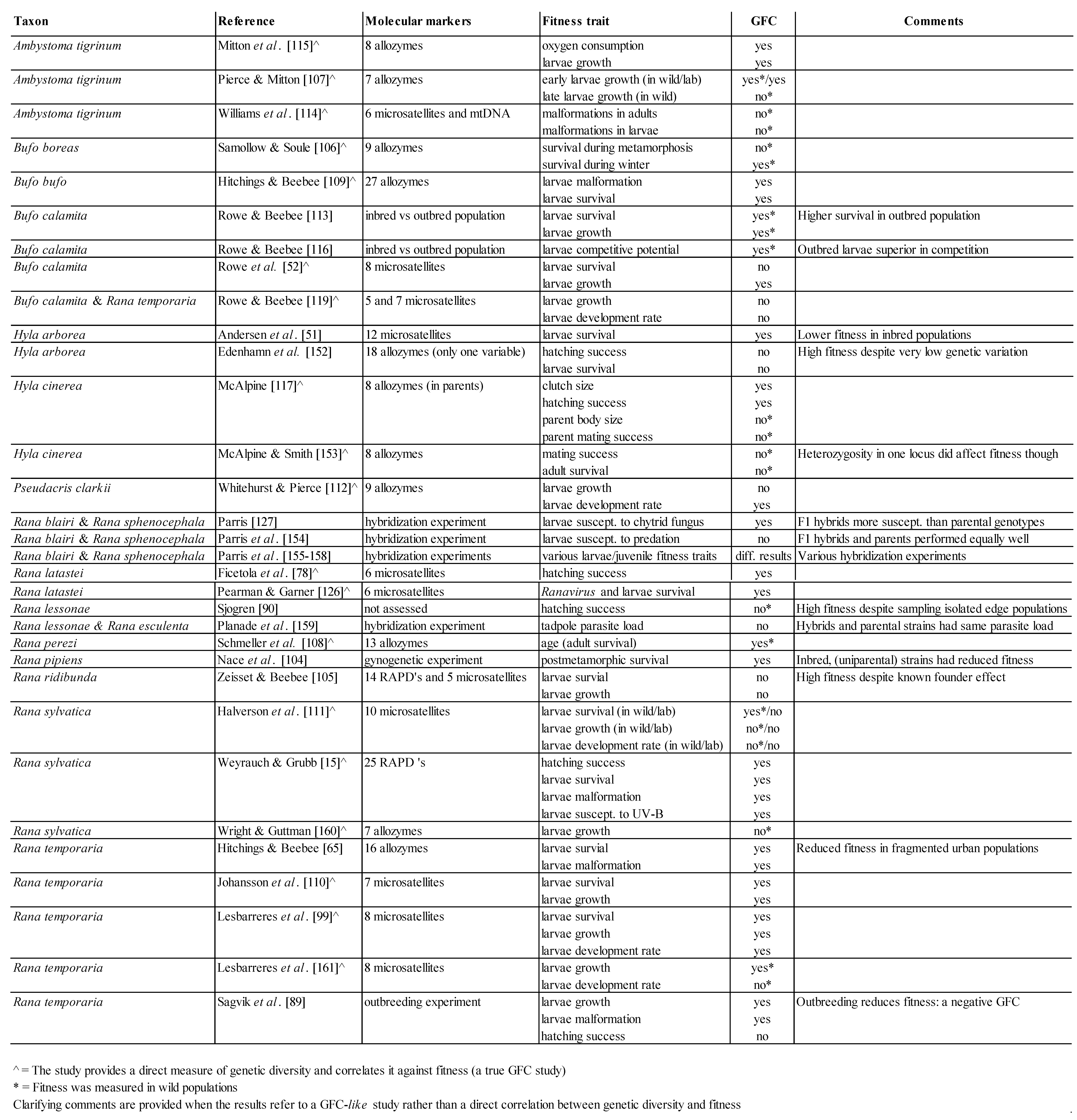
2.2. Review of GFCs in Amphibians
2.2.1. Measures of fitness
2.2.2. Limitations
2.2.3. GFCs and synergisms
3. Conclusions
4. Textbox
4.1. Genetic Drift and Inbreeding—Big ‘Players’ in Small Population Genetics
Acknowledgements
References and Notes
- Wake, D.B.; Vredenburg, V.T. Are we in the midst of the sixth mass extinction? A view from the world of amphibians. Proc. Nat. Acad. Sci. USA 2008, 105, 11466–11473. [Google Scholar] [CrossRef] [PubMed]
- Pimm, S.L.; Russell, G.J.; Gittleman, J.L.; Brooks, T.M. The future of biodiversity. Science 1995, 269, 347–350. [Google Scholar] [CrossRef] [PubMed]
- Chapin, F.S.; Zavaleta, E.S.; Eviner, V.T.; Naylor, R.L.; Vitousek, P.M.; Reynolds, H.L.; Hooper, D.U.; Lavorel, S.; Sala, O.E.; Hobbie, S.E.; Mack, M.C.; Diaz, S. Consequences of changing biodiversity. Nature 2000, 405, 234–242. [Google Scholar] [CrossRef] [PubMed]
- Schellnhuber, H.J.; Kokott, J.; Beese, F.O.; Fraedrich, K.; Klemmer, P.; Kruse-Graumann, L.; Neumann, C.; Renn, O.; Schulze, E.D.; Tilzer, M.; Velsinger, P.; Zimmermann, H. World in Transition: Conservation and Sustainable use of the Biosphere; Earthscan Publications Ltd: London, UK, 2001. [Google Scholar]
- Stuart, S.N.; Chanson, J.S.; Cox, N.A.; Young, B.E.; Rodrigues, A.S.L.; Fischman, D.L.; Waller, R.W. Status and trends of amphibian declines and extinctions worldwide. Science 2004, 306, 1783–1786. [Google Scholar] [CrossRef] [PubMed]
- McCallum, M.L. Amphibian decline or extinction? Current declines dwarf background extinction rate. J. Herpetol. 2007, 41, 483–491. [Google Scholar] [CrossRef]
- Beebee, T.J.; Wilkinson, J.; Buckley, J. Amphibian declines are not uniquely high amongst the vertebrates: trend determination and the British perspective. Diversity 2009, 1, 67–88. [Google Scholar] [CrossRef]
- Wind, E. Effects of habitat fragmentation on amphibians: what do we know and where do we go from here? In Proceedings of the Biology and Management of Species and Habitats at Risk; Darling, L.M., Ed.; B.C. Ministry of Environment, Lands and Parks: Victoria B.C. and University College of the Cariboo, Kamloops B.C.. , 1999; pp. 885–894. [Google Scholar]
- Cushman, S.A. Effects of habitat loss and fragmentation on amphibians: a review and prospectus. Biol. Conserv. 2006, 128, 231–240. [Google Scholar] [CrossRef]
- Gallant, A.L.; Klaver, R.W.; Casper, G.S.; Lannoo, M.J. Global rates of habitat loss and implications for amphibian conservation. Copeia 2007, 4, 967–979. [Google Scholar] [CrossRef]
- Gardner, T.A.; Barlow, J.; Peres, C.A. Paradox, presumption and pitfalls in conservation biology: the importance of habitat change for amphibians and reptiles. Biol. Conserv. 2007, 138, 166–179. [Google Scholar] [CrossRef]
- Carey, C.; Alexander, M.A. Climate change and amphibian declines: is there a link? Divers. Distrib. 2003, 9, 111–121. [Google Scholar] [CrossRef]
- Corn, P. S. Climate change and amphibians. Anim. Biodivers. Conserv. 2005, 28, 59–67. [Google Scholar]
- Licht, L.E.; Grant, K.P. The effects of ultraviolet radiation on the biology of amphibians. Amer. Zool. 1997, 37, 137–145. [Google Scholar]
- Weyrauch, S.L.; Grubb, T.C. Effects of the interaction between genetic diversity and UV-B radiation on wood frog fitness. Biol. Conserv. 2006, 20, 802–810. [Google Scholar] [CrossRef]
- Blaustein, A.R.; Kiesecker, J.M. Complexity in conservation: lessons from the global decline of amphibian populations. Ecol. Lett. 2002, 5, 597–608. [Google Scholar] [CrossRef]
- Kats, L.B.; Ferrer, R.P. Alien predators and amphibian declines: review of two decades of science and the transition to conservation. Divers. Distrib. 2003, 9, 99–110. [Google Scholar] [CrossRef]
- Carey, C.; Bryant, C.J. Possible interrelations among environmental toxicants, amphibian development, and decline of amphibian populations. Environ. Health Perspect. 1995, 103, 13–17. [Google Scholar] [CrossRef] [PubMed]
- Marco, A.; Quilchano, C.; Blaustein, A.R. Sensitivity to nitrate and nitrite in pond-breeding amphibians from the Pacific northwest, USA. Environ. Toxicol. Chem. 1999, 18, 2836–2839. [Google Scholar] [CrossRef]
- Hels, T.; Buchwald, E. The effect of road kills on amphibian populations. Biol. Conserv. 2001, 99, 331–340. [Google Scholar] [CrossRef]
- Gibbs, J.P.; Shriver, W.G. Can road mortality limit populations of pool-breeding amphibians? Wetlands Ecol. Manag. 2005, 13, 281–289. [Google Scholar]
- Warkentin, I.G.; Bickford, D.; Sodhi, N.S.; Bradshaw, C.J.A. Eating frogs to extinction. Conserv. Biol. 2009, 23, 1056–1059. [Google Scholar] [CrossRef] [PubMed]
- Daszak, P.; Cunningham, A.A.; Hyatt, A.D. Infectious disease and amphibian population declines. Divers. Distrib. 2003, 9, 141–150. [Google Scholar] [CrossRef]
- James, T.Y.; Litvintseva, A.P.; Vilgalys, R.; Morgan, J.A.T.; Taylor, J.W.; Fisher, M.C.; Berger, L.; Weldon, C.; du Preez, L.; Longcore, J.E. Rapid global expansion of the fungal disease chytridiomycosis into declining and healthy amphibian populations. PLoS Pathog. 2009, 5, e1000458. [Google Scholar] [CrossRef] [PubMed]
- Rödder, D.; Kielgast, J.; Bielby, J.; Schmidtlein, S.; Bosch, J.; Garner, T.W.; Veith, M.; Walker, S.; Fisher, M.; Lötters, S. Global amphibian extinction risk assessment for the panzootic chytrid fungus. Diversity 2009, 1, 52–66. [Google Scholar] [CrossRef]
- Voyles, J.; Young, S.; Berger, L.; Campbell, C.; Voyles, W.F.; Dinudom, A.; Cook, D.; Webb, R.; Alford, R.A.; Skerratt, L.F.; Speare, R. Pathogenesis of chytridiomycosis, a cause of catastrophic amphibian declines. Science 2009, 326, 582–585. [Google Scholar] [CrossRef] [PubMed]
- Collins, J.P.; Storfer, A. Global amphibian declines: sorting the hypotheses. Divers. Distrib. 2003, 9, 89–98. [Google Scholar] [CrossRef]
- Blaustein, A.R.; Bancroft, B.A. Amphibian population declines: Evolutionary considerations. Bioscience 2007, 57, 437–444. [Google Scholar] [CrossRef]
- Pounds, J.A.; Bustamante, M.R.; Coloma, L.A.; Consuegra, J.A.; Fogden, M.P.L.; Foster, P.N.; La Marca, E.; Masters, K.L.; Merino-Viteri, A.; Puschendorf, R.; Ron, S.R.; Sanchez-Azofeifa, G.A.; Still, C.J.; Young, B.E. Widespread amphibian extinctions from epidemic disease driven by global warming. Nature 2006, 439, 161–167. [Google Scholar] [CrossRef] [PubMed]
- Kiesecker, J.M.; Blaustein, A.R. Synergism between UV-B radiation and a pathogen magnifies amphibian embryo mortality in nature. Proc. Nat. Acad. Sci. USA 1995, 92, 11049–11052. [Google Scholar] [CrossRef] [PubMed]
- Rohr, J.R.; Schotthoefer, A.M.; Raffel, T.R.; Carrick, H.J.; Halstead, N.; Hoverman, J.T.; Johnson, C.M.; Johnson, L.B.; Lieske, C.; Piwoni, M.D.; Schoff, P.K.; Beasley, V.R. Agrochemicals increase trematode infections in a declining amphibian species. Nature 2008, 455, 1235–1239. [Google Scholar] [CrossRef] [PubMed]
- Shaffer, M.L. Population viability analysis. Conserv. Biol. 1990, 4, 39–40. [Google Scholar] [CrossRef]
- Frankel, O.H.; Soulé, M.E. Conservation and Evolution; Cambridge University Press: Cambridge, UK, 1981. [Google Scholar]
- Willi, Y.; Van Buskirk, J.; Hoffmann, A.A. Limits to the adaptive potential of small populations. Ann. Rev. Ecol. Evol. Syst. 2006, 37, 433–458. [Google Scholar] [CrossRef]
- Frankham, R.; Ballou, J.D.; Briscoe, D.A. Introduction to Conservation Genetics; Cambridge University Press: Cambridge, UK, 2002. [Google Scholar]
- Funk, W.C.; Tallmon, D.A.; Allendorf, F.W. Small effective population size in the long-toed salamander. Mol. Ecol. 1999, 8, 1633–1640. [Google Scholar] [CrossRef] [PubMed]
- Beebee, T.J.C.; Griffiths, R.A. The amphibian decline crisis: a watershed for conservation biology? Biol. Conserv. 2005, 125, 271–285. [Google Scholar] [CrossRef]
- Schmeller, D.S.; Merila, J. Demographic and genetic estimates of effective population and breeding size in the amphibian Rana temporaria. Conserv. Biol. 2007, 21, 142–151. [Google Scholar] [CrossRef] [PubMed]
- Scribner, K.T.; Arntzen, J.W.; Burke, T. Effective number of breeding adults in Bufo bufo estimated from age-specific variation at minisatellite loci. Mol. Ecol. 1997, 6, 701–712. [Google Scholar] [CrossRef] [PubMed]
- Rowe, G.; Beebee, T.J.C. Reconciling genetic and demographic estimators of effective population size in the anuran amphibian Bufo calamita. Conserv. Genet. 2004, 5, 287–298. [Google Scholar] [CrossRef]
- Pechmann, J.H.K.; Scott, D.E.; Semlitsch, R.D.; Caldwell, J.P.; Vitt, L.J.; Gibbons, J.W. Declining amphibian populations—the problem of separating human impacts from natural fluctuations. Science 1991, 253, 892–895. [Google Scholar] [CrossRef] [PubMed]
- Richter, S.C.; Young, J.E.; Johnson, G.N.; Seigel, R.A. Stochastic variation in reproductive success of a rare frog, Rana sevosa: implications for conservation and for monitoring amphibian populations. Biol. Conserv. 2003, 111, 171–177. [Google Scholar] [CrossRef]
- Alford, R.A.; Richards, S.J. Global amphibian declines: a problem in applied ecology. Ann. Rev. Ecol. Syst. 1999, 30, 133–165. [Google Scholar] [CrossRef]
- Ficetola, G.F.; Padoa-Schioppa, E.; Wang, J.; Garner, T.W.J. Polygyny, census and effective population size in the threatened frog, Rana latastei. Anim. Conserv. 2009. [Google Scholar] [CrossRef]
- Vucetich, J.A.; Waite, T.A. Erosion of heterozygosity in fluctuating populations. Conserv. Biol. 1999, 13, 860–868. [Google Scholar] [CrossRef]
- Cabe, P.R.; Page, R.B.; Hanlon, T.J.; Aldrich, M.E.; Connors, L.; Marsh, D.M. Fine-scale population differentiation and gene flow in a terrestrial salamander (Plethodon cinereus) living in continuous habitat. Heredity 2007, 98, 53–60. [Google Scholar] [CrossRef] [PubMed]
- Driscoll, D.A. Genetic structure of the frogs Geocrinia lutea and Geocrinia rosea reflects extreme population divergence and range changes, not dispersal barriers. Evolution 1998, 52, 1147–1157. [Google Scholar] [CrossRef]
- Dubois, A. Developmental pathway, speciation and supraspecific taxonomy in amphibians 1. Why are there so many frog species in Sri Lanka? Alytes 2004, 22, 19–37. [Google Scholar]
- Sodhi, N.S.; Bickford, D.; Diesmos, A.C.; Lee, T.M.; Koh, L.P.; Brook, B.W.; Sekercioglu, C.H.; Bradshaw, C.J.A. Measuring the meltdown: drivers of global amphibian extinction and decline. PLoS One 2008, 3. Article No.: e1636. [Google Scholar]
- Frankham, R. Relationship of genetic variation to population size in wildlife. Conserv. Biol. 1996, 10, 1500–1508. [Google Scholar] [CrossRef]
- Andersen, L.W.; Fog, K.; Damgaard, C. Habitat fragmentation causes bottlenecks and inbreeding in the European tree frog (Hyla arborea). Proc. Roy. Soc. London Ser. B 2004, 271, 1293–1302. [Google Scholar] [CrossRef]
- Rowe, G.; Beebee, T.J.C.; Burke, T. Microsatellite heterozygosity, fitness and demography in natterjack toads Bufo calamita. Anim. Conserv. 1999, 2, 85–92. [Google Scholar] [CrossRef]
- Marsh, D.M.; Trenham, P.C. Metapopulation dynamics and amphibian conservation. Conserv. Biol. 2001, 15, 40–49. [Google Scholar] [CrossRef]
- Hanski, I. Metapopulation dynamics. Nature 1998, 396, 41–49. [Google Scholar] [CrossRef]
- Fischer, M. Species loss after habitat fragmentation. Trend. Ecol. Evol. 2000, 15, 396–396. [Google Scholar] [CrossRef]
- Harrison, S.; Bruna, E. Habitat fragmentation and large-scale conservation: what do we know for sure? Ecography 1999, 22, 225–232. [Google Scholar] [CrossRef]
- Pimm, S.L.; Raven, P. Biodiversity—Extinction by numbers. Nature 2000, 403, 843–845. [Google Scholar] [CrossRef] [PubMed]
- Tilman, D.; May, R.M.; Lehman, C.L.; Nowak, M.A. Habitat destruction and the extinction depth. Nature 1994, 371, 65–66. [Google Scholar] [CrossRef]
- DiBattista, J.D. Patterns of genetic variation in anthropogenically impacted populations. Conserv. Genet. 2008, 9, 141–156. [Google Scholar] [CrossRef]
- Toro, M.A.; Caballero, A. Characterization and conservation of genetic diversity in subdivided populations. Phil. Trans. Roy. Soc. B-Biol. Sci. 2005, 360, 1367–1378. [Google Scholar]
- Young, A.G.; Clarke, G.M. Genetics, Demography and Viability of Fragmented Populations; Cambridge University Press: Cambridge, UK, 2000. [Google Scholar]
- Allentoft, M.E.; Siegismund, H.R.; Briggs, L.; Andersen, L.W. Microsatellite analysis of the natterjack toad (Bufo calamita) in Denmark: populations are islands in a fragmented landscape. Conserv. Genet. 2009, 10, 15–28. [Google Scholar] [CrossRef]
- Richter, S.C.; Crother, B.I.; Broughton, R.E. Genetic consequences of population reduction and geographic isolation in the critically endangered frog, Rana sevosa. Copeia 2009, 801–808. [Google Scholar]
- Arens, P.; van der Sluis, T.; van't Westende, W.P.C.; Vosman, B.; Vos, C.C.; Smulders, M.J.M. Genetic population differentiation and connectivity among fragmented Moor frog (Rana arvalis) populations in The Netherlands. Landscape Ecol. 2007, 22, 1489–1500. [Google Scholar] [CrossRef]
- Hitchings, S.P.; Beebee, T.J.C. Genetic substructuring as a result of barriers to gene flow in urban Rana temporaria (common frog) populations: implications for biodiversity conservation. Heredity 1997, 79, 117–127. [Google Scholar] [CrossRef] [PubMed]
- Lesbarreres, D.; Primmer, C.R.; Lode, T.; Merila, J. The effects of 20 years of highway presence on the genetic structure of Rana dalmatina populations. Ecoscience 2006, 13, 531–538. [Google Scholar] [CrossRef]
- Reh, W.; Seitz, A. The influence of land use on the genetic structure of populations of the common frog Rana temporaria. Biol. Conserv. 1990, 54, 239–249. [Google Scholar] [CrossRef]
- Vos, C.C.; Antonisse-De Jong, A.G.; Goedhart, P.W.; Smulders, M.J.M. Genetic similarity as a measure for connectivity between fragmented populations of the moor frog (Rana arvalis). Heredity 2001, 86, 598–608. [Google Scholar] [CrossRef] [PubMed]
- Spear, S.F.; Storfer, A. Landscape genetic structure of coastal tailed frogs (Ascaphus truei) in protected vs. managed forests. Mol. Ecol. 2008, 17, 4642–4656. [Google Scholar]
- Curtis, J.M.R.; Taylor, E.B. The genetic structure of coastal giant salamanders (Dicamptodon tenebrosus) in a managed forest. Biol. Conserv. 2004, 115, 45–54. [Google Scholar] [CrossRef]
- Dixo, M.; Metzger, J.P.; Morgante, J.S.; Zamudio, K.R. Habitat fragmentation reduces genetic diversity and connectivity among toad populations in the Brazilian Atlantic Coastal Forest. Biol. Conserv. 2009, 142, 1560–1569. [Google Scholar] [CrossRef]
- Telles, M.P.de C.; Diniz-Filho, J.A.F.; Bastos, R.P.; Soares, T.N.; Guimaraes, L.D.; Lima, L.P. Landscape genetics of Physalaemus cuvieri in Brazilian cerrado: correspondence between population structure and patterns of human occupation and habitat loss. Biol. Conserv. 2007, 139, 37–46. [Google Scholar]
- Burns, E.L.; Eldridge, M.D.B.; Houlden, B.A. Microsatellite variation and population structure in a declining Australian hylid Litoria aurea. Mol. Ecol. 2004, 13, 1745–1757. [Google Scholar] [CrossRef] [PubMed]
- Hazell, D. Frog ecology in modified Australian landscapes: a review. Wildlife Res. 2003, 30, 193–205. [Google Scholar] [CrossRef]
- Funk, W.C.; Blouin, M.S.; Corn, P.S.; Maxell, B.A.; Pilliod, D.S.; Amish, S.; Allendorf, F.W. Population structure of Columbia spotted frogs (Rana luteiventris) is strongly affected by the landscape. Mol. Ecol. 2005, 14, 483–496. [Google Scholar] [CrossRef] [PubMed]
- Garner, T.W.J.; Pearman, P.B.; Angelone, S. Genetic diversity across a vertebrate species' range: a test of the central-peripheral hypothesis. Mol. Ecol. 2004, 13, 1047–1053. [Google Scholar] [CrossRef] [PubMed]
- Beebee, T.J.C.; Rowe, G. Microsatellite analysis of natterjack toad Bufo calamita Laurenti populations: consequences of dispersal from a Pleistocene refugium. Biol. J. Linn. Soc. 2000, 69, 367–381. [Google Scholar] [CrossRef]
- Ficetola, G.F.; Garner, T.W.J.; De Bernardi, F. Genetic diversity, but not hatching success, is jointly affected by postglacial colonization and isolation in the threatened frog, Rana latastei. Mol. Ecol. 2007, 16, 1787–1797. [Google Scholar] [CrossRef] [PubMed]
- Blaustein, A.R.; Wake, D.B.; Sousa, W.P. Amphibian declines―judging stability, persistence, and susceptibility of populations to local and global extinctions. Conserv. Biol. 1994, 8, 60–71. [Google Scholar] [CrossRef]
- Tallmon, D.A.; Funk, W.C.; Dunlap, W.W.; Allendorf, F.W. Genetic differentiation among Long-Toed Salamander (Ambystoma macrodactylum) populations. Copeia 2000, 27–35. [Google Scholar] [CrossRef]
- Kraaijeveld-Smit, F.J.L.; Beebee, T.J.C.; Griffiths, R.A.; Moore, R.D.; Schley, L. Low gene flow but high genetic diversity in the threatened Mallorcan midwife toad Alytes muletensis. Mol. Ecol. 2005, 14, 3307–3315. [Google Scholar] [CrossRef] [PubMed]
- Spear, S.F.; Peterson, C.R.; Matocq, M.D.; Storfer, A. Landscape genetics of the blotched tiger salamander (Ambystoma tigrinum melanostictum). Mol. Ecol. 2005, 14, 2553–2564. [Google Scholar] [CrossRef] [PubMed]
- Sinsch, U. Migration and orientation in anuran amphibians. Ethol. Ecol. Evol. 1990, 2, 65–79. [Google Scholar] [CrossRef]
- Rothermel, B.B.; Semlitsch, R.D. An experimental investigation of landscape resistance of forest versus old-field habitats to emigrating juvenile amphibians. Conserv. Biol. 2002, 16, 1324–1332. [Google Scholar] [CrossRef]
- Trenham, P.C.; Shaffer, H.B. Amphibian upland habitat use and its consequences for population viability. Ecol. Appl. 2005, 15, 1158–1168. [Google Scholar] [CrossRef]
- Smith, M.A.; Green, D.M. Dispersal and the metapopulation paradigm in amphibian ecology and conservation: are all amphibian populations metapopulations? Ecography 2005, 28, 110–128. [Google Scholar] [CrossRef]
- Phillips, B.L.; Shine, R. Spatial and temporal variation in the morphology (and thus, predicted impact) of an invasive species in Australia. Ecography 2006, 29, 205–212. [Google Scholar] [CrossRef]
- Bowne, D.R.; Bowers, M.A. Interpatch movements in spatially structured populations: a literature review. Landscape Ecol. 2004, 19, 1–20. [Google Scholar] [CrossRef]
- Sagvik, J.; Uller, T.; Olsson, M. Outbreeding depression in the common frog, Rana temporaria. Conserv. Genet. 2005, 6, 205–211. [Google Scholar] [CrossRef]
- Sjogren, P. Extinction and isolation gradients in metapopulations - the case of the pool frog (Rana lessonae). Biol. J. Linn. Soc. 1991, 42, 135–147. [Google Scholar] [CrossRef]
- Vos, C.C.; Chardon, J.P. Effects of habitat fragmentation and road density on the distribution pattern of the moor frog Rana arvalis. J. Appl. Ecol. 1998, 35, 44–56. [Google Scholar] [CrossRef]
- Carr, L.W.; Fahrig, L. Effect of road traffic on two amphibian species of differing vagility. Conserv. Biol. 2001, 15, 1071–1078. [Google Scholar] [CrossRef]
- Funk, W.C.; Greene, A.E.; Corn, P.S.; Allendorf, F.W. High dispersal in a frog species suggests that it is vulnerable to habitat fragmentation. Biol. Lett. 2005, 1, 13–16. [Google Scholar] [CrossRef] [PubMed]
- Crnokrak, P.; Roff, D.A. Inbreeding depression in the wild. Heredity 1999, 83, 260–270. [Google Scholar] [CrossRef] [PubMed]
- Hedrick, P.W.; Kalinowski, S.T. Inbreeding depression in conservation biology. Ann. Rev. Ecol. Syst. 2000, 31, 139–162. [Google Scholar] [CrossRef]
- Hansson, B.; Westerberg, L. On the correlation between heterozygosity and fitness in natural populations. Mol. Ecol. 2002, 11, 2467–2474. [Google Scholar] [CrossRef] [PubMed]
- DeWoody, Y.D.; DeWoody, J.A. On the estimation of genome-wide heterozygosity using molecular markers. J. Hered. 2005, 96, 85–88. [Google Scholar] [CrossRef] [PubMed]
- Balloux, F.; Amos, W.; Coulson, T. Does heterozygosity estimate inbreeding in real populations? Mol. Ecol. 2004, 13, 3021–3031. [Google Scholar]
- Lesbarreres, D.; Primmer, S.R.; Laurila, A.; Merila, J. Environmental and population dependency of genetic variability-fitness correlations in Rana temporaria. Mol. Ecol. 2005, 14, 311–323. [Google Scholar] [CrossRef] [PubMed]
- Reed, D.H.; Frankham, R. Correlation between fitness and genetic diversity. Conserv. Biol. 2003, 17, 230–237. [Google Scholar] [CrossRef]
- Coltman, D.W.; Slate, J. Microsatellite measures of inbreeding: a meta-analysis. Evolution 2003, 57, 971–983. [Google Scholar] [CrossRef] [PubMed]
- Chapman, J.R.; Nakagawa, S.; Coltman, D.W.; Slate, J.; Sheldon, B.C. A quantitative review of heterozygosity-fitness correlations in animal populations. Mol. Ecol. 2009, 18, 2746–2765. [Google Scholar] [CrossRef] [PubMed]
- Gower, D.J.; Wilkinson, M. Conservation biology of caecilian amphibians. Conserv. Biol. 2005, 19, 45–55. [Google Scholar] [CrossRef]
- Nace, G.W.; Richards, C.M.; Asher, J.H. Parthenogenesis and genetic variability.1. Linkage and inbreeding estimations in the frog, Rana pipiens. Genetics 1970, 66, 349–368. [Google Scholar]
- Zeisset, I.; Beebee, T.J.C. Population genetics of a successful invader: the marsh frog Rana ridibunda in Britain. Mol. Ecol. 2003, 12, 639–646. [Google Scholar] [CrossRef] [PubMed]
- Samollow, P.B.; Soule, M.E. A case of stress related heterozygote superiority in nature. Evolution 1983, 37, 646–649. [Google Scholar] [CrossRef]
- Pierce, B.A.; Mitton, J.B. Allozyme heterozygosity and growth in the tiger salamander, Ambostyma tigrinum. J. Hered. 1982, 73, 250–253. [Google Scholar] [PubMed]
- Schmeller, D.S.; Schregel, J.; Veith, M. The importance of heterozygosity in a frog's life. Naturwissenschaften 2007, 94, 360–366. [Google Scholar] [CrossRef] [PubMed]
- Hitchings, S.P.; Beebee, T.J.C. Loss of genetic diversity and fitness in Common Toad (Bufo bufo) populations isolated by inimical habitat. J. Evol. Biol. 1998, 11, 269–283. [Google Scholar]
- Johansson, M.; Primmer, C.R.; Merila, J. Does habitat fragmentation reduce fitness and adaptability? A case study of the common frog (Rana temporaria). Mol. Ecol. 2007, 16, 2693–2700. [Google Scholar] [CrossRef] [PubMed]
- Halverson, M.A.; Skelly, D.K.; Caccone, A. Inbreeding linked to amphibian survival in the wild but not in the laboratory. J. Hered. 2006, 97, 499–507. [Google Scholar] [CrossRef] [PubMed]
- Whitehurst, P.H.; Pierce, B.A. The relationship between allozyme variation and life history traits of the spotted chorus frog Pseudacris clarkii. Copeia 1991, 1032–1039. [Google Scholar] [CrossRef]
- Rowe, G.; Beebee, T.J.C. Population on the verge of a mutational meltdown? Fitness costs of genetic load for an amphibian in the wild. Evolution 2003, 57, 177–181. [Google Scholar] [CrossRef] [PubMed]
- Williams, R.N.; Bos, D.H.; Gopurenko, D.; DeWoody, J.A. Amphibian malformations and inbreeding. Biol. Lett. 2008, 4, 549–552. [Google Scholar] [CrossRef] [PubMed]
- Mitton, J.B.; Carey, C.; Kocher, T.D. The relation of enzyme heterozygosity to standard and active oxygen-consumption and body size of tiger salamanders, Ambystoma tigrinum. Physiol. Zool. 1986, 59, 574–582. [Google Scholar]
- Rowe, G.; Beebee, T.J.C. Intraspecific competition disadvantages inbred natterjack toad (Bufo calamita) genotypes over outbred ones in a shared pond environment. J. Anim. Ecol. 2005, 74, 71–76. [Google Scholar] [CrossRef]
- McAlpine, S. Genetic heterozygosity and reproductive success in the green treefrog, Hyla cinerea. Heredity 1993, 70, 553–558. [Google Scholar] [CrossRef]
- Slate, J.; David, P.; Dodds, K.G.; Veenvliet, B.A.; Glass, B.C.; Broad, T.E.; McEwan, J.C. Understanding the relationship between the inbreeding coefficient and multilocus heterozygosity: theoretical expectations and empirical data. Heredity 2004, 93, 255–265. [Google Scholar] [CrossRef] [PubMed]
- Rowe, G.; Beebee, T.J.C. Fitness and microsatellite diversity estimates were not correlated in two outbred anuran populations. Heredity 2001, 87, 558–565. [Google Scholar] [CrossRef] [PubMed]
- Slate, J.; Pemberton, J. Does reduced heterozygosity depress sperm quality in wild rabbits (Oryctolagus cuniculus)? Curr. Biol. 2006, 16, R790–R791. [Google Scholar]
- Baer, B.; Schmid-Hempel, P. Experimental variation in polyandry affects parasite loads and fitness in a bumble-bee. Nature 1999, 397, 151–154. [Google Scholar] [CrossRef]
- Acevedo-Whitehouse, K.; Gulland, F.; Greig, D.; Amos, W. Disease susceptibility in California sea lions. Nature 2003, 422, 35–35. [Google Scholar] [CrossRef] [PubMed]
- Hughes, W.O.H.; Boomsma, J.J. Genetic diversity and disease resistance in leaf-cutting ant societies. Evolution 2004, 58, 1251–1260. [Google Scholar] [CrossRef] [PubMed]
- Spielman, D.; Brook, B.W.; Briscoe, D.A.; Frankham, R. Does inbreeding and loss of genetic diversity decrease disease resistance? Conserv. Genet. 2004, 5, 439–448. [Google Scholar]
- Daszak, P. Emerging infectious diseases of wildlife―threats to biodiversity and human health. Science 2000, 287, 1756–1756. [Google Scholar] [CrossRef]
- Pearman, P.B.; Garner, T.W.J. Susceptibility of Italian agile frog populations to an emerging strain of Ranavirus parallels population genetic diversity. Ecol. Lett. 2005, 8, 401–408. [Google Scholar] [CrossRef]
- Parris, M.J. Hybrid response to pathogen infection in interspecific crosses between two amphibian species (Anura: Ranidae). Evol. Ecol. Res. 2004, 6, 457–471. [Google Scholar]
- Bridges, C.M.; Semlitsch, R.D. Genetic variation in insecticide tolerance in a population of southern leopard frogs (Rana sphenocephala): Implications for amphibian conservation. Copeia 2001, 7–13. [Google Scholar] [CrossRef]
- Pierce, B.A.; Sikand, N. Variation in acid tolerance of Connecticut wood frogs―genetic and maternal effects. Can. J. Zool. 1985, 63, 1647–1651. [Google Scholar] [CrossRef]
- Semlitsch, R.D.; Bridges, C.M.; Welch, A.M. Genetic variation and a fitness tradeoff in the tolerance of gray treefrog (Hyla versicolor) tadpoles to the insecticide carbaryl. Oecologia 2000, 125, 179–185. [Google Scholar] [CrossRef] [PubMed]
- Laurila, A.; Karttunen, S.; Merila, J. Adaptive phenotypic plasticity and genetics of larval life histories in two Rana temporaria populations. Evolution 2002, 56, 617–627. [Google Scholar] [CrossRef] [PubMed]
- Laugen, A.T.; Laurila, A.; Rasanen, K.; Merila, J. Latitudinal countergradient variation in the common frog (Rana temporaria) development rates―evidence for local adaptation. J. Evol. Biol. 2003, 16, 996–1005. [Google Scholar] [CrossRef] [PubMed]
- Weitere, M.; Tautz, D.; Neumann, D.; Steinfartz, S. Adaptive divergence vs. environmental plasticity: tracing local genetic adaptation of metamorphosis traits in salamanders. Mol. Ecol. 2004, 13, 1665–1677. [Google Scholar]
- Blaustein, A.R.; Edmond, B.; Kiesecker, J.M.; Beatty, J.J.; Hokit, D.G. Ambient ultraviolet-radiation causes mortality in salamander eggs. Ecol. Appl. 1995, 5, 740–743. [Google Scholar] [CrossRef]
- Hays, J.B.; Blaustein, A.R.; Kiesecker, J.M.; Hoffman, P.D.; Pandelova, I.; Coyle, D.; Richardson, T. Developmental responses of amphibians to solar and artificial UVB sources: A comparative study. Photochem. Photobiol. 1996, 64, 449–456. [Google Scholar] [CrossRef] [PubMed]
- Anzalone, C.R.; Kats, L.B.; Gordon, M.S. Effects of solar UV-B radiation on embryonic development in Hyla cadaverina, Hyla regilla, and Taricha torosa. Conserv. Biol. 1998, 12, 646–653. [Google Scholar] [CrossRef]
- Brook, B.W.; Tonkyn, D.W.; Q'Grady, J.J.; Frankham, R. Contribution of inbreeding to extinction risk in threatened species. Conserv. Ecol. 2002, 6. [Google Scholar]
- Spielman, D.; Brook, B.W.; Frankham, R. Most species are not driven to extinction before genetic factors impact them. Proc. Nat. Acad. Sci. USA 2004, 101, 15261–15264. [Google Scholar] [CrossRef] [PubMed]
- Lande, R. Genetics and demography in biological conservation. Science 1988, 241, 1455–1460. [Google Scholar] [CrossRef] [PubMed]
- Hedrick, P. W. Genetics of Populations; Jones & Bartlett Publishers: Boston, 2004. [Google Scholar]
- Kimura, M.; Ohta, T. Theoretical Aspects of Population Genetics; Princeton University Press: New Jersey, USA, 1971. [Google Scholar]
- Lacy, R.C. Loss of genetic diversity from managed populations: interacting effects of drift, mutation, immigration, selection and population subdivision. Conserv. Biol. 1987, 1, 143–158. [Google Scholar] [CrossRef]
- Keller, L.F.; Waller, D.M. Inbreeding effects in wild populations. Trend. Ecol. Evol. 2002, 17, 230–241. [Google Scholar] [CrossRef]
- Caughley, G. Directions in conservation biology. J. Anim. Ecol. 1994, 63, 215–244. [Google Scholar] [CrossRef]
- Hedrick, P.W. Purging inbreeding depression and the probability of extinction - full sib mating. Heredity 1994, 73, 363–372. [Google Scholar] [CrossRef] [PubMed]
- Leberg, P.L.; Firmin, B.D. Role of inbreeding depression and purging in captive breeding and restoration programmes. Mol. Ecol. 2008, 17, 334–343. [Google Scholar] [CrossRef] [PubMed]
- Charlesworth, B. The effect of synergistic epistasis on the inbreeding load. Genet. Res. 1998, 71, 85–89. [Google Scholar] [CrossRef] [PubMed]
- Hughes, A.L.; Nei, M. Pattern of nucleotide substitution at major histocompatibility complex class I loci reveals overdominant selection. Nature 1988, 335, 167–170. [Google Scholar] [CrossRef] [PubMed]
- Radwan, J.; Biedrzycka, A.; Babik, W. Does reduced MHC diversity decrease viability of vertebrate populations? Biol. Conserv. 2010, in press. [Google Scholar]
- Beauclerc, K.; Johnson, B.; White, B. Genetic rescue of an inbred captive population of the critically endangered Puerto Rican crested toad (Peltophryne lemur) by mixing lineages. Conserv. Genet. Available online. [CrossRef]
- Edmands, S. Between a rock and a hard place: evaluating the relative risks of inbreeding and outbreeding for conservation and management. Mol. Ecol. 2007, 16, 463–475. [Google Scholar] [CrossRef] [PubMed]
- Edenhamn, P.; Hoggren, M.; Carlson, A. Genetic diversity and fitness in peripheral and central populations of the European tree frog Hyla arborea. Hereditas 2000, 133, 115–122. [Google Scholar] [CrossRef] [PubMed]
- McAlpine, S.; Smith, M.H. Genetic correlates of fitness in the green treefrog, Hyla cinerea. Herpetologica 1995, 51, 393–400. [Google Scholar]
- Parris, M.J.; Laird, C.W.; Semlitsch, R.D. Differential predation on experimental populations of parental and hybrid leopard frog (Rana blairi and Rana sphenocephala) larvae. J. Herpetol. 2001, 35, 479–485. [Google Scholar] [CrossRef]
- Parris, M.J. Hybridization in leopard frogs (Rana pipiens complex): larval fitness components in single-genotype populations and mixtures. Evolution 1999, 53, 1872–1883. [Google Scholar] [CrossRef]
- Parris, M.J. Experimental analysis of hybridization in leopard frogs (Anura: Ranidae): larval performance in desiccating environments. Copeia 2000, 2000, 11–19. [Google Scholar] [CrossRef]
- Parris, M.J. Hybridization in leopard frogs (Rana pipiens complex): terrestrial performance of newly metamorphosed hybrid and parental genotypes in field enclosures. Can. J. Zool. 2001, 79, 1552–1558. [Google Scholar] [CrossRef]
- Parris, M.J. High larval performance of leopard frog hybrids: effects of environment-dependent selection. Ecology 2001, 82, 3001–3009. [Google Scholar]
- Planade, B.; Lena, J.-P.; Li, H.; Plenet, S.; Guegan, J.-F.; Thomas, F.; Hurtrez-Bousses, S.; Renaud, F.; Joly, P. Tracking a heterosis effect in the field: tadpole resistance to parasites in the water frog hybridogenetic complex. Parasitology 2009, 136, 1003–1013. [Google Scholar] [CrossRef] [PubMed]
- Wright, M.F.; Guttman, S.I. Lack of an association between heterozygosity and growth rate in the wood frog, Rana sylvatica. Can. J. Zool. 1995, 73, 569–575. [Google Scholar] [CrossRef]
- Lesbarreres, D.; Schmeller, D.S.; Primmer, C.R.; Merila, J. Genetic variability predicts common frog (Rana temporaria) size at metamorphosis in the wild. Heredity 2007, 99, 41–46. [Google Scholar]
© 2010 by the authors; licensee Molecular Diversity Preservation International, Basel, Switzerland. This article is an open-access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Allentoft, M.E.; O’Brien, J. Global Amphibian Declines, Loss of Genetic Diversity and Fitness: A Review. Diversity 2010, 2, 47-71. https://doi.org/10.3390/d2010047
Allentoft ME, O’Brien J. Global Amphibian Declines, Loss of Genetic Diversity and Fitness: A Review. Diversity. 2010; 2(1):47-71. https://doi.org/10.3390/d2010047
Chicago/Turabian StyleAllentoft, Morten E., and John O’Brien. 2010. "Global Amphibian Declines, Loss of Genetic Diversity and Fitness: A Review" Diversity 2, no. 1: 47-71. https://doi.org/10.3390/d2010047
APA StyleAllentoft, M. E., & O’Brien, J. (2010). Global Amphibian Declines, Loss of Genetic Diversity and Fitness: A Review. Diversity, 2(1), 47-71. https://doi.org/10.3390/d2010047



