Discovery and Potential of SNP Markers in Characterization of Tunisian Olive Germplasm
Abstract
:1. Introduction
2. Results and Discussion
2.1. Targeted Sampling
| Gene | SNP position* | Type |
|---|---|---|
| OEW | 28 | C/T |
| 31 | C/G | |
| 36 | C/T | |
| 159 | C/T | |
| 250 | C/G | |
| 289 | G/T | |
| OEX | 1 | G/T |
| 33 | C/T | |
| 155 | C/T |
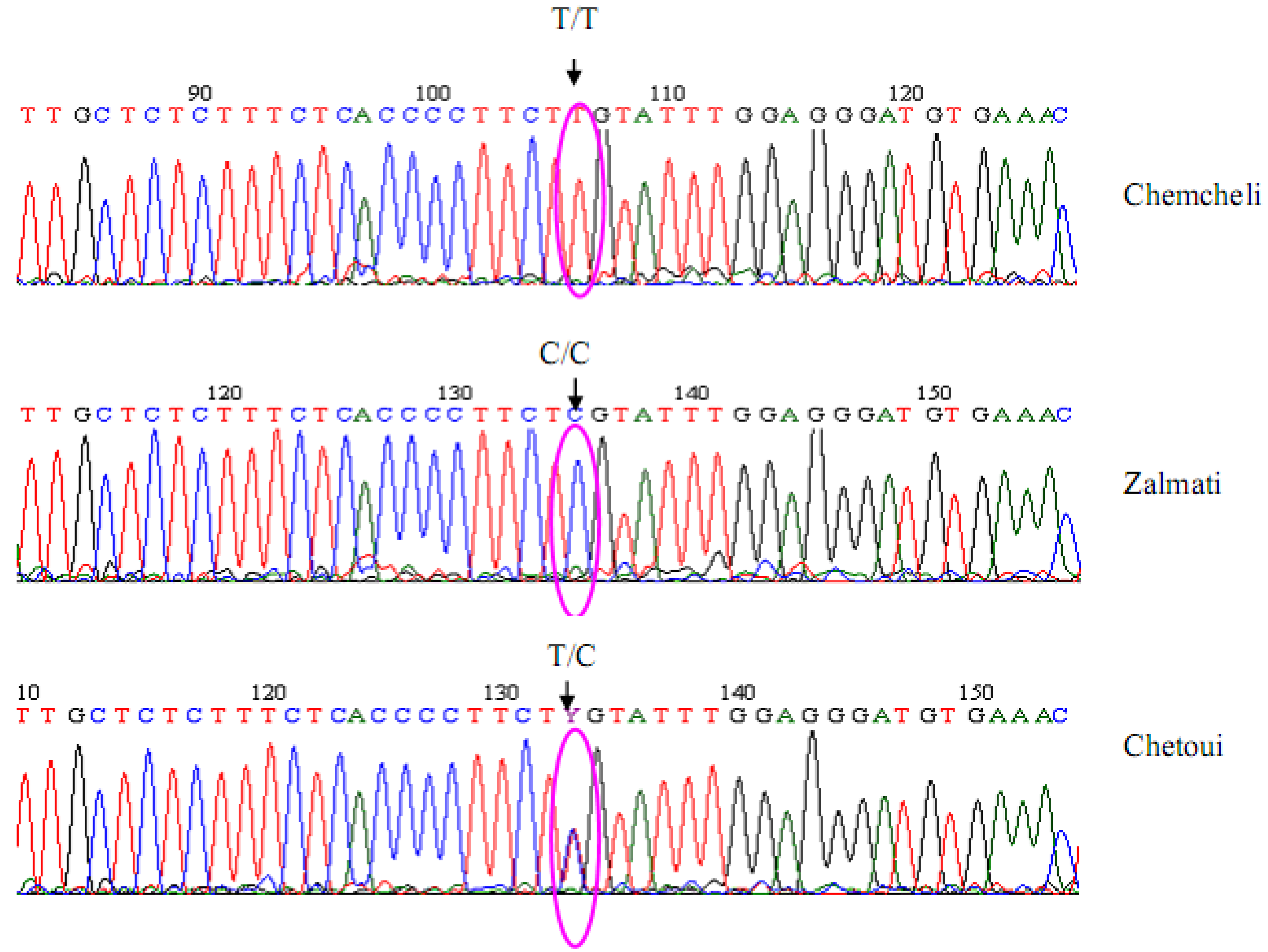
2.2. Marker Characteristics
| Genes | SNP | Observed heterozygosity (%) | Power of discrimination (PD) (%) | Genotypic frequencies (%) | Allele frequencies (%) | |||
|---|---|---|---|---|---|---|---|---|
| OEW | oew28 | 29.2 | 59 | 54.2 T/T | 16.6 C/C | 29.2C/T | 68.7:T | 31.3:C |
| oew31 | 45.8 | 63 | 37.5 G/G | 12.5 C/C | 45.8 C/G | 60.4:G | 39.6:C | |
| oew36 | 70.8 | 43 | 4.2 T/T | 25.0 C/C | 70.8 C/T | 60.4:C | 39.6:T | |
| oew159 | 33.3 | 61 | 50.0 T/T | 16.7 C/C | 33.3 C/T | 66.0:T | 34.0:C | |
| oew250 | 37.5 | 51 | 58.3G/G | 4.16 C/C | 37.5 G/C | 77.1:G | 22.9:C | |
| oew289 | 37.5 | 59 | 12.5 T/T | 50.0G/G | 37.5 G/T | 68.7:G | 31.3:T | |
| OEX | oex1 | 45.8 | 60 | 41.7 T/T | 12.5 G/G | 45.8 T/G | 64.6:T | 35.4:G |
| oex33 | 58.3 | 48 | 41.7 T/T | 0 C/C | 58.3 C/T | 70.8:T | 29.2:C | |
| oex155 | 29.2 | 64 | 25.0 T/T | 45.8 C/C | 29.2 C/T | 60.4:C | 39.6:T | |
2.3. Genetic Diversity Levels
2.4. Patterns of Genetic Diversity
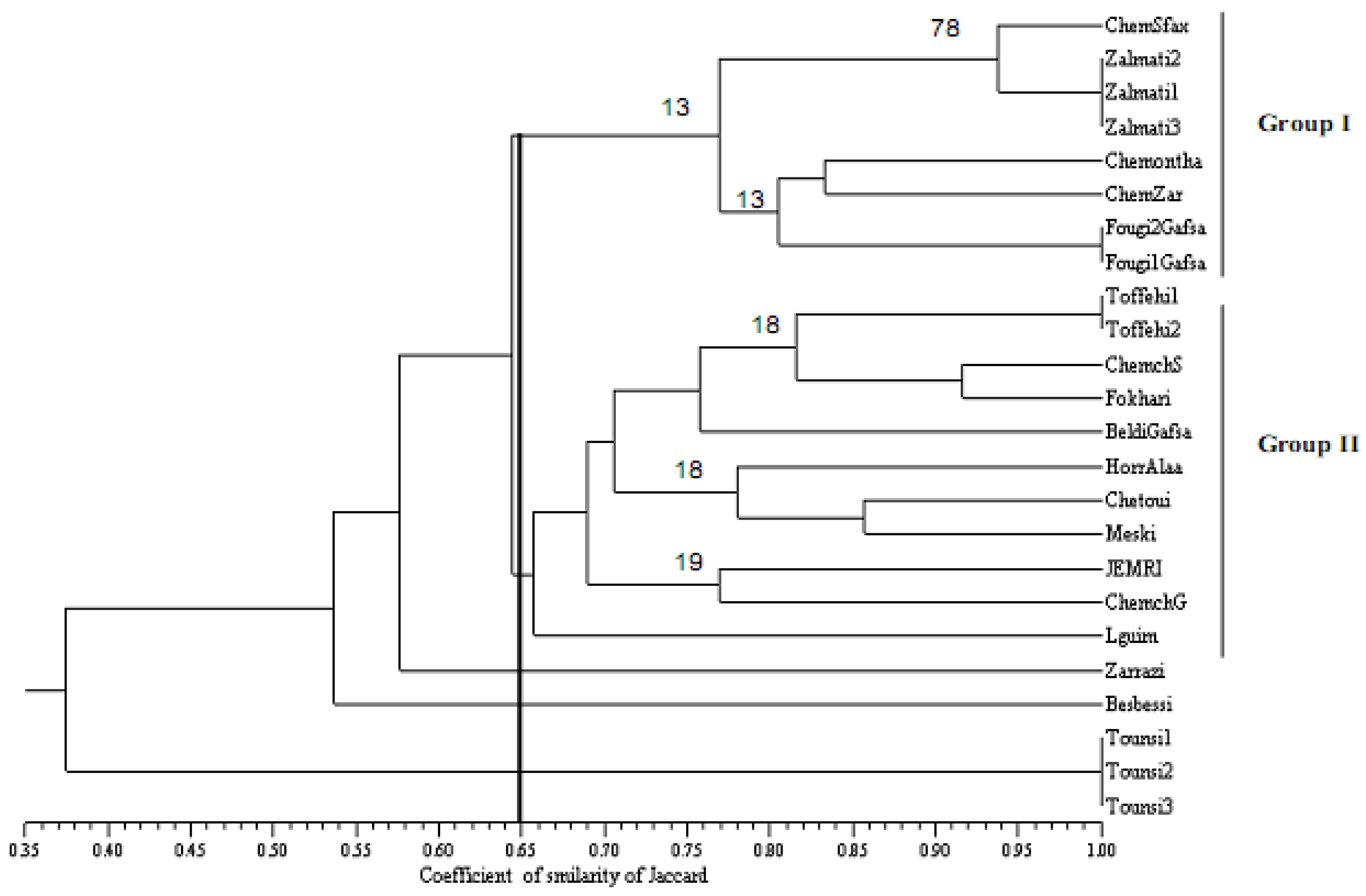
2.5. Concensus Dendrogram Based for SSR and SNP Markers
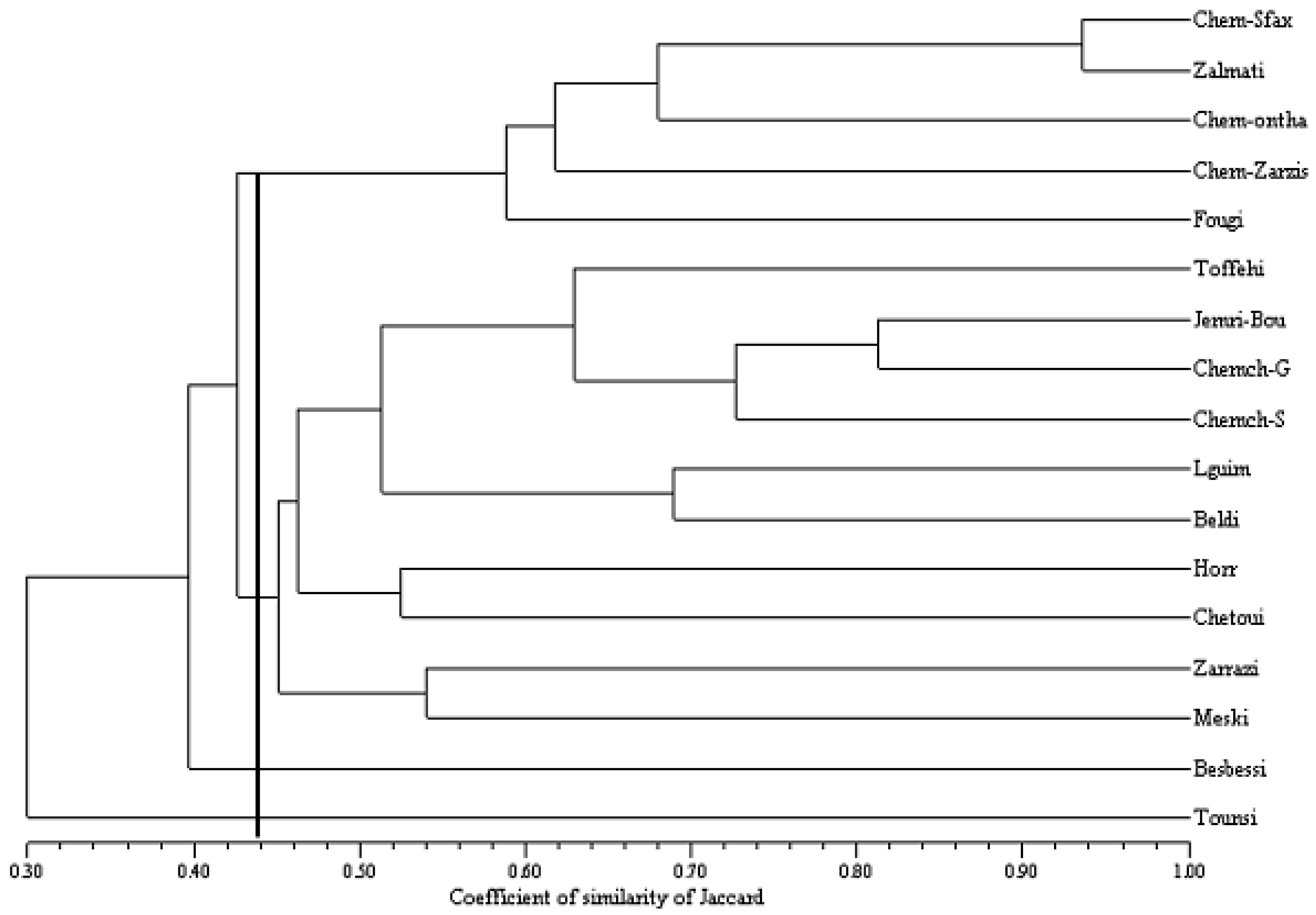
2.6. Selecting a Subset of Highly Discriminative Markers
3. Experimental Section
3.1. Plant Material
| Cultivar name | Name in tree | Growing Regiona |
|---|---|---|
| ‘Beldi’ | BeldiGafsa | Gafsa |
| ‘Besbessi’ | Besbessi | Tunis |
| ‘Chemcheli’ | Chemcheli-S | Sfax |
| ‘Chemcheli’ | Chemcheli-G | Gafsa |
| ‘Chemlali ontha’ | Chem-ontha | Zarzis |
| ‘Chemlali’ | Chem-Sfax1 | Sfax (Sfax) |
| ‘Chemlalizarzis’ | Chem-Zar | Zarzis |
| ‘Chetoui’ | Chetoui1 | Borj el Amri |
| ‘Chetoui’ | Chetoui2 | Mohsen el Imem |
| ‘Fougi’ | FougiGafsa | Gafsa |
| ‘Fougi’ | Fougi2Gafsa | Gafsa |
| ‘Horr’ | Horr | Kairouan |
| ‘Lguim’ | Lguim | Gafsa |
| ‘Toffehi’ | Toffehi | Zarzis |
| ‘Tounsi’ | Tounsi | Gafsa |
| ‘Tounsi’ | Tounsi | Gafsa |
| ‘Tounsi’ | Tounsi | Gafsa |
| ‘Zalmati’ | Zalmati1 | Zarzis |
| ‘Zalmati’ | Zalmati2 | Zarzis |
| ‘Zalmati’ | Zalmati3 | Zarzis |
| ‘Zarrazi’ | Zarrazi | Zarzis |
3.2. DNA Extraction from Leaves
3.3. Primer Desing
- OEW F: 5’ TACTAATCTTGAAGGTAATCG 3’
- OEW R: 5’ TCTTATAACATGGCACTTAC 3’
- OEX F: 5’CTATTCAAGCATTGGGAGCA 3’
- OEX R: 5’CCCAAACCATGTCCCATAAG 3’.
3.4. PCR and Sequencing
3.5. Marker Discovery Strategies
3.6. Genetic Diversity and Data Analyses
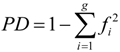
4. Conclusions
Acknowledgements
References and Notes
- Barranco, D.; Rallo, L. Olive cultivars in Spain. Hortech 2000, 10, 107–110. [Google Scholar]
- Besnard, G.; Bervillé, A. Multiple origins for mediterranean olive (Olea europaea L. subsp. europaea) based upon mitochondrial DNA polymorphisms. CR Acad. Sci Ser. III 2000, 19, 81–100. [Google Scholar]
- Grati-Kamoun, N.; Ayadi, M.; Khlif, M.; Tridui, A.; Karray, B.; Rekik, H.; Rekik, B.; Hamdi, T.; Arous, M.N. Pomological and chemical characterization of Tunisia olive tree (Olea europaea L.). In Proceedings of the 4th International Olive Growing Symposium CIHEAM-IAM; Bari, Italy, September 25–30 2000. [Google Scholar]
- Angiolillo, A.; Mencuccini, M.; Baldoni, L. Olive genetic diversity assessed using amplified fragment length polymorphisms. Theoret. Appl. Genet. 1999, 10, 411–421. [Google Scholar] [CrossRef]
- Bandelj, D.; Jakse, J.; Javornik, B. DNA fingerprinting of olive varieties by microsatellite markers. Food Technol. Biotechnol. 2002, 5, 185–190. [Google Scholar]
- Garant, D.; Kruuk, L.E. How to use molecular marker data to measure evolutionary parameters in wild populations. Mol. Ecol. 2005, 15, 1843–1859. [Google Scholar] [CrossRef]
- Morgante, M.; Salamini, F. From plant genomics to breeding practice. Curr. Opin. Biotechnol. 2003, 5, 214–219. [Google Scholar] [CrossRef]
- Dalmasso, A.; Fontanella, E.; Piatti, P.; Civera, T.; Rosati, S.; Bottero, M.T. A multiplex PCR assay for the identification of animal species in feedstuffs. Mol. Cell. Probes 2004, 6, 81–87. [Google Scholar]
- Hernandez, M.; Esteve, T.; Pla, M. Real-time polymerase chain reaction based assays for quantitative detection of barley, rice, sunflower, and wheat. J. Agric. Food Chem. 2005, 6, 7003–7009. [Google Scholar] [CrossRef]
- Matsunagaa, T.; Chikuni, K.; Tanabeb, R.; Muroyab, S.; Shibata, K.; Yamadaa, J.; Shinmuraa, Y. A quick and simple method for the identification of meat species and meat products by PCR assay. Meat. Sci. 1999, 5, 143–148. [Google Scholar]
- Belaj, A.; Trujillo, I.; ee la Rosa, R.; Rallo, L. Polymorphism and discrimination capacity of randomly amplified polymorphic markers in an olive germplasm bank. J. Amer. Soc. Hort. Sci. 2001, 7, 64–71. [Google Scholar]
- Carriero, F.; Fontanazza, G.; Cellini, F.; Giorio, G. Identification of simple sequence repeats (SSRs) in olive (Olea europaea L.). Theor. Appl. Genet. 2002, 6, 301–307. [Google Scholar]
- Grati-Kamoun, N.; Mahmoud, F.L.; Rebai, A.; Gargouri, A.; Panaud, O.; Saar, A. Genetic diversity of Tunisian olive tree (Olea europaea L.) cultivars assessed by AFLP markers. Genet. Resour. Crop Ev. 2006, 10, 265–275. [Google Scholar]
- Pasqualone, A.; Caponio, F. Inter-simple sequence repeat DNA markers for identification of drupes from different Olea europaea L. cultivars. Eur. Food. Res. Technol. 2001, 3, 240–243. [Google Scholar] [CrossRef]
- Rekik, I.; Salimonti, A.; Grati-Kammoun, N.; Muzzaluppo, I.; Lepais, O.; Gerber, S.; Perri, E.; Rebai, A. Characterisation and identification of tunisian olive tree varieties by microsatellite markers. Hortscience 2008, 5, 1371–1376. [Google Scholar]
- Baldoni, L.; Pellegrini, M.; Mencuccini, M.; Mulas, M.; Angiolillo, A. Genetic relationships among cultivated and wild olives revealed by AFLP markers. Acta Hortic. 2000, 8, 275–283. [Google Scholar]
- Consolandi, C.; Palmieri, L.; Doveri, S.; Maestri, E.; Marmiroli, N.; Reale, S.; Lee, D.; Baldoni, L.; Tosti, N.; Severgnini, M.; de Bellis, G.; Castiglioni, B. Olive variety identification by ligation detection reaction in a universal array format. Afr. J. Biotechnol. 2007, 9, 565–574. [Google Scholar]
- Reale, S.; Doveri, S.; Díaz, A.; Angiolillo, A.; Lucentini, L.; Pilla, F.; Martín, A.; Donini, P.; Lee, D. SNP-based markers for discriminating olive (Olea europaea L.) cultivars. Genome 2006, 15, 1193–1209. [Google Scholar]
- Palmieri, L.; Vietina, M.; Bottura, C.; Donini, P.; Nelson, M. SNP development in olive and their application on varietal determination in olive oil. In Proceedings of the XLVIII Italian Society of Agricultural Genetics-SIFV-SIGA Joint Meeting; Lecce, Italy, September 15–18 2004. [Google Scholar]
- Shibuya, M.; Zhang, H.; Endo, A.; Shishikura, K.; Kushiro, T.; Ebizuka, Y. Two branches of the lupeol synthase gene in the molecular evolution of plant oxidosqualene cyclases. Eur. J. Biochem. 1999, 36, 266–302. [Google Scholar]
- Tatusova, T.A.; Madden, L.T. BLAST 2 sequences. A new tool for comparing protein and nucleotide sequences. FEMS Microbiol. Lett. 1999, 3, 247–250. [Google Scholar] [CrossRef]
- Fabbri, A.; Hormaza, J.I.; Polito, V.S. Random amplified polymorphic DNA analysis of olive (Olea europaea L) cultivars. J. Amer. Soc. Hort. Sci. 1995, 4, 538–542. [Google Scholar]
- Rozen, S.; Skaletsky, H. Primer3 on the WWW for general users and for biologist programmers. Methods Mol. Biol. 2000, 21, 365–386. [Google Scholar]
- Thompson, J.D.; Higgins, D.G.; Gibson, T.J. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids. Res. 1994, 22, 673–695. [Google Scholar]
- Kloosterman, A.D.; Budowle, B.; Daselaar, P. PCR-amplification and detection of the human D1S80 VNTR locus. Amplification conditions, population genetics and application in forensic analysis. Int. J. Legal Med. 1993, 7, 257–264. [Google Scholar] [CrossRef]
- Jaccard, P. Nouvelles recherches sur la distribution florale. Bull. Soc. Vaud. Sci. Nat. 1908, 44, 223–270. [Google Scholar]
- Rohlf, F.J. NTSYS-PC: Numerical Taxonomy and Multivariate Analysis System (version 2.1); Exeter Software: Setauket, NY, USA, 1999. [Google Scholar]
© 2010 by the authors; licensee Molecular Diversity Preservation International, Basel, Switzerland. This article is an open-access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Hakim, I.R.; Kammoun, N.G.; Makhloufi, E.; Rebaï, A. Discovery and Potential of SNP Markers in Characterization of Tunisian Olive Germplasm. Diversity 2010, 2, 17-27. https://doi.org/10.3390/d2010017
Hakim IR, Kammoun NG, Makhloufi E, Rebaï A. Discovery and Potential of SNP Markers in Characterization of Tunisian Olive Germplasm. Diversity. 2010; 2(1):17-27. https://doi.org/10.3390/d2010017
Chicago/Turabian StyleHakim, Imen Rekik, Naziha Grati Kammoun, Emna Makhloufi, and Ahmed Rebaï. 2010. "Discovery and Potential of SNP Markers in Characterization of Tunisian Olive Germplasm" Diversity 2, no. 1: 17-27. https://doi.org/10.3390/d2010017
APA StyleHakim, I. R., Kammoun, N. G., Makhloufi, E., & Rebaï, A. (2010). Discovery and Potential of SNP Markers in Characterization of Tunisian Olive Germplasm. Diversity, 2(1), 17-27. https://doi.org/10.3390/d2010017




