Genetic Diversity of Five Pelodiscus sinensis Sub-Populations in the Dongting Lake Basin Based on Cytb and 12S rRNA Markers
Abstract
1. Introduction
2. Materials and Methods
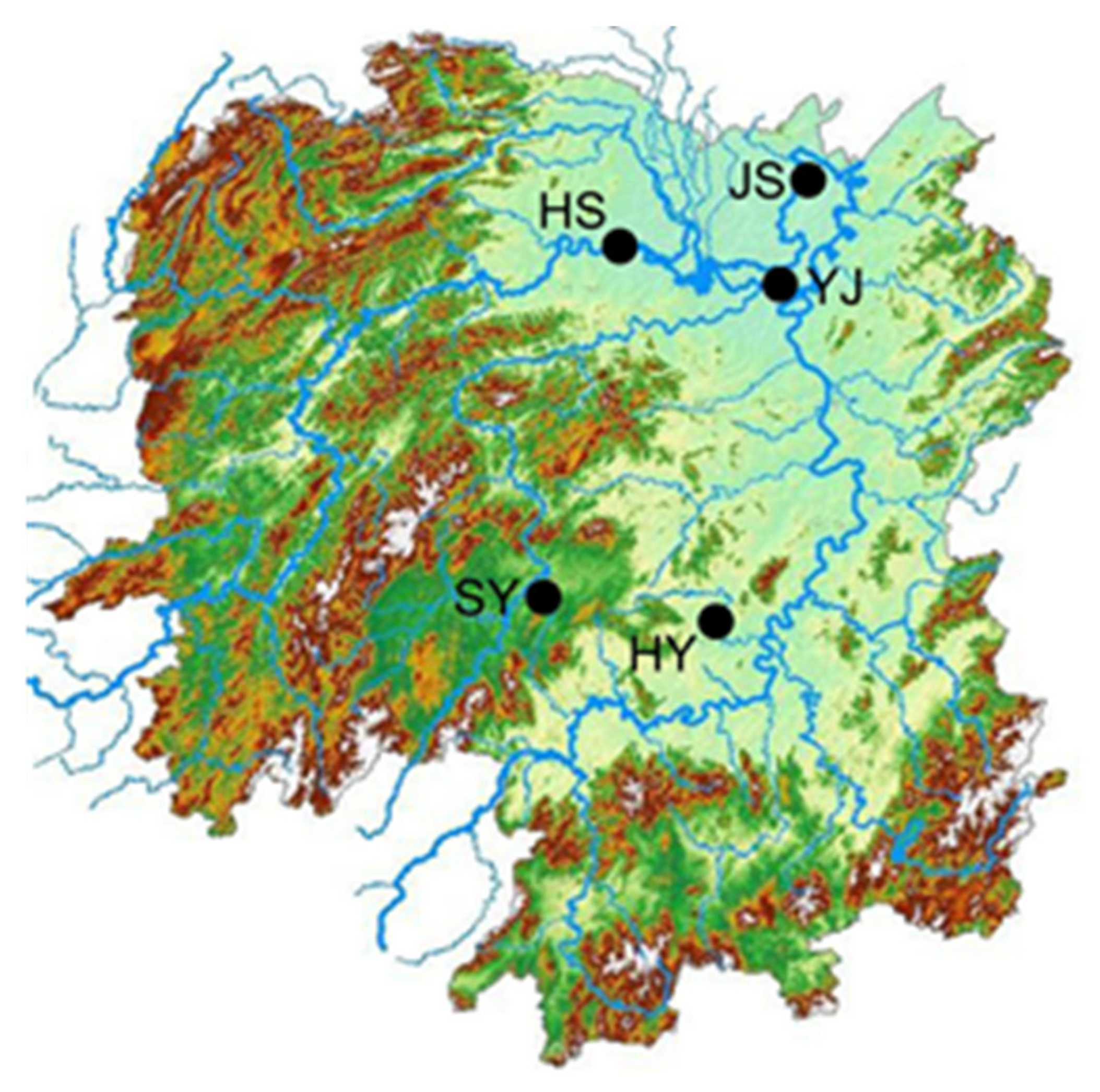
2.1. Sample Collection
2.2. DNA Extraction, PCR Amplification and Sequencing
2.3. Data Analysis
3. Results
3.1. Sequence Characteristics of Cytb and 12S rRNA Genes
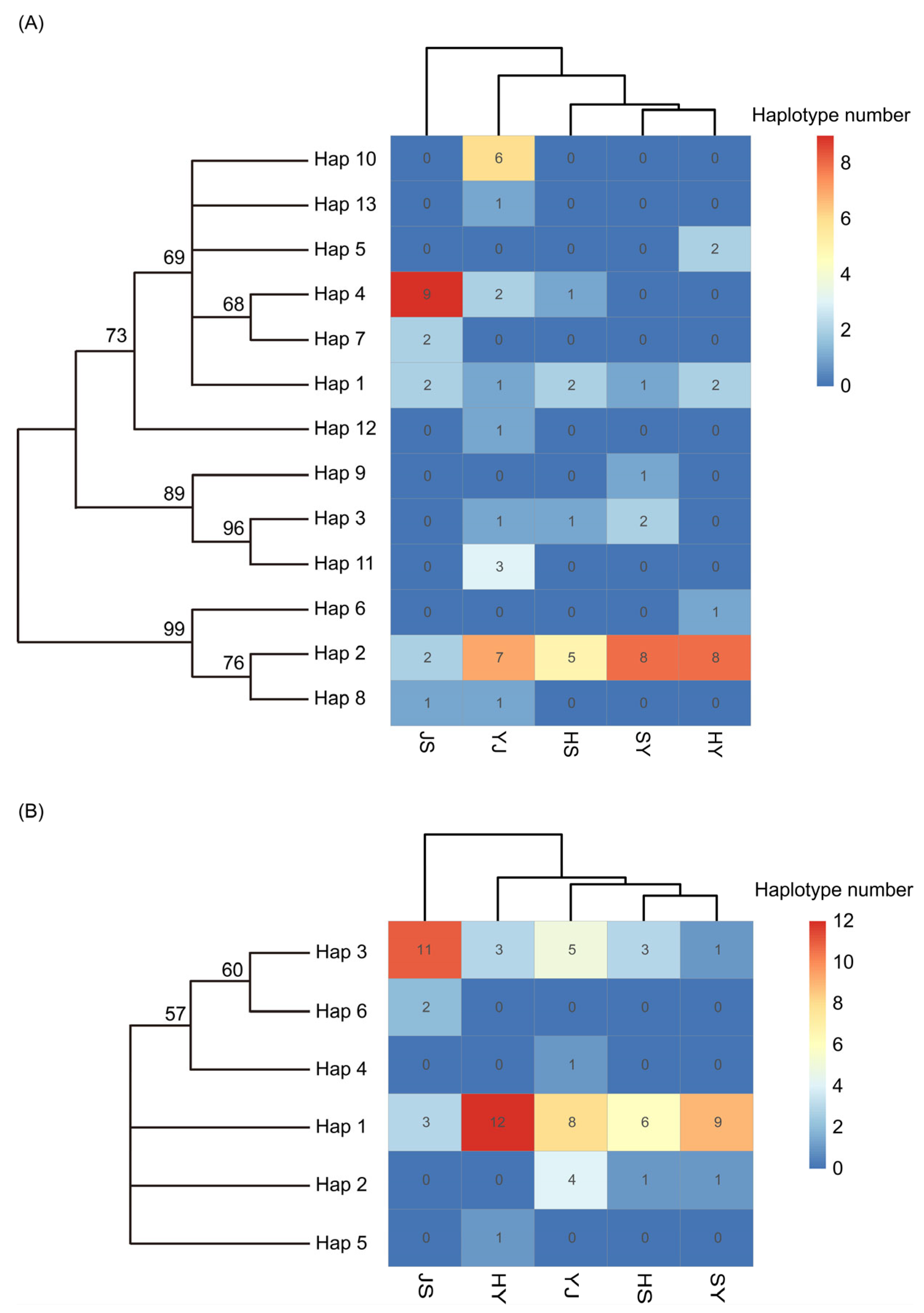
3.2. Genetic Diversity of P. sinensis Sub-Population
3.3. Genetic Structure of P. sinensis Sub-Population
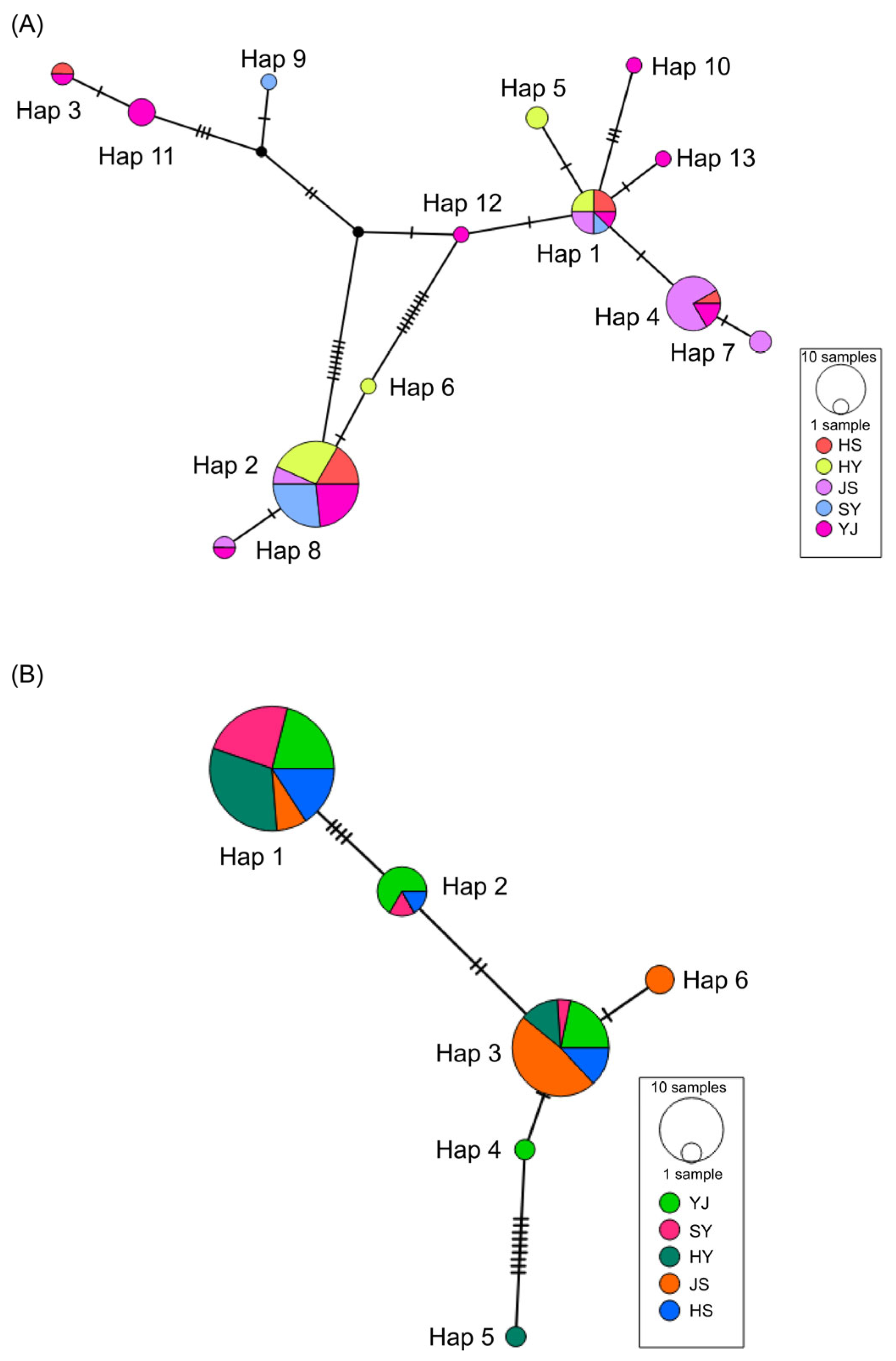
3.4. Phylogenetic Tree and Haplotype Network Diagram of P. sinensis Sub-Populations
3.5. Historical Dynamics of P. sinensis Sub-Populations
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Zeng, D.; Xiong, G.; Wang, X.; Zhou, X.; Wang, P.; Peng, N.; Luo, Z.; Li, X. Transcriptome Analysis and Identification of Genes and Single-Nucleotide Polymorphisms Associated with Growth Traits in the Chinese Soft-Shelled Turtle, Pelodiscus sinensis. J. World Aquac. Soc. 2021, 52, 913–931. [Google Scholar] [CrossRef]
- Feng, R.; Zhang, Z.; Guan, Y. Physiological and Transcriptional Analysis of Chinese Soft-Shelled Turtle (Pelodiscus sinensis) in Response to Acute Nitrite Stress. Aquat. Toxicol. 2021, 237, 105899. [Google Scholar] [CrossRef] [PubMed]
- Wu, B.; Huang, L.; Chen, J.; Zhang, Y.; Wang, J.; He, J. Gut Microbiota of Homologous Chinese Soft-Shell Turtles (Pelodiscus sinensis) in Different Habitats. BMC Microbiol. 2021, 21, 142. [Google Scholar] [CrossRef] [PubMed]
- Fisheries and Fishery Administration of the Ministry of Agriculture and Rural Affairs Bureau of China. 2024 China Fishery Statistics Yearbook; Agricultural Press of China: Beijing, China, 2024. [Google Scholar]
- Wang, P.; Zeng, D.; Xiong, G.; Zhou, X.; Jiang, H.; Hu, Y.; Ge, L.; Wang, X. Integrated Analysis of MRNA-Seq and MicroRNA-Seq Depicts the Potential Roles of miRNA-mRNA Networks in Pigmentation of Chinese Soft-Shelled Turtle (Pelodiscus sinensis). Aquac. Rep. 2021, 20, 100686. [Google Scholar] [CrossRef]
- Zhang, C.; Zhang, H.; Xu, X.; He, Z.; Wang, C. Comparative Analyses of 16S rRNA Genetic Diversity in Newly Cultured Varieties of Pelodiscus sinensis. J. Fish. Sci. China 2014, 21, 398–404. [Google Scholar]
- Nong, Z.; Wei, L.; Ye, X.; Liu, K.; Lu, Y.; Yan, X.; Zhang, S. Nutrient Composition Analysis and Quality Evaluation of Muscle of Different Populations of Pelodiscus luteus in Guangxi. Feed Ind. 2024, 45, 91–97. [Google Scholar]
- Lu, Y.; Liang, Y.; Wan, G.; Wang, P.; Zeng, J.; Wang, X.; Hu, Y.; Li, X. Morphological, Growth and Genetic Diversity of Changyong Population of Chinese Soft-Shelled Turtle (Pelodiscus sinensis). J. Hunan Agric. Univ. (Nat. Sci.) 2025, 51, 98–106. [Google Scholar]
- Zhu, X.; Chen, C.; Huang, Q.; Ke, Y. Chinese Soft-Shell Turtle “Zhushui No. 1.”. Chinese Fish. 2021, 5, 92–97. [Google Scholar]
- Zhang, J.; Zhou, Q.; Yang, X.; Yu, P.; Zhou, W.; Gui, Y.; Ouyang, X.; Wan, Q. Characterization of the Complete Mitochondrial Genome and Phylogenetic Analysis of Pelodiscus sinensis, a Mutant Chinese Soft-Shell Turtle. Conserv. Genet. Resour. 2019, 11, 279–282. [Google Scholar] [CrossRef]
- Zhang, J.; Wang, F.; Jiang, Y.-L.; Hou, G.-J.; Cheng, Y.-S.; Chen, H.-L.; Li, X. Modern Greenhouse Culture of Juvenile Soft-Shelled Turtle, Pelodiscus sinensis. Aquac. Int. 2017, 25, 1607–1624. [Google Scholar] [CrossRef]
- Zeng, D.; Li, X.; Wang, X.; Xiong, G. Development of SNP Markers Associated with Growth-Related Genes of Pelodiscus sinensis. Conserv. Genet. Resour. 2020, 12, 87–92. [Google Scholar] [CrossRef]
- Li, T.; Li, W.; Zhao, J.; Shi, Y.; Hong, X.; Wang, Y.; Zhu, X. Correlation Analysis of the Microsatellite DNA Markers and Growth Traits of Chinese Soft Shell Turtle (Trionyx sinensis). Genomics Appl. Biol. 2016, 35, 63–71. [Google Scholar]
- Jia, M.; Li, Q.; Zhang, T.; Dong, B.; Liang, X.; Fu, S.; Yu, J. Genetic Diversity Analysis of the Chinese Daur Ethnic Group in Heilongjiang Province by Complete Mitochondrial Genome Sequencing. Front. Genet. 2022, 13, 919063. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Chen, M.; Chen, C.; Fang, Y.; Cui, W.; Lei, F.; Zhu, B. Genetic Background of Kirgiz Ethnic Group from Northwest China Revealed by Mitochondrial DNA Control Region Sequences on Massively Parallel Sequencing. Front. Genet. 2022, 13, 729514. [Google Scholar] [CrossRef]
- Chen, J.; Jiao, J.; Yuan, X.; Huang, X.; Huang, L.; Lin, L.; Yin, W.; Yao, J.; Zhang, H. Complete Mitochondrial Genomes of Four Pelodiscus sinensis Strains and Comparison with Other Trionychidae Species. Biology 2023, 12, 406. [Google Scholar] [CrossRef] [PubMed]
- Zhong, L.; Wang, M.; Li, D.; Tang, S.; Zhang, T.; Bian, W.; Chen, X. Complete Mitochondrial Genome of Freshwater Goby Rhinogobius cliffordpopei (Perciformes gobiidae): Genome Characterization and Phylogenetic Analysis. Genes Genom. 2018, 40, 1137–1148. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Wu, P.-D.; Zhang, D.-Z.; Zhang, H.-B.; Tang, B.-P.; Liu, Q.-N.; Dai, L.-S. Mitochondrial Genome of the Yellow Catfish Pelteobagrus fulvidraco and Insights into Bagridae Phylogenetics. Genomics 2019, 111, 1258–1265. [Google Scholar] [CrossRef]
- Chen, X.; Guo, L.; Li, M.; Meng, Z.; Lin, H. The Molecular Phylogenetic Relationships of 13 Epinepheline Species in Southern Coastal Provinces of China. Acta Sci. Nat. Univ. Sunyatseni 2014, 53, 123–130. [Google Scholar]
- Liu, Y.; Liu, Y.; Liu, B.; Yu, Y.; Song, C.; Wang, C. Identification Analysis of DNA Barcoding Based on Mitochondrial Cyt b and 12S rRNA Genes in Myctophidae Fishes. J. Fish. Sci. China 2023, 30, 1112–1126. [Google Scholar]
- Xiao, F.; Li, W.; Hong, X.; Zhu, A.; Zhu, X.; Shi, Y.; Huang, Q. Growth Comparison of Three Breeding Populations of Pelodiscus sinensis. Guangdong Agric. Sci. 2014, 41, 107–110. [Google Scholar]
- Xiong, G.; Wang, X.-Q.; Zhou, X.-W.; Zeng, D.; Chen, Z.-N.; Wang, P.; Kang, L. Genetic Variation in the Chinese Soft-Shell Turtles (Pelodiscus Spp.) Revealed by Sequences of Mitochondrial Cytb Gene. Mitochondrial DNA Part A DNA Mapp. Seq. Anal. 2019, 30, 874–879. [Google Scholar] [CrossRef]
- Liang, Y.; Tian, P.; Lu, Y.; Qin, Q.; Wang, Z.; Xiong, G.; Wang, X.; Hu, Y. Establishment and Population Genetic Analysis of SNP Fingerprinting of Chinese Soft-Shelled Turtle (Pelodiscus sinensis). Aquac. Reports 2024, 38, 102340. [Google Scholar] [CrossRef]
- Liang, Y.; Huang, C.; Wang, P.; Xiao, H.; Wang, Z.; Zeng, J.; Wang, X.; Xiong, S.; Hu, Y.; Qin, Q. Research on SSR Genetic Molecular Markers and Morphological Differences of Different Pelodiscus sinensis Populations. Genes 2025, 16, 318. [Google Scholar] [CrossRef] [PubMed]
- Engstrom, T.N.; Edwards, T.; Osentoski, M.F.; Myers, E.M. A Compendium of PCR Primers for MtDNA, Microsatellite, and Other Nuclear Loci for Freshwater Turtles and Tortoises. Chelonian Res. Monogr. 2007, 4, 124–141. [Google Scholar]
- Tedesco, P.; Caffara, M.; Davidovich, N.; Luci, V.; Cantori, A.; Fioravanti, M.L.; Gustinelli, A. Morphological and molecular data on acuariid nematodes in European great cormorants (Phalacrocorax carbo sinensis) and pygmy cormorants (Microcarbo pygmaeus). Sci. Rep. 2024, 14, 13712. [Google Scholar] [CrossRef] [PubMed]
- Larkin, M.A.; Blackshields, G.; Brown, N.P.; Chenna, R.; McGettigan, P.A.; McWilliam, H.; Valentin, F.; Wallace, I.M.; Wilm, A.; Lopez, R.; et al. Clustal W and Clustal X version 2.0. Bioinformatics 2007, 23, 2947–2948. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular evolutionary genetics analysis version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef]
- Librado, P.; Rozas, J. DnaSP v5: A software for comprehensive analysis of DNA polymorphism data. Bioinformatics 2009, 25, 1451–1452. [Google Scholar] [CrossRef]
- Leigh, J.W.; Bryant, D. POPART: Full-feature software for haplotype network construction. Methods Ecol. Evol. 2015, 6, 1110–1116. [Google Scholar] [CrossRef]
- Excoffier, L.; Lischer, H.E.L. Arlequin suite ver 3.5: A new series of programs to perform population genetics analyses under Linux and Windows. Mol. Ecol. Resour. 2010, 10, 564–567. [Google Scholar] [CrossRef]
- Sha, H.; Luo, X.Z.; Wang, D.; Li, X.; Zou, G.W.; Liang, H.W. New Insights to Protection and Utilization of Silver Carp (Hypophthalmichthys molitrix) in Yangtze River Based on Microsatellite Analysis. Fish. Res. 2021, 241, 105997. [Google Scholar] [CrossRef]
- Chen, K.; Wang, W.; Chen, X.; Zhu, W.; Wang, H. Analysis of Genetic Diversity among Five Populations of Lutraria maxima Based on MtDNA Cytb and D-Loop Sequences. Mar. Fish. 2020, 42, 149–160. [Google Scholar] [CrossRef]
- Tang, Q.; Xie, J.; Xia, Z.; Cai, M.; Wu, Y.; Bai, L.; Du, H.; Li, J.; Yang, G. Genetic Diversity of the Breeding Populations of Giant Freshwater Prawn Macrobrachium rosenbergii. Acta Hydrobiol. Sin. 2020, 44, 1097–1104. [Google Scholar] [CrossRef]
- Li, D.; Liu, Y.; Liu, Y.; Tang, S.; Gu, X.; Yin, J.; Jiang, Q.; Zhang, T. Genetic Diversity Analysis of Corbicula fluminea in the Tai Lake Based on Mitochondrial COI and Cytb Gene Sequences. J. Biol. 2023, 40, 69–73. [Google Scholar] [CrossRef]
- Grant, W.A.S.; Bowen, B.W. Shallow Population Histories in Deep Evolutionary Lineages of Marine Fishes: Insights from Sardines and Anchovies and Lessons for Conservation. J. Hered. 1998, 89, 415–426. [Google Scholar] [CrossRef]
- Li, L.; Tan, S.; Wang, B.; Xu, J.; Han, X. Genetic Diversity of Three Different Populations of Soft Shelled Turtle Trionyx sinensis Using Mitochondrial D-Loop Gene. Chin. J. Fish. 2020, 33, 7–11. [Google Scholar]
- Huang, X.; Qian, G.; Li, C. The Genetic Diversity of Mitochondrial DNA D-Loop Region of Three Geographical Populations in Trionyx sinensis. J. Fish. China 2012, 36, 17–24. [Google Scholar] [CrossRef]
- Xiao, H.; Hu, Y.; Qin, Q.; Xu, D.; Liang, Y.; Liu, X.; Wang, X. Genetic Diversity Analysis of Pelodiscus sinensis and Their Hybrid from Two Geographical Populations of Using Cyt b Sequence. J. Hunan Agric. Univ. (Nat. Sci.) 2023, 49, 486–490. [Google Scholar]
- Li, S.; Lv, G.; Li, C.; Ma, D.; Lin, Z.; Hu, G. MtDNA Polymorphism Analysis of Local Populations of Soft-Shelled Turtle from East China. J. Fish. Sci. China 1997, 4, 2–7. [Google Scholar]
- Yu, F.; Zhang, Q.; Tang, Y.; Zhou, J. Preliminary Study on Genetic Variation of Mitochondrial DNA Cytochrome b Gene of Pelodiscus sinensis. Guangdong Agric. Sci. 2011, 38, 5–7. [Google Scholar]
- Liu, Y.; Shi, Y.; Zhu, X.; Zhao, J.; Zhou, G.; Hong, X. Genetic Diversity in Five Populations of Trionyx sinensis Revealed by Microsatellite Markers. Genomics Appl. Biol. 2012, 31, 141–146. [Google Scholar] [CrossRef]
- Li, Y.; Luo, M.; Feng, B.; Zhu, W.; Fu, J.; Liang, Z.; Xie, X.; Miao, L.; Dong, Z. Genetic Diversity Analysis of Seven American Shad (Alosa sapidissima) Populations in Southern Jiangsu, China. J. Shanghai Ocean Univ. 2024, 33, 285–296. [Google Scholar]
- Wang, H.; Guo, X.; Zhang, R.; Wang, L.; Li, T.; Xing, X. Population Structure and Origin Evolution Analysis of Dwarf Sika Deer Population Based on Four Genes/Regions of Mitochondrial DNA. Acta Agric. Boreali-Occident. Sin. 2018, 27, 1723–1730. [Google Scholar]
- Zhang, H.; Chen, Z.; Li, G.; Tang, Y.; Wen, Y.; Li, X.; Wang, M.; Liu, J.; Peng, L.; Xiao, Y.; et al. Cloning and Genetic Diversity Analysis of Mitochondrial Cytochrome b in Hanshou Trionyx sinensis. Acta Laser Biol. Sin. 2018, 27, 359–366. [Google Scholar]
- Qin, N.; Zhang, L.; Xiao, M.; Zhou, X.; Zhang, G.; Ma, X.; Wei, K. Population Genetic Diversity and Genetic Structure of Zacco Platypus in the Western Wuling Mountains and Adjacent Areas. J. Fish. Sci. China 2024, 31, 301–315. [Google Scholar]
- Andrews, K.R.; Good, J.M.; Miller, M.R.; Luikart, G.; Hohenlohe, P.A. Harnessing the Power of RADseq for Ecological and Evolutionary Genomics. Nat. Rev. Genet. 2016, 17, 81–92. [Google Scholar] [CrossRef]
- Kundu, S.; Kumar, V.; Tyagi, K.; Chakraborty, R.; Singha, D.; Rahaman, I.; Pakrashi, A.; Chandra, K. Complete Mitochondrial Genome of Black Soft-Shell Turtle (Nilssonia nigricans) and Comparative Analysis with Other Trionychidae. Sci. Rep. 2018, 8, 17378. [Google Scholar] [CrossRef]
- Xu, X.; Zhang, H.; He, Z. Sequence Composition of Mitochondrial 12S rRNA Genes between Two Varieties of Pelodiscus sinensis. J. Econ. Anim. 2012, 16, 163–167. [Google Scholar]
- Nei, M. Molecular Population Genetics and Evolution; North-Holland Publishing Company: Amsterdam, The Netherlands, 1975. [Google Scholar]
- Wright, S. Evolution and the Genetics of Populations: A Treatise in Four Volumes, Volume 4 Variability Within and Among Natural Populations; The University of Chicago Press: Chicago, IL, USA, 1978. [Google Scholar]
- Luo, Y.; Fang, D.; Zhou, Y.; Xu, D.; Peng, Y.; Peng, F.; Zhang, G.; Liu, K.; You, Y. Genetic Diversity of Silver Carp (Hypophthalmichthys molitrix) in Lower Reaches of Yangtze River Based on Microsatellite Markers. South China Fish. Sci. 2021, 17, 48–57. [Google Scholar]
- Xu, X.; Cai, X.; Bai, S. Genetic Diversity of the Black Grouse Tetrao tetrix baikalensis at Daxinganling Based on the MtDNA D-Loop Sequences. Chin. J. Zool. 2013, 48, 673–679. [Google Scholar]
- Han, X.; Gu, Y.; Xu, M.; Sha, Y.; Xu, J.; Han, Y. A Comparative Analysis of Mitochondrial Genome Sequence from Different Populations of Pelodiscus sinensis in Taihu Basin. J. Chang. Inst. Technol. (Nat. Sci.) 2017, 31, 91–94. [Google Scholar]
- Xiong, F.; Liu, H.; Zhai, D.; Duan, X.; Tian, H.; Chen, D. Population Genetic Structure of Pelteobagrus vachelli in the Upper Yangtze River Based on Genome Re-Sequencing. Biodivers. Sci. 2023, 31, 22391. [Google Scholar] [CrossRef]
- Zhu, C.; Fen, W.; Xin, D.; Chang, M.S.; Lin, Z.; Wang, J.; Xiang, Z.; Song, G.; Salin, K.R.; Wang, H.; et al. Genetic Diversity Variations of Soft-Shelled Turtle (Pelodiscus sinensis) Inferred from Microsatellite Approaches. Pak. J. Zool. 2025, 57, 1225. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhang, H.; He, Z.; Xu, X.; Li, Z.; Du, J. Sequence Variation and Population Genetic Structure of Five Populations in Pelodiscus sinensis Based on Cytochrome b Gene. Oceanol. Limnol. Sin. 2008, 39, 234–239. [Google Scholar]
- Wang, J.; Zhu, P.; Wang, J.; Gui, Y.; Li, D. Genetic Diversity Analysis of Corbicula fluminea in Dongting Lake Based on the Mitochondrial Cyt b Gene. J. Hydroecol. 2018, 39, 89–94. [Google Scholar] [CrossRef]
- Tajima, F. Statistical Method for Testing the Neutral Mutation Hypothesis by DNA Polymorphism. Genetics 1989, 123, 585–595. [Google Scholar] [CrossRef]
- Márcia Barbosa, A.; Real, R.; Muñoz, A.R.; Brown, J.A. New Measures for Assessing Model Equilibrium and Prediction Mismatch in Species Distribution Models. Divers. Distrib. 2013, 19, 1333–1338. [Google Scholar] [CrossRef]
| Gene | Base Content (%) | |||||
|---|---|---|---|---|---|---|
| T | C | A | G | A+T | C+G | |
| Cytb | 31.1 | 11.9 | 27.5 | 29.5 | 58.6 | 41.4 |
| 12S rRNA | 22.2 | 22.6 | 37.7 | 17.6 | 59.9 | 40.1 |
| Gene | Sub-Population | N | S | H | Hd | Pi | K | Tajima’s D | P | Fu’s Fs | P |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Cytb | HS | 10 | 24 | 6 | 0.889 | 0.015 | 9.778 | 0.698 | 0.752 | 4.094 | 0.978 |
| SY | 11 | 20 | 3 | 0.378 | 0.008 | 5.400 | −0.744 | 0.219 | 3.905 | 0.965 | |
| HY | 16 | 11 | 4 | 0.615 | 0.010 | 5.000 | 1.660 | 0.959 | 3.946 | 0.954 | |
| JS | 16 | 19 | 7 | 0.750 | 0.009 | 5.617 | 0.147 | 0.638 | 2.408 | 0.892 | |
| YJ | 18 | 30 | 10 | 0.876 | 0.016 | 10.706 | 0.853 | 0.786 | 0.986 | 0.740 | |
| Total | 71 | 25 | 13 | 0.750 | 0.014 | 6.547 | 0.523 | 0.671 | 3.068 | 0.906 | |
| 12S rRNA | HS | 10 | 9 | 5 | 0.844 | 0.010 | 4.089 | 1.845 | 0.982 | 3.167 | 0.957 |
| SY | 11 | 10 | 4 | 0.491 | 0.006 | 2.4 | −0.726 | 0.262 | 1.705 | 0.839 | |
| HY | 16 | 19 | 5 | 0.600 | 0.011 | 4.283 | −0.756 | 0.199 | 4.502 | 0.986 | |
| JS | 16 | 7 | 3 | 0.508 | 0.006 | 2.183 | 0.123 | 0.545 | 3.109 | 0.936 | |
| YJ | 18 | 7 | 4 | 0.712 | 0.009 | 3.15 | 2.444 | 0.995 | 2.985 | 0.929 | |
| Total | 71 | 14 | 6 | 0.609 | 0.010 | 3.23 | 0.586 | 0.597 | 3.094 | 0.929 |
| Gene | Sub-Population | HS | SY | HY | JS | YJ |
|---|---|---|---|---|---|---|
| Cytb | HS | - | 0.073/8.54 | −0.019/−4.74 | 0.180 */0.99 | −0.063/−4.36 |
| SY | 0.012 | - | −0.033/−8.51 | 0.514 */0.23 | 0.113/1.83 | |
| HY | 0.013 | 0.010 | - | 0.378 */0.42 | 0.051/4.10 | |
| JS | 0.015 | 0.018 | 0.016 | - | 0.177 */1.02 | |
| YJ | 0.015 | 0.014 | 0.015 | 0.016 | - | |
| 12S rRNA | HS | - | 0.023/17.46 | −0.026/−9.74 | 0.288 */0.68 | −0.053/−5.97 |
| SY | 0.009 | - | −0.047/−8.11 | 0.558 */0.19 | 0.152/1.36 | |
| HY | 0.011 | 0.009 | - | 0.424 */0.34 | 0.085/2.82 | |
| JS | 0.012 | 0.014 | 0.002 | - | 0.189 */1.08 | |
| YJ | 0.010 | 0.010 | 0.012 | 0.011 | - |
| Gene | Source of Variation | df | Sum of Squares | Variance Component | Percentage of Variation (%) | Fixation Index |
|---|---|---|---|---|---|---|
| Cytb | Among sub-populations | 4 | 40.456 | 0.552 Va | 16.35 | |
| Within sub-populations | 62 | 175.067 | 2.824 Vb | 83.65 | ||
| Total | 66 | 215.522 | 3.376 | 100 | 0.164 | |
| 12S rRNA | Among sub-populations | 4 | 25.045 | 0.351 Va | 20.85 | |
| Within sub-populations | 66 | 87.997 | 1.333 Vb | 79.15 | ||
| Total | 70 | 113.042 | 1.685 | 100 | 0.209 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zuo, Z.; Xiao, H.; Wu, Q.; Tian, L.; Gao, F.; Li, C.; Ni, J.; Peng, Z.; Xiang, J. Genetic Diversity of Five Pelodiscus sinensis Sub-Populations in the Dongting Lake Basin Based on Cytb and 12S rRNA Markers. Diversity 2025, 17, 575. https://doi.org/10.3390/d17080575
Zuo Z, Xiao H, Wu Q, Tian L, Gao F, Li C, Ni J, Peng Z, Xiang J. Genetic Diversity of Five Pelodiscus sinensis Sub-Populations in the Dongting Lake Basin Based on Cytb and 12S rRNA Markers. Diversity. 2025; 17(8):575. https://doi.org/10.3390/d17080575
Chicago/Turabian StyleZuo, Zhiliang, Hewei Xiao, Qifan Wu, Lu Tian, Feng Gao, Cheng Li, Jiajia Ni, Zhitao Peng, and Jin Xiang. 2025. "Genetic Diversity of Five Pelodiscus sinensis Sub-Populations in the Dongting Lake Basin Based on Cytb and 12S rRNA Markers" Diversity 17, no. 8: 575. https://doi.org/10.3390/d17080575
APA StyleZuo, Z., Xiao, H., Wu, Q., Tian, L., Gao, F., Li, C., Ni, J., Peng, Z., & Xiang, J. (2025). Genetic Diversity of Five Pelodiscus sinensis Sub-Populations in the Dongting Lake Basin Based on Cytb and 12S rRNA Markers. Diversity, 17(8), 575. https://doi.org/10.3390/d17080575






