Monitoring Genetic Diversity in Lithuanian Riverine Populations of Stuckenia pectinata Using SSR and ISSR Markers
Abstract
1. Introduction
2. Materials and Methods
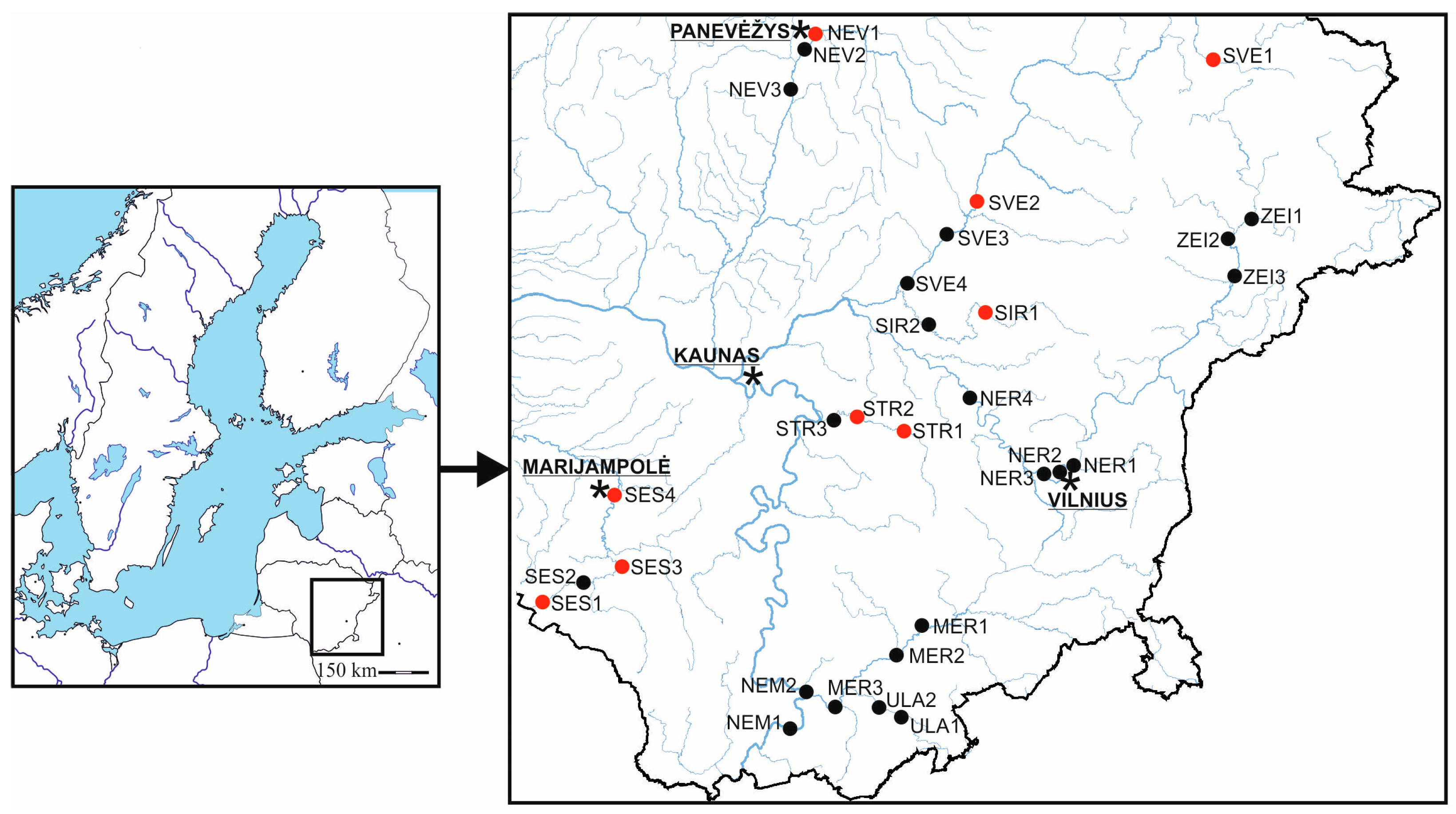
2.1. Plant Material
2.2. DNA Analysis
2.3. Data Analysis
3. Results
3.1. SSR Analysis of S. pectinata Populations
3.2. ISSR Analysis of S. pectinata Populations
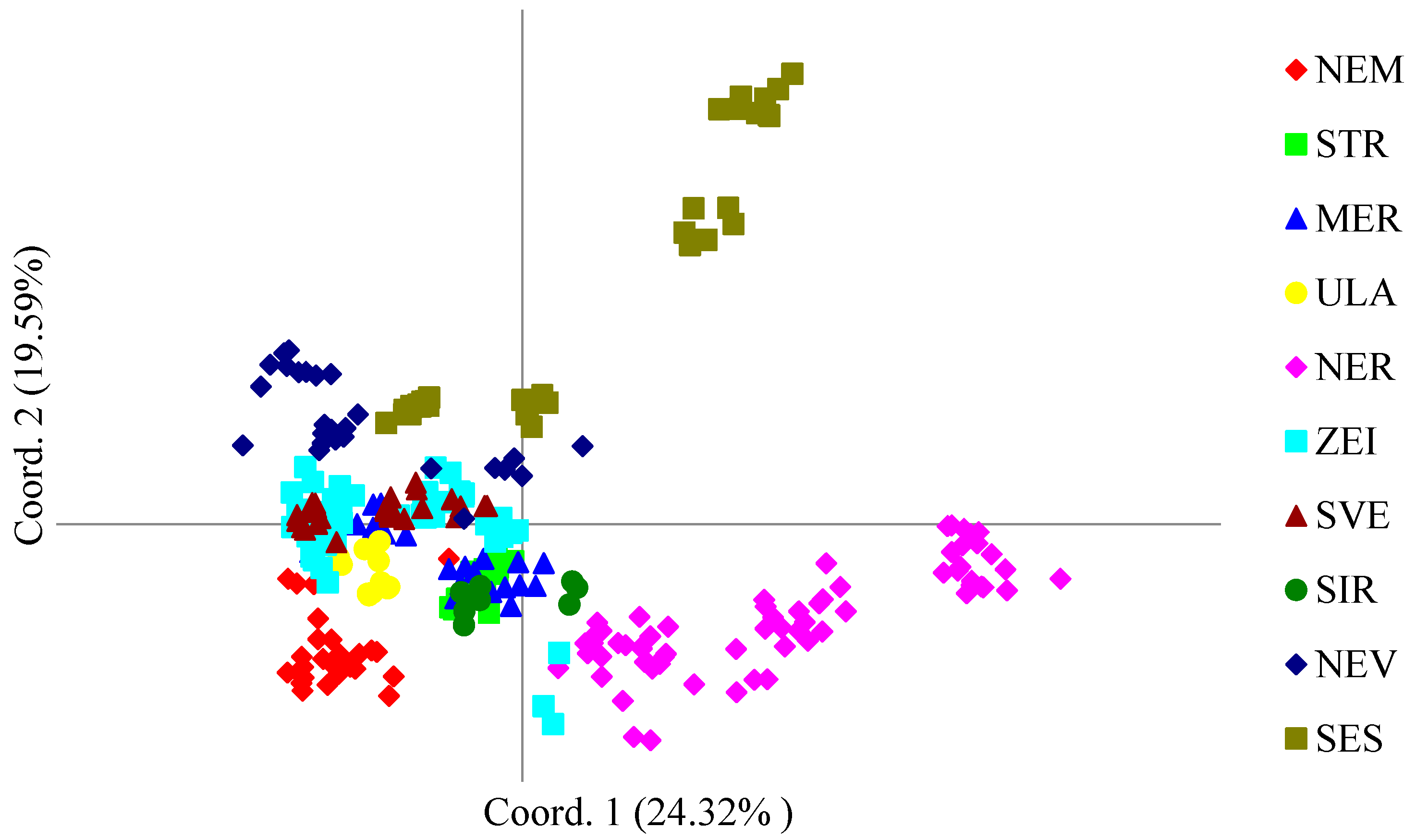
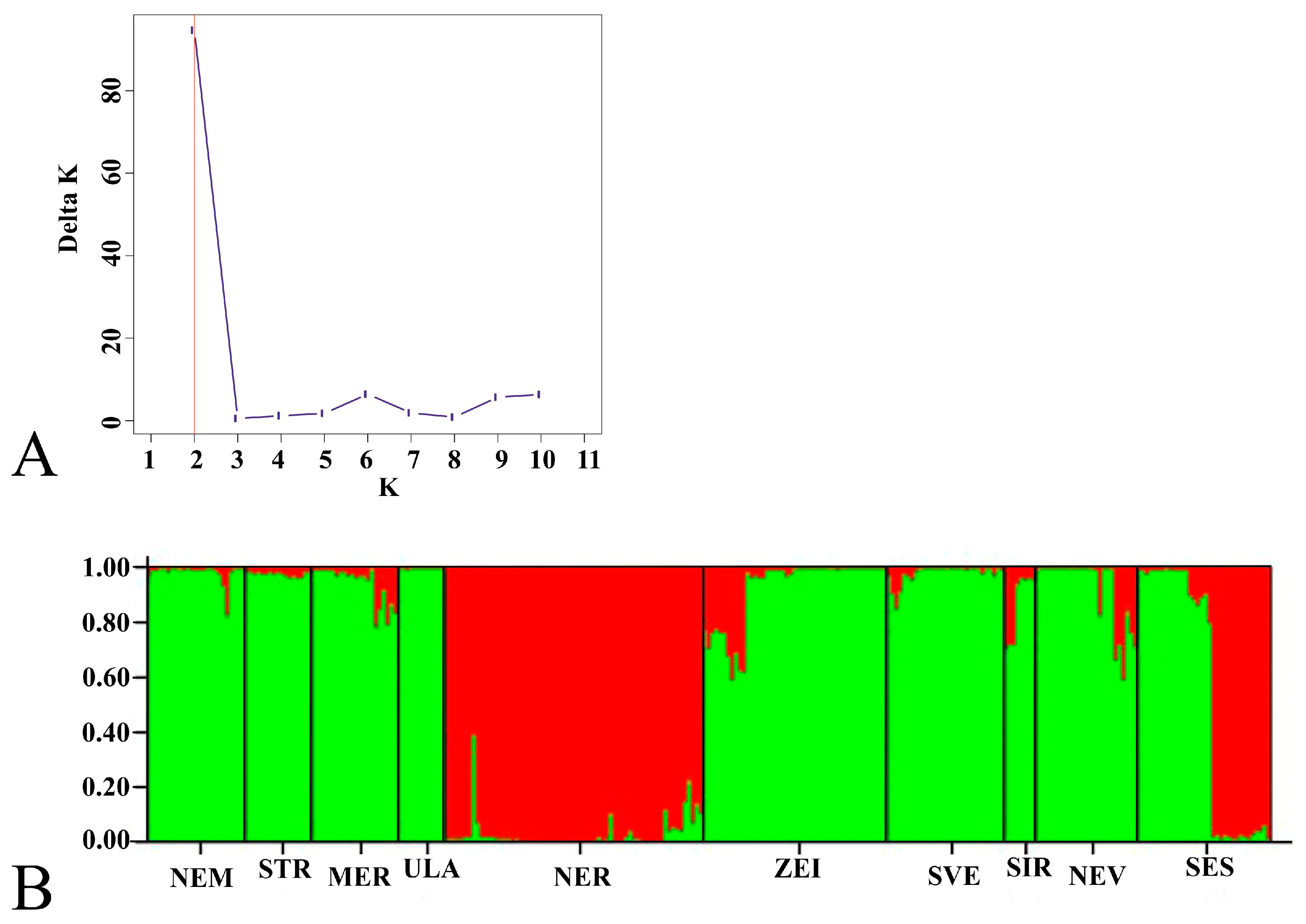
3.3. Population Genetic Structure
3.4. A Comparison of Genetic Diversity and Habitat Parameters
4. Discussion
4.1. Genetic Diversity
4.2. Genetic Differentiation and Population Structure
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ramsar Convention Secretariat. Wise use of wetlands: Concepts and approaches for the wise use of wetlands. In Ramsar Handbooks for the Wise Use of Wetlands, 4th ed.; Ramsar Convention Secretariat: Gland, Switzerland, 2010; Volume 1. [Google Scholar]
- Anderson, N.O.; Krokaitė-Kudakienė, E.; Jocienė, L.; Rekašius, T.; Chernyagina, O.A.; Paulauskas, A.; Kupčinskienė, E. Genetic Differentiation of Reed Canarygrass (Phalaris arundinacea L.) within Eastern Europe and Eurasia. Genes 2024, 15, 734. [Google Scholar] [CrossRef]
- Barrat-Segretain, M.H. Strategies of reproduction, dispersion, and competition in river plants: A Review. Vegetatio 1996, 123, 13–37. [Google Scholar] [CrossRef]
- Lone, P.A.; Bhardwaj, A.K.; Bahar, F.A. A Study of Comparative Purification Efficiency of Two Species of Potamogeton (Submerged Macrophyte) In Wastewater Treatment. IJSRP 2013, 3, 1–5. [Google Scholar]
- Gao, Y.N.; Dong, J.; Fu, Q.Q.; Wang, Y.P.; Chen, C.; Li, J.H.; Li, R.; Zhou, C.J. Allelopathic effects of submerged macrophytes on phytoplankton. Allelopath. J. 2017, 40, 1–22. [Google Scholar] [CrossRef]
- Poikane, S.; Portielje, R.; Denys, L.; Elferts, D.; Kelly, M.; Kolada, A.; Mäemets, H.; Phillips, G.; Søndergaard, M.; Willby, N.; et al. Macrophyte assessment in European lakes: Diverse approaches but convergent views of ‘good’ ecological status. Ecol. Indic. 2018, 94, 185–197. [Google Scholar] [CrossRef]
- Malea, L.; Nakou, K.; Papadimitriou, A.; Exadactylos, A.; Orfanidis, S. Physiological Responses of the Submerged Macrophyte Stuckenia pectinata to High Salinity and Irradiance Stress to Assess Eutrophication Management and Climatic Effects: An Integrative Approach. Water 2021, 13, 1706. [Google Scholar] [CrossRef]
- Ganie, A.H.; Reshi, Z.A.; Wafai, B.A. Reproductive ecology of Potamogeton pectinatus L. (= Stuckenia pectinata (L.) Börner) in relation to its spread and abundance in freshwater ecosystems of the Kashmir Valley, India. Trop. Ecol. 2016, 57, 787–803. [Google Scholar]
- Misteli, B.; Pannard, A.; Labat, F.; Fosso, L.K.; Baso, N.C.; Harpenslager, S.F.; Motitsoe, S.N.; Thiebaut, G.; Piscart, C. How Invasive Macrophytes Affect Macroinvertebrate Assemblages and Sampling Efficiency: Results from a Multinational Survey. Limnologica 2022, 96, 125998. [Google Scholar] [CrossRef]
- Swanepoel, J.H.; Vermaak, J.F. Preliminary results on the uptake and release of 32P by Potamogeton pectinatus. J. Limnol. Soc. S. Afr. 1977, 3, 63–65. [Google Scholar] [CrossRef]
- Janauer, G.A. Veränderungen organischer und anorganischer Inhaltsstoffe in Potamogeton pectinatus L. bei steigender Gewasserbelastung. Metabolic changes in Potamogeton pectinatus L. by enhanced eutrophication. Flora 1979, 168, 344–351. [Google Scholar] [CrossRef]
- Van Wijk, R.J. Ecological studies on Potamogeton pectinatus L. III. Reproductive strategies and germination ecology. Aquat. Bot. 1989, 33, 271–299. [Google Scholar] [CrossRef]
- Wiegleb, G.; Brux, H.; Herr, W. Human impact on the ecological performance of Potamogeton species in northwestern Germany. Vegetatio 1991, 97, 161–172. [Google Scholar] [CrossRef]
- Bernez, I.; Chicouène, D.; Haury, J. Changes of Potamogeton pectinatus clumps under variable, artificially flooded river water regimes. Belg. J. Bot. 2007, 140, 51–59. [Google Scholar]
- Nolet, B.A.; Langevoord, O.; Bevan, R.M.; Engelaar, K.R.; Klaassen, M.; Mulder, R.J.W.; Van Dijk, S. Spatial variation in tuber depletion by swans explained by differences in net intake rates. Ecology 2001, 82, 1655–1667. [Google Scholar] [CrossRef]
- Søndergaard, M.; Johansson, L.S.; Lauridsen, T.L.; Jørgensen, T.B.; Liboriussen, L.; Jeppesen, E. Submerged macrophytes as indicators of the ecological quality of lakes. Freshw. Biol. 2010, 55, 893–908. [Google Scholar] [CrossRef]
- Hettiarachchi, P.; Triest, L. Isozyme polymorphism in the genus Potamogeton (Potamogetonaceae). In Isozymes in Water Plants; Opera Botanica Belgica; Triest, L., Ed.; National Botanical Garden of Belgium: Meise, Belgium, 1991; Volume 4, pp. 89–116. ISBN 9789072619037. [Google Scholar]
- Gornall, R.; Hollingsworth, P.; Preston, C. Evidence for spatial structure and directional gene flow in a population of an aquatic plant, Potamogeton coloratus. Heredity 1998, 80, 414–421. [Google Scholar] [CrossRef]
- Mader, E.; van Viersen, W.; Schwenk, K. Clonal diversity in the submerged macrophyte Potamogeton pectinatus L. inferred from nuclear and cytoplasmic variation. Aquat. Bot. 1998, 62, 147–160. [Google Scholar] [CrossRef]
- Hangelbroek, H.H.; Ouborg, N.J.; Santamaria, L.; Schwenk, K. Clonal diversity and structure within a population of the pondweed Potamogeton pectinatus foraged by Bewick’s Swans. Mol. Ecol. 2002, 11, 2137–2150. [Google Scholar] [CrossRef]
- King, R.A.; Gornall, R.J.; Preston, C.D.; Croft, J.M. Population differentiation of Potamogeton pectinatus in the Baltic Sea with reference to waterfowl dispersal. Mol. Ecol. 2002, 11, 1947–1956. [Google Scholar] [CrossRef]
- Nies, G.; Reusch, T.B.H. Evolutionary divergence and possible incipient speciation in post-glacial populations of a cosmopolitan aquatic plant. J. Evol. Biol. 2005, 18, 19–26. [Google Scholar] [CrossRef]
- Triest, L.; Fénart, S. Clonal diversity and spatial genetic structure of Potamogeton pectinatus in managed pond and river populations. Hydrobiologia 2014, 737, 145–161. [Google Scholar] [CrossRef]
- Han, Q.; Wang, G.; Li, W.; Liu, F. Genetic diversity of Potamogeton pectinatus L. in relation to species diversity in a pair of lakes of contrasting trophic levels. Biochem. Syst. Ecol. 2014, 57, 60–66. [Google Scholar] [CrossRef]
- Volkova, P.A.; Kipriyanova, L.M.; Maltseva, S.Y.; Bobrov, A.A. In search of speciation: Diversification of Stuckenia pectinata s.l. (Potamogetonaceae) in southern Siberia (Asian Russia). Aquat. Bot. 2017, 143, 25–32. [Google Scholar] [CrossRef]
- Abbasi, S.; Afsharzadeh, S.; Saeidi, H.; Triest, L. Strong Genetic Differentiation of Submerged Plant Populations across Mountain Ranges: Evidence from Potamogeton pectinatus in Iran. PLoS ONE 2016, 11, e0161889. [Google Scholar] [CrossRef]
- Abbasi, S.; Afsharzadeh, S.; Saeidi, H. Genetic diversity of Potamogeton pectinatus L. in Iran as revealed by ISSR markers. Acta Bot. Croat. 2017, 76, 177–182. [Google Scholar] [CrossRef]
- Fehrer, J.; Nagy Nejedlá, M.; Hellquist, C.B.; Bobrov, A.A.; Kaplan, Z. Evolutionary history and patterns of geographical variation, fertility, and hybridization in Stuckenia (Potamogetonaceae). Front. Plant Sci. 2022, 13, 1042517. [Google Scholar] [CrossRef]
- Triest, L.; Bossaer, L.; Hailu, A.B.; Mäemets, H.; Terer, T.; Tóth, V.R.; Sierens, T.A. Pleistocene legacy of gene pools, ecodemes and admixtures of Stuckenia pectinata (L.) Börner as evidenced from microsatellites, complete chloroplast genomes and ribosomal RNA cistron (Europe, Africa). Aquat. Bot. 2025, 196, 103825. [Google Scholar] [CrossRef]
- Vyšniauskienė, R.; Rančelienė, V.; Naugžemys, D.; Rudaitytė-Lukošienė, E.; Patamsytė, J.; Butkauskas, D.; Kupčinskienė, E.; Žvingila, D. Genetic diversity of Nuphar lutea in Lithuanian river populations. Aquat. Bot. 2020, 161, 103–173. [Google Scholar] [CrossRef]
- Jablonskis, J.; Kovalenkovienė, M.; Tamkevičienė, A. Channel network of the Lithuanian rivers and small streams. Ann. Geograph. 2007, 40, 46–56. [Google Scholar]
- Stanley, E.; Fisher, H.S.; Grimm, N.B. Ecosystem expansion and contraction: A desert stream perspective. BioScience 1997, 47, 427–435. [Google Scholar] [CrossRef]
- Anderson, D.; Moggridge, H.; Warren, P.; Shucksmith, J. The Impacts of ‘run-of-river’ Hydropower on the Physical and Ecological Condition of Rivers. Water Environ. J. 2015, 29, 268–276. [Google Scholar] [CrossRef]
- Arroita, M.; Flores, L.; Larrañaga, A.; Martínez, A.; Martínez-Santos, M.; Pereda, O.; Ruiz-Romera, E.; Salagaistua, L.; Elosegi, A. Water Abstraction Impacts Stream Ecosystem Functioning via Wetted-Channel Contraction. Freshw. Biol. 2017, 62, 243–257. [Google Scholar] [CrossRef]
- Bunn, S.; Arthington, A. Basic Principles and Ecological Consequences of Altered Flow Regimes for Aquatic Biodiversity. Environ. Manag. 2002, 30, 492–507. [Google Scholar] [CrossRef]
- Liu, L.; Wang, J.; Ma, X.; Li, M.; Guo, X.; Yin, M.; Cai, Y.; Yu, X.; Du, N.; Wang, R.; et al. Impacts of the Yellow River and Qingtongxia dams on genetic diversity of Phragmites australis in Ningxia Plain, China. Aquat. Bot. 2021, 169, 103341. [Google Scholar] [CrossRef]
- Blanch, S.J.; Ganf, G.G.; Walker, K.F. Tolerance of riverine plants to flooding and exposure indicated by water regime. Regul. Rivers Res. Manag. 1999, 15, 43–62. [Google Scholar] [CrossRef]
- Naugžemys, D.; Lambertini, C.; Patamsytė, J.; Butkuvienė, J.; Khasdan, V.; Žvingila, D. Genetic diversity patterns in Phragmites australis populations in straightened and in natural river sites in Lithuania. Hydrobiologia 2021, 848, 3317–3330. [Google Scholar] [CrossRef]
- Kingsford, R.T. Ecological impacts of dams, water diversions and river management on floodplain wetlands in Australia. Austral. Ecol. 2000, 25, 109–127. [Google Scholar] [CrossRef]
- Erős, T.; Kuehne, L.; Dolezsai, A.; Sommerwerk, N.; Wolter, C. A systematic review of assessment and conservation management in large floodplain rivers—Actions postponed. Ecol. Indic. 2019, 98, 453–461. [Google Scholar] [CrossRef]
- Krokaitė, E.; Shakeneva, D.; Juškaitytė, E.; Rekašius, T.; Nemaniūtė-Gužienė, J.; Butkuvienė, J.; Patamsytė, J.; Rančelienė, V.; Vyšniauskienė, R.; Duchovskienė, L.; et al. Nitrogen concentration of the aquatic plant species in relation to land cover type and other variables of the environment. Zemdirb. Agric. 2019, 106, 203–212. [Google Scholar] [CrossRef]
- Wiegleb, G.; Kaplan, Z. An account of the species of Potamogeton L. (Potamogetonaceae). Folia Geobot. 1998, 33, 241–316. [Google Scholar] [CrossRef]
- Doyle, J.J.; Doyle, J.J. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem. Bull. 1987, 19, 11–15. [Google Scholar]
- Nies, G.; Reusch, T.B.H. Nine polymorphic microsatellite loci for the fennel Pondweed Potamogeton pectinatus L. Mol. Ecol. Notes 2004, 4, 563–565. [Google Scholar] [CrossRef]
- Patamsytė, J.; Naugžemys, D.; Čėsnienė, T.; Kleizaitė, V.; Demina, O.N.; Mikhailova, S.I.; Agafonov, V.A.; Žvingila, D. Evaluation and comparison of the genetic structure of Bunias orientalis populations in their native range and two non-native ranges. Plant Ecol. 2018, 219, 101–114. [Google Scholar] [CrossRef]
- Van Oosterhout, C.; Hutchinson, W.F.; Wills, D.P.M.; Shipley, P. MICRO-CHECKER: Software for identifying and correcting genotyping errors in microsatellite data. Mol.Ecol. Notes 2004, 4, 535–538. [Google Scholar] [CrossRef]
- Patamsytė, J.; Rančelis, V.; Čėsnienė, T.; Kleizaitė, V.; Tunaitienė, V.; Naugžemys, D.; Vaitkūnienė, V.; Žvingila, D. Clonal structure and reduced diversity of the invasive alien plant Erigeron annuus in Lithuania. Cent. Eur. J. Biol. 2013, 8, 898–911. [Google Scholar] [CrossRef]
- Peakall, R.; Smouse, P.E. GenAlEx 6.5: Genetic analysis in Excel. Population genetic software for teaching and research—An update. Bioinformatics 2012, 28, 2537–2539. [Google Scholar] [CrossRef]
- Goudet, J. FSTAT (Version 2.9.4), a Program to Estimate and Test Population Genetics Parameters. 2003. Available online: http://www2.unil.ch/popgen/softwares/fstat.htm (accessed on 20 July 2025).
- Coart, E.; Van Glabeke, S.; Petit, R.J.; Van Bockstaele, E.; Roldán-Ruiz, I. Range-wide versus local patterns of genetic diversity in hornbeam (Carpinus betulus L.). Conserv. Genet. 2005, 6, 259–273. [Google Scholar] [CrossRef]
- Dorken, M.E.; Eckert, C.G. Severely reduced sexual reproduction in northern populations of a clonal plant, Decodon verticillatus (Lythraceae). J. Ecol. 2001, 89, 339–350. [Google Scholar] [CrossRef]
- Falush, D.; Stephens, M.; Pritchard, K.J. Inference of population structure using multilocus genotype data: Dominant markers and null alleles. Mol. Ecol. Notes 2007, 7, 574–578. [Google Scholar] [CrossRef]
- Evanno, G.; Regnaut, S.; Goudet, J. Detecting the number of clusters of individuals using the software STRUCTURE: A simulation study. Mol. Ecol. 2005, 14, 2611–2620. [Google Scholar] [CrossRef]
- Earl, D.A.; von Holdt, B.M. STRUCTURE HARVESTER: A website and program for visualizing STRUCTURE output and implementing the Evanno method. Conserv. Genet. Resour. 2012, 4, 359–361. [Google Scholar] [CrossRef]
- Nybom, H. Comparison of different nuclear DNA markers for estimating intraspecific genetic diversity in plants. Mol. Ecol. 2004, 13, 1143e1155. [Google Scholar] [CrossRef]
- Bettencourt, S.X.; Mendonça, D.; Lopes, M.S.; Rocha, S.; Monjardino, P.; Monteiro, L.; da Câmara Machado, A. Genetic diversity and population structure of the endemic Azorean juniper, Juniperus brevifolia (Seub.) Antoine, inferred from SSRs and ISSR markers. Biochem. Syst. Ecol. 2015, 59, 314–324. [Google Scholar] [CrossRef]
- Li, Y.C.; Fahima, T.; Krugman, T.; Beiles, A.; Röder, M.S.; Korol, A.B.; Nevo, E. Parallel microgeographic patterns of genetic diversity and divergence revealed by allozyme, RAPD, and microsatellites in Triticum dicoccoides at Ammiad, Israel. Conserv. Genet. 2000, 1, 191–207. [Google Scholar] [CrossRef]
- Mariette, S.; Chagné, D.; Lézier, C.; Pastuszka, P.; Raffin, A.; Plomion, C.; Kremer, A. Genetic diversity within and among Pinus pinaster populations: Comparison between AFLP and microsatellite markers. Heredity 2001, 86, 469–479. [Google Scholar] [CrossRef]
- Crinò, P.; Pagnotta, M.A. Phenotyping, Genotyping, and Selections within Italian Local Landraces of Romanesco Globe Artichoke. Diversity 2017, 9, 14. [Google Scholar] [CrossRef]
- Amom, T.; Tikendra, L.; Apana, N.; Goutam, M.; Sonia, P.; Koijam, A.S.; Potshangbam, A.M.; Rahaman, H.; Nongdam, P. Efficiency of RAPD, ISSR, iPBS, SCoT and phytochemical markers in the genetic relationship study of five native and economical important bamboos of North-East India. Phytochemistry 2020, 174, 112330. [Google Scholar] [CrossRef] [PubMed]
- Krokaite, E.; Janulioniene, R.; Jociene, L.; Rekašius, T.; Rajackaite, G.; Paulauskas, A.; Marozas, V.; Kupčinskiene, E. Relating Invasibility and Invasiveness: Case Study of Impatiens parviflora. Front. Ecol. Evol. 2022, 10, 845947. [Google Scholar] [CrossRef]
- Collevatti, R.G.; Grattapaglia, D.; Hay, J.D. Population genetic structure of the endangered tropical tree species Caryocar brasiliense, based on variability at microsatellite loci. Mol. Ecol. 2001, 10, 349–356. [Google Scholar] [CrossRef]
- Santamaria, L. Why are most aquatic plants widely distributed? Dispersal, clonal growth and small-scale heterogeneity in a stressful environment. Acta Oecol. 2002, 23, 137–154. [Google Scholar] [CrossRef]
- Ma, S.; Shen, Y.; Li, M.; Jiang, R.; Cai, L.; Wu, T.; Gao, L.; Wu, M.; He, P. Establishment of Novel Simple Sequence Repeat Markers in Phragmites australis and Application in Wetlands of Nanhui Dongtan, Shanghai. Biology 2025, 14, 356. [Google Scholar] [CrossRef]
- Li, X.; Wang, X.; Cui, Z.; Shi, W.; Huang, J.; Wang, J. Development of Polymorphic Microsatellite Markers and Identification of Applications for Wild Walnut (Juglans regia L.) in Middle Asia. Diversity 2023, 15, 1073. [Google Scholar] [CrossRef]
- Hazelton, E.L.G.; McCormick, M.K.; Sievers, M.; Kettenring, K.M.; Whigham, D.F. Stand age is associated with clonal diversity, but not vigor, community structure, or insect herbivory in Chesapeake Bay Phragmites australis. Wetlands 2015, 35, 877–888. [Google Scholar] [CrossRef]
- Anderson, N.O.; Jocienė, L.; Krokaitė, E.; Rekašius, T.; Paulauskas, A.; Kupčinskienė, E. Genetic diversity of Phalaris arundinacea populations in relation to river regulation in the Merkys basin, Lithuania. River Res. Applic. 2018, 34, 300–309. [Google Scholar] [CrossRef]
- Hoban, S.; Archer, F.I.; Bertola, L.D.; Bragg, J.G.; Breed, M.F.; Bruford, M.W.; Coleman, M.A.; Ekblom, R.; Funk, W.C.; Grueber, C.E.; et al. Global genetic diversity status and trends: Towards a suite of Essential Biodiversity Variables (EBVs) for genetic composition. Biol. Rev. 2022, 97, 1511–1538. [Google Scholar] [CrossRef]
| Locus | Primer Sequence (5′ ⟶ 3′) | Repeat | Na | S | Ho | He |
|---|---|---|---|---|---|---|
| Potpect 24 | F Cy3 TCAGTGAAAGAAAGCCAGGA R GGGCTTATGGCGTTATCAA | (GA)n | 11 | 160–188 | 0.944 | 0.584 |
| Potpect 26 | F 6-Fam GTATAGGCGAGGTGCGAGAG R CTTCATGTCGACCACCTTCC | (CT)n | 14 | 229–275 | 0.882 | 0.570 |
| Potpect 28 | F 6-Fam TCGTTTCCTCCATTCGTAGG R AATAAAAAGGGCCCAGACC | (GA)n | 5 | 161–175 | 0.689 | 0.485 |
| Potpect 32 | F Hex CAGCAAACGAAACAACCAAA R AAAAGAAGCCGTTGTTTACAGAG | (GA)n | 10 | 221–239 | 0.596 | 0.470 |
| Potpect 34 | F 6-Fam GTAAGGCAAGCAGCGTCAAC R GTTTGTGAGCTAGCGGGAAG | (GA)n | 11 | 222–244 | 0.850 | 0.592 |
| Potpect 37 | F Hex CACTTCCTCTGTGCTGCTTG R GCGTGCTCTTCCTGAGTTCT | (CT)n | 6 | 142–172 | 0.721 | 0.468 |
| Potpect 39 | F Hex TCACAACACCTCACCCAGAA R CCATTTCCATTCCTCACTGC | (GA)n | 6 | 142–172 | 0.768 | 0.525 |
| Potpect 40 | F Cy3 AAATCTCCAAATATTTCCACTGTTG R CAAAGATTGAGCTCCCCAAA | (GA)n | 9 | 187–209 | 0.551 | 0.398 |
| Potpec 42 | F Cy3 TTAGCAAGTGGGTGGGTTTC R TGCACTCGTGTGTCTCTTCC | (CT)n | 6 | 192–206 | 0.591 | 0.447 |
| Total | 78 | |||||
| Mean | 8.67 | 0.732 | 0.504 | |||
| SE | 1.03 | 0.047 | 0.022 | |||
| Population | Plants Studied | GR | Genotypes | ||
|---|---|---|---|---|---|
| Name | Code | Unique | Multiclonal | ||
| Nemunas | NEM | 15 | 0.643 | 9 | 1 |
| Merkys | MER | 16 | 0.667 | 7 | 4 |
| Ūla | ULA | 16 | 0.333 | 3 | 3 |
| Neris | NER | 24 | 0.522 | 9 | 4 |
| Žeimena | ZEI | 16 | 0.533 | 8 | 1 |
| Šventoji | SVE | 16 | 0 | 0 | 1 |
| Širvinta | SIR | 16 | 0.533 | 4 | 5 |
| Nevėžis | NEV | 16 | 0.400 | 5 | 2 |
| Šešupė | SES | 16 | 0.667 | 7 | 4 |
| Total | 151 | 52 | 25 | ||
| Average | 16.78 | 0.478 | 5.78 | 2.78 | |
| SE | 0.91 | 0.070 | 1.01 | 0.52 | |
| Pop | N | P, % | Nd | Ne | I | Ho | He | Fis | Private Alleles | |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | NEM | 10 | 100 | 4.111 | 2.850 | 1.126 | 0.756 | 0.615 | −0.208 | 0.778 |
| 2 | MER | 11 | 100 | 3.556 | 2.241 | 0.919 | 0.778 | 0.532 | −0.441 | 0.444 |
| 3 | ULA | 6 | 88.89 | 1.889 | 1.707 | 0.538 | 0.648 | 0.375 | −0.630 | 0 |
| 4 | NER | 13 | 100 | 4.000 | 2.771 | 1.065 | 0.821 | 0.589 | −0.392 | 0.889 |
| 5 | ZEI | 9 | 100 | 3.000 | 2.274 | 0.898 | 0.642 | 0.539 | −0.164 | 0.333 |
| 6 | SIR | 9 | 100 | 3.111 | 2.691 | 1.021 | 0.778 | 0.608 | −0.325 | 0.778 |
| 7 | NEV | 7 | 88.89 | 3.000 | 2.188 | 0.826 | 0.651 | 0.491 | −0.316 | 0.444 |
| 8 | SES | 11 | 100 | 3.444 | 2.699 | 1.055 | 0.667 | 0.594 | −0.129 | 0.667 |
| Average | 9.50 | 97.22 | 3.264 | 2.428 | 0.931 | 0.718 | 0.533 | −0.326 | 0.542 | |
| SE | 0.80 | 1.81 | 0.248 | 0.139 | 0.066 | 0.026 | 0.031 | 0.058 | 0.104 |
| Pop | N | P, % | Br [8] | Nd | Na | Ne | I | He | |
|---|---|---|---|---|---|---|---|---|---|
| 1 | NEM | 25 | 41.51 | 1.281 | 1.277 | 1.415 | 1.217 | 0.197 | 0.130 |
| 2 | STR | 17 | 18.24 | 1.159 | 0.950 | 1.182 | 1.105 | 0.092 | 0.062 |
| 3 | MER | 22 | 50.31 | 1.448 | 1.377 | 1.503 | 1.360 | 0.289 | 0.199 |
| 4 | ULA | 12 | 20.13 | 1.193 | 0.937 | 1.201 | 1.134 | 0.116 | 0.078 |
| 5 | NER | 66 | 68.55 | 1.526 | 1.642 | 1.686 | 1.411 | 0.354 | 0.238 |
| 6 | ZEI | 47 | 59.12 | 1.448 | 1.453 | 1.591 | 1.347 | 0.305 | 0.204 |
| 7 | SVE | 30 | 37.11 | 1.302 | 1.233 | 1.371 | 1.230 | 0.196 | 0.132 |
| 8 | SIR | 8 | 21.38 | 1.214 | 1.006 | 1.214 | 1.155 | 0.128 | 0.088 |
| 9 | NEV | 26 | 49.69 | 1.390 | 1.384 | 1.497 | 1.292 | 0.258 | 0.172 |
| 10 | SES | 34 | 51.57 | 1.458 | 1.415 | 1.516 | 1.351 | 0.290 | 0.198 |
| Average | 28.70 | 41.76 | 1.342 | 1.267 | 1.418 | 1.260 | 0.223 | 0.150 | |
| SE | 5.44 | 5.49 | 0.041 | 0.075 | 0.055 | 0.034 | 0.029 | 0.019 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2026 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license.
Share and Cite
Patamsytė, J.; Butkuvienė, J.; Naugžemys, D.; Žvingila, D. Monitoring Genetic Diversity in Lithuanian Riverine Populations of Stuckenia pectinata Using SSR and ISSR Markers. Diversity 2026, 18, 26. https://doi.org/10.3390/d18010026
Patamsytė J, Butkuvienė J, Naugžemys D, Žvingila D. Monitoring Genetic Diversity in Lithuanian Riverine Populations of Stuckenia pectinata Using SSR and ISSR Markers. Diversity. 2026; 18(1):26. https://doi.org/10.3390/d18010026
Chicago/Turabian StylePatamsytė, Jolanta, Jurgita Butkuvienė, Donatas Naugžemys, and Donatas Žvingila. 2026. "Monitoring Genetic Diversity in Lithuanian Riverine Populations of Stuckenia pectinata Using SSR and ISSR Markers" Diversity 18, no. 1: 26. https://doi.org/10.3390/d18010026
APA StylePatamsytė, J., Butkuvienė, J., Naugžemys, D., & Žvingila, D. (2026). Monitoring Genetic Diversity in Lithuanian Riverine Populations of Stuckenia pectinata Using SSR and ISSR Markers. Diversity, 18(1), 26. https://doi.org/10.3390/d18010026






