The Splendour and Misery of European Pink Salmon, Oncorhynchus gorbuscha: Abundant Odd Lineage vs. Depressed Even Lineage—Insights from cytb Gene Analysis
Abstract
1. Introduction
2. Materials and Methods
2.1. Sampling
2.2. Genetic Analysis
2.3. Data Analysis
3. Results
3.1. Odd-Year Spawning Lineage
3.2. Even-Year Spawning Lineage
3.3. Odd-Year Versus Even-Year Pink Salmon Lineages
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Brykov, V.A.; Polyakova, N.; Skurikhina, L.A.; Kukhlevsky, A.D. Geographical and temporal mitochondrial DNA variability in populations of pink salmon. J. Fish Biol. 1996, 48, 899–909. [Google Scholar] [CrossRef]
- Churikov, D.; Gharrett, A.J. Comparative phylogeography of the two pink salmon broodlines: An analysis based on a mitochondrial DNA genealogy. Mol. Ecol. 2002, 11, 1077–1101. [Google Scholar] [CrossRef]
- Glubokovskii, M.K.; Zhivotovskii, L.A. Population structure of pink salmon: A system of fluctuating stocks. Biol. Mor. 1986, 12, 39–44. [Google Scholar]
- Beacham, T.D.; Withler, R.E.; Murray, C.B.; Barner, L.W. Variation in body size, morphology, egg size, and biochemical genetics of pink salmon in British Columbia. Trans. Am. Fish. Soc. 1988, 117, 109–126. [Google Scholar] [CrossRef]
- Gritsenko, O.F. Population structure of the pink salmon Oncorhynchus gorbuscha (Walbaum). Vopr. Ikhtiol. 1981, 21, 787–799. [Google Scholar]
- Oke, K.B.; Cunningham, C.J.; Quinn, T.P.; Hendry, A.P. Independent lineages in a common environment: The roles of determinism and contingency in shaping the migration timing of even- versus odd-year pink salmon over broad spatial and temporal scales. Ecol. Lett. 2019, 22, 1547–1556. [Google Scholar] [CrossRef] [PubMed]
- Sato, S.; Urawa, S. Genetic variation of Japanese pink salmon populations inferred from nucleotide sequence analysis of the mitochondrial DNA control region. Environ. Biol. Fish 2017, 100, 1355–1372. [Google Scholar] [CrossRef]
- Podlesnykh, A.V.; Brykov, V.A.; Ponomareva, E.V.; Kinas, N.M.; Klovach, N.V. Development of Pink Salmon Introduced into European Waters Based on the Results of Genetic Research. Russ. J. Mar. Biol. 2020, 46, 360–367. [Google Scholar]
- Zelenina, D.A.; Zhivotovsky, L.A.; Soshnina, V.A.; Vilkova, O.Y.; Glubokovsky, M.K. Intraspecific Differentiation of Asian Pink Salmon Inferred from the Mitochondrial cytb Gene Sequences. Rus. J. Genet. 2022, 58, 1323–1333. [Google Scholar] [CrossRef]
- Salmenkova, E.A.; Omelchenko, V.T.; Rekubratsky, A.V.; Gordeeva, N.V.; Yan, Y.Y.; Radchenko, O.A. Allozyme variation in populations of pink salmon, Oncorhynchus gorbuscha (Walbaum), from some regions of the species range. Russ. J. Genet. 2006, 42, 155–167. [Google Scholar]
- Beacham, T.D.; McIntosh, B.; MacConnachie, C.; Spilsted, B.; White, B.A. Population structure of pink salmon (Oncorhynchus gorbuscha) in British Columbia and Washington, determined with microsatellites. Fish. Bull. 2012, 110, 242–256. [Google Scholar]
- Tarpey, C.M.; Seeb, J.E.; McKinney, G.J.; Seeb, L.W. Single-nucleotide polymorphism data describe contemporary population structure and diversity in allochronic lineages of pink salmon (Oncorhynchus gorbuscha). Can. J. Fish. Aq. Sci. 2018, 75, 987–997. [Google Scholar] [CrossRef]
- Christensen, K.A.; Rondeau, E.B.; Sakhrani, D.; Biagi, C.A.; Johnson, H.; Joshi, J.; Flores, A.-M.; Leelakumari, S.; Moore, R.; Pandoh, P.K.; et al. The pink salmon genome: Uncovering the genomic consequences of a two-year life cycle. PLoS ONE 2021, 16, e0255752. [Google Scholar] [CrossRef] [PubMed]
- Yakovenko, M.Y. The pink salmon as an object of fishery in the White Sea basin. In Problems of Study, Rational Exploitation and Conservation of Natural Resources of the White Sea. Proceedings of the 6th Regional Meeting; Alimov, A.F., Ed.; Zoological Institute RAS: St. Petersburg, Russia, 1995; pp. 35–38. (In Russian) [Google Scholar]
- Gordeeva, N.V.; Salmenkova, E.A.; Altukhov, Y.P.; Makhrov, A.A.; Pustovoit, S.P. Genetic changes in pink salmon Oncorhynchus gorbuscha Walbaum during acclimatization in the White Sea basin. Russ. J. Genet. 2003, 39, 322–332. [Google Scholar] [CrossRef]
- Bakshtansky, E.L. The introduction of pink salmon into the Kola Peninsula. In Salmon Ranching; Academic Press: London, UK, 1980; Volume 245. [Google Scholar]
- Gordeeva, N.V.; Salmenkova, E.A. Experimental microevolution: Transplantation of pink salmon into the European North. Evol. Ecol. 2011, 25, 657–679. [Google Scholar] [CrossRef]
- Gordeeva, N.V.; Salmenkova, E.A.; Altukhov, Y.P. Acclimatization of pink salmon, Oncorhynchus gorbuscha Walbaum in the European North: Data of restriction analysis of mtDNA. Russ. J. Genet. 2004, 40, 393–400. [Google Scholar] [CrossRef]
- Bjerknes, V.; Vaag, A.B. Migration and capture of pink salmon, Oncorhynchus gorbuscha Walbaum in Finnmark, North Norway. J. Fish Biol. 1980, 16, 291–297. [Google Scholar] [CrossRef]
- Alekseev, M.Y.; Tkachenko, A.V.; Zubchenko, A.V.; Shkatelov, A.P.; Nikolaev, A.M. Distribution, Spawning and the Possibility of Fishery of Introduced Pink Salmon (Oncorhynchus gorbusha Walbaum) in Rivers of Murmansk Oblast. Russ. J. Biol. Invasions 2019, 10, 109–117. (In Russian) [Google Scholar] [CrossRef]
- Lennox, R.J.; Berntsen, H.H.; Garseth, A.H.; Ninch, S.G.; Hindar, K.; Ugedal, O.; Utne, K.R.; Vollset, K.W.; Whoriskey, F.G.; Thorstad, E.B. Prospects for the future of pink salmon in three oceans: From the native Pacific to the novel Arctic and Atlantic. Fish Fish. 2023, 24, 759–776. [Google Scholar] [CrossRef]
- Bogdanov, V.D.; Kizhevatov, Y.A. Pink salmon (Oncorhynchus gorbuscha Walbaum, 1792) in water bodies and watercourses of the Yamalo-Nenets Autonomous Okrug. Sci. Bull. Yamalo-Nenets Auton. Okrug 2007, 6, 3–4. [Google Scholar]
- Skora, M.E.; Jones, J.I.; Youngson, A.F.; Robertson, S.; Wells, A.; Lauridsen, R.B.; Copp, G.H. Evidence of potential establishment of pink salmon Oncorhynchus gorbuscha in Scotland. J. Fish Biol. 2023, 102, 721–726. [Google Scholar] [CrossRef]
- Guay, J.D.; Lennox, R.J.; Thorstad, E.B.; Vollset, K.W.; Stensland, S.; Erkinaro, J.; Nguyen, V.M. Recreational anglers in Norway report widespread dislike of invasive pink salmon. People Nat. 2024, 6, 41–53. [Google Scholar] [CrossRef]
- Gordeev, I.I.; Tkachenko, A.V.; Tortsev, A.M.; Studionov, I.I.; Genrikh, E.A.; Kanzeparova, A.N.; Belyaev, V.A. Pink salmon fishery in the European part of Russia: Results for 2023. In Bulletin on the Study of Pacific Salmon in the Far East; TINRO: Vladivostok, Russia, 2024; no. 18; pp. 123–131, (In Russian). [Google Scholar] [CrossRef]
- Thorstad, E.; Staveley, T.; Fiske, P. Pink salmon in rivers: Current knowledge, overlap, and potential interactions with Atlantic salmon. In Management of Pink Salmon in the North Atlantic and Their Potential Threats to Wild Atlantic Salmon; North Atlantic Salmon Conservation Organization: Edinburgh, UK, 2024. [Google Scholar]
- Dunmall, K.M.; Bean, C.W.; Berntsen, H.H.; Ensing, D.; Erkinaro, J.; Irvine, J.R.; Utne, K.R. Invading and range-expanding pink salmon inform management actions for marine species on the move. ICES J. Mar. Sci. 2025, 82, fsae199. [Google Scholar] [CrossRef]
- Belyaev, V.A.; Ponomareva, E.V.; Malyutina, A.M.; Mel’nikova, M.N.; Stroganov, A.N. On the Origins of the Negative Impact of Invasive Pink Salmon Oncorhynchus gorbuscha on Natural Populations of Atlantic Salmon Salmo salar (Salmonidae). J. Ichthyol. 2025, 65, 116–128. [Google Scholar] [CrossRef]
- Gilbey, J.; Soshnina, V.A.; Volkov, A.A.; Zelenina, D.A. Comparative genetic variability of pink salmon from different parts of their range: Native Pacific, artificially introduced White Sea and naturally invasive Atlantic Scottish rivers. J. Fish Biol. 2022, 100, 549–560. [Google Scholar] [CrossRef] [PubMed]
- Ivanova, N.V.; deWaard, J.; Hebert, P.D.N. An inexpensive, automation-friendly protocol for recovering high-quality DNA. Mol. Ecol. Notes 2006, 6, 998–1002. [Google Scholar] [CrossRef]
- Zelenina, D.A.; Soshnina, V.A.; Sergeev, A.A. Phylogeography and mitochondrial polymorphism of Asian coho salmon. Mol. Biol. 2020, 54, 876–883. [Google Scholar] [CrossRef]
- Kearse, M.; Moir, R.; Wilson, A.; Stones-Havas, S.; Cheung, M.; Sturrock, S.; Buxton, S.; Cooper, A.; Markowitz, S.; Duran, C.; et al. Geneious Basic: An integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 2012, 28, 1647–1649. [Google Scholar] [CrossRef] [PubMed]
- Leigh, J.W.; Bryant, D. PopART: Full-feature software for haplotype network construction. Methods Ecol. Evol. 2015, 6, 1110–1116. [Google Scholar] [CrossRef]
- Excoffier, L.; Lischer, H.E.L. Arlequin suite ver 3.5: A new series of programs to perform population genetics analyses under Linux and Windows. Mol. Ecol. Res. 2010, 10, 564–567. [Google Scholar] [CrossRef]
- Raymond, M.; Rousset, F. An Exact Test for Population Differentiation. Evolution 1995, 49, 1280–1283. [Google Scholar] [CrossRef]
- Zelenina, D.A.; (Russian Federal Research Institute of Fisheries and Oceanography, Moscow 105187, Russia); Soshnina, V.A.; (Russian Federal Research Institute of Fisheries and Oceanography, Moscow 105187, Russia). Unpublished Work. 2025. [Google Scholar]
- Aspinwall, N. Genetic Analysis of North American Populations of the Pink Salmon, Oncorhynchus gorbuscha, Possible Evidence for the Neutral Mutation-Random Drift Hypothesis. Evolution 1974, 28, 295–305. [Google Scholar] [CrossRef]
- Shevlyakov, E.А.; Dederer, N.A. Dynamics of abundance and intrapopulation structure of pink salmon at the western and northeastern coasts of Kamchatka. Izv. Tikhookean. Nauchno-Issled. Inst. Rybn. Khoz. Okeanogr. 2022, 202, 369–389. [Google Scholar] [CrossRef]
- Kaev, A.M. Some Results of Studies on Pink Salmon Oncorhynchus gorbuscha’s Number Dynamics on the Northeastern Coasts of Sakhalin Island. J. Ichthyol. 2019, 59, 885–894. [Google Scholar] [CrossRef]
- Kwain, W.; Lawrie, A.H. Pink salmon in the Great Lakes. Fisheries 1981, 6, 2–6. [Google Scholar] [CrossRef]
- Gharrett, A.J.; Thomason, M.A. Genetic changes in pink salmon (Oncorhynchus gorbuscha) following their introduction into the Great Lakes. Can. J. Fish. Aquat. Sci. 1987, 44, 787–792. [Google Scholar] [CrossRef]
- Sparks, M.M.; Schraidt, C.E.; Yin, X.; Seeb, L.W.; Christie, M.R. Rapid genetic adaptation to a novel ecosystem despite a large founder event. Mol. Ecol. 2024, 33, e17121. [Google Scholar] [CrossRef]
- Bagdovitz, M.S.; Taylor, W.W.; Wagner, W.C.; Nicolette, J.P.; Spangler, G.R. Pink salmon populations in the U.S. waters of Lake Superior, 1981–1984. J. Great Lakes Res. 1986, 12, 72–81. [Google Scholar] [CrossRef]
- Anas, R.E. Three-year-old Pink Salmon. J. Fish. Res. Board Can. 1959, 16, 91–94. [Google Scholar] [CrossRef]
- Turner, C.E.; Bilton, H.T. Another Pink Salmon (Oncorhynchus gorbuscha) in its Third Year. J. Fish. Res. Board Can. 1968, 25, 1993–1996. [Google Scholar] [CrossRef]
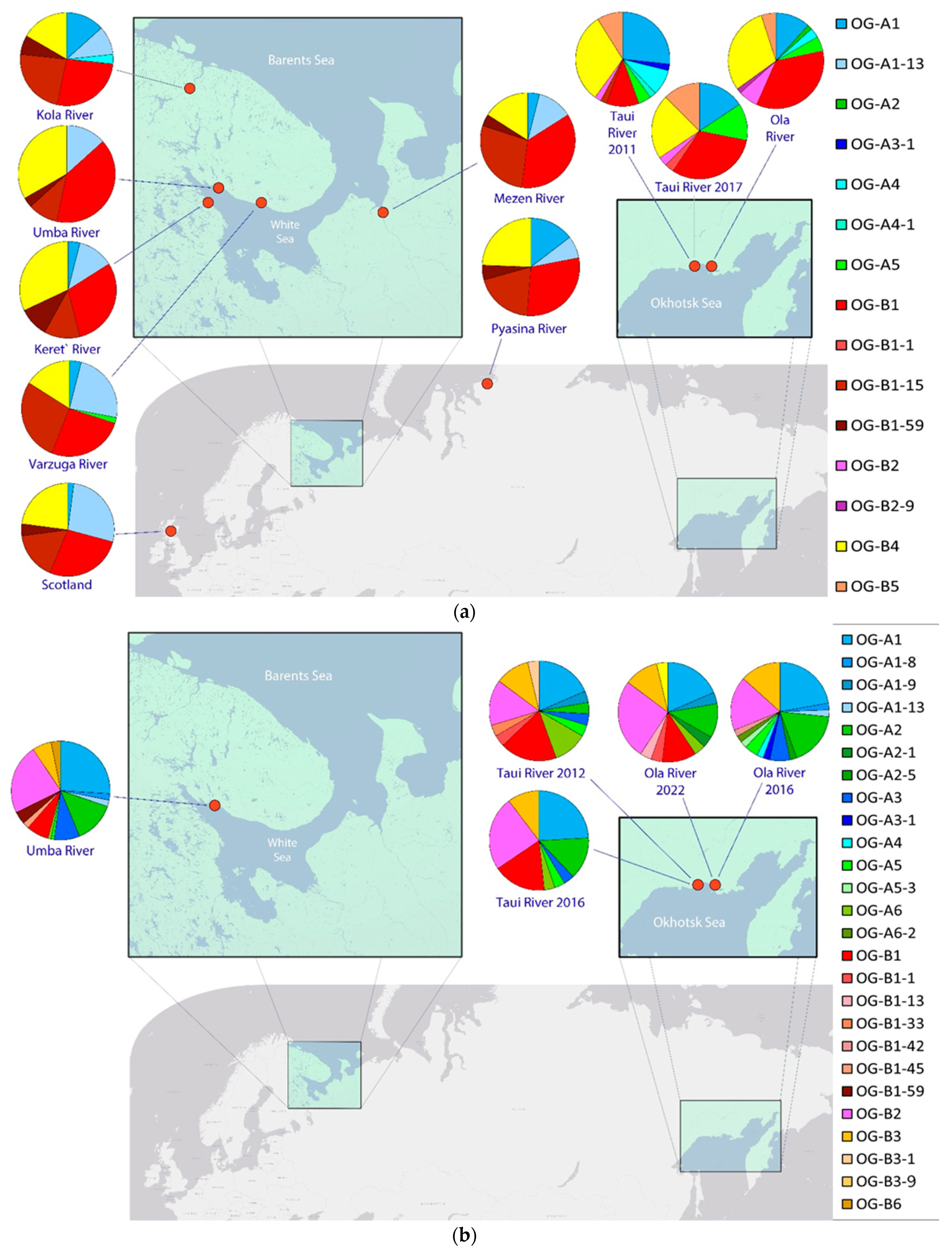

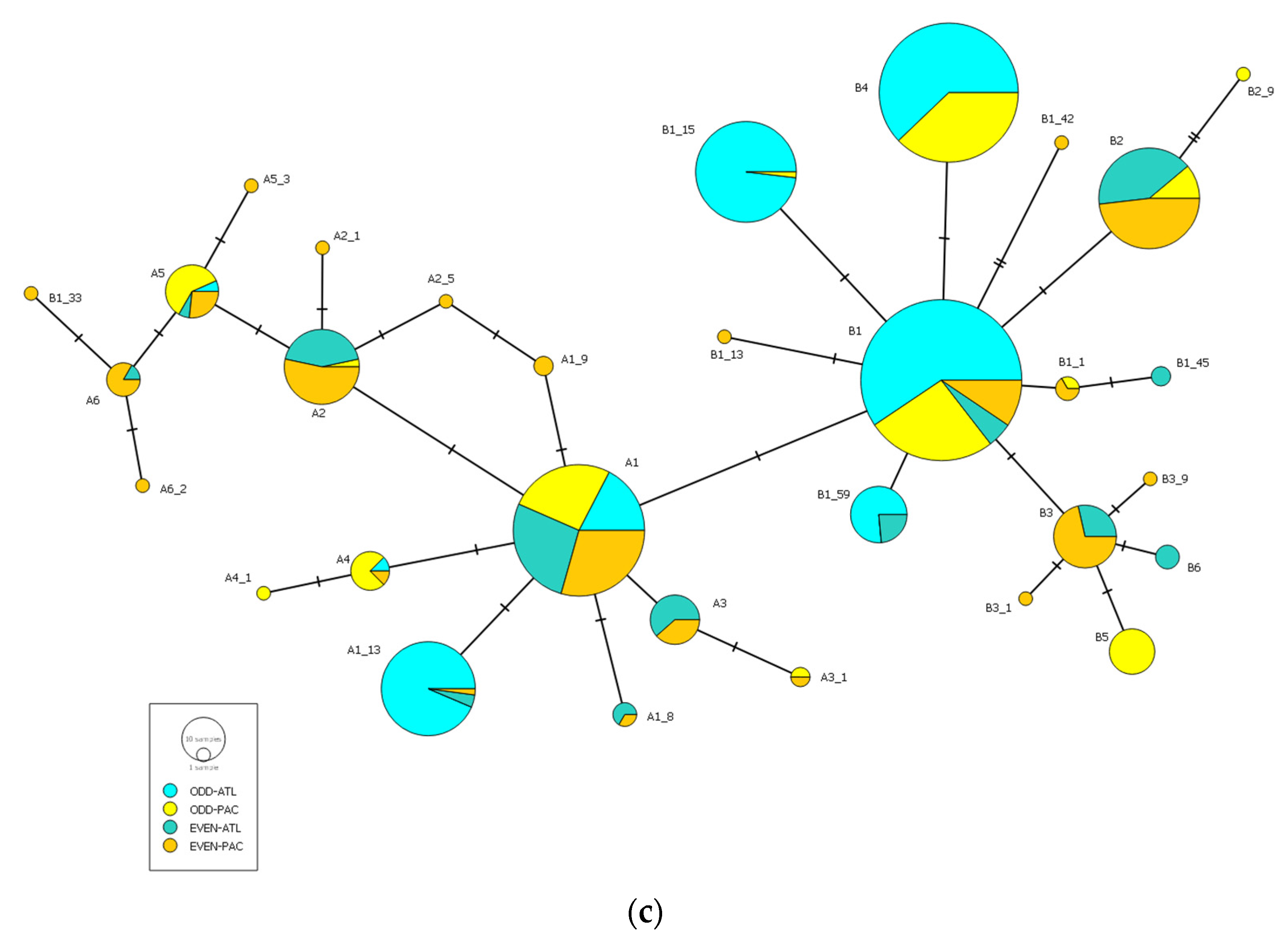

| Sea Basin | Sampling Location | Odd-Year Spawning Lineage | Even-Year Spawning Lineage | ||||
|---|---|---|---|---|---|---|---|
| Abbr. | Date of Collection | Sample Size | Abbr. | Date of Collection | Sample Size | ||
| North Sea | Scotland | SCOT17 * | Summer 2017 | 48 | |||
| Barents Sea | Kola River | KOLA19 | July 2019 | 30 | |||
| White Sea | Varzuga River | VARZ23 | July 2023 | 50 | |||
| Umba River | UMBA21 | Summer 2021 | 30 | UMBA18 | August–September 2018 | 96 | |
| Keret River | KER17 * | Summer 2017 | 50 | ||||
| Mezen River | MEZ23 | August 2023 | 25 | ||||
| Kara Sea | Pyasina River | PYAS17 | Summer 2017 | 41 | |||
| Okhotsk Sea | Ola River | OLA09 * | July 2009 | 60 | OLA16 | July 2016 | 45 |
| OLA22 | July–August 2022 | 27 | |||||
| Taui River | TAUI11 * | July 2011 | 45 | TAUI12 | July 2012 | 27 | |
| TAUI17 | July 2017 | 32 | TAUI16 | July 2016 | 29 | ||
| Odd-Lineage Samples | |||||||
| Sample | Ocean Basin | N | H | S | π × 100 | h | D |
| SCOT17 | A | 48 | 6 | 6 | 0.1604 | 0.7872 | 0.5336 |
| KOLA19 | A | 30 | 7 | 7 | 0.1545 | 0.8414 | −0.3253 |
| VARZ23 | A | 50 | 6 | 6 | 0.1539 | 0.7845 | 0.4314 |
| UMBA21 | A | 30 | 5 | 6 | 0.1235 | 0.7241 | −0.4843 |
| KER17 | A | 50 | 6 | 6 | 0.149 | 0.7829 | 0.3368 |
| MEZ23 | A | 25 | 6 | 6 | 0.1336 | 0.78 | −0.4311 |
| PYAS17 | A | 41 | 6 | 6 | 0.1356 | 0.8073 | −0.0414 |
| OLA09 | P | 60 | 9 | 10 | 0.145 | 0.7757 | −0.8629 |
| TAUI11 | P | 45 | 10 | 12 | 0.184 | 0.8182 | −0.9571 |
| TAUI17 | P | 32 | 7 | 8 | 0.1767 | 0.8226 | −0.2834 |
| ODD ATLANTIC/ARCTIC | A | 274 | 8 | 9 | 0.1471 | 0.7899 | 0.0656 |
| ODD PACIFIC | P | 137 | 13 | 15 | 0.1664 | 0.8102 | −1.0098 |
| ALL ODD | 411 | 15 | 18 | 0.1576 | 0.8157 | −1.0118 | |
| Even-Lineage Samples | |||||||
| Sample | Ocean Basin | N | H | S | π × 100 | h | D |
| UMBA18 | A | 96 | 13 | 12 | 0.1846 | 0.8498 | −0.5223 |
| OLA16 | P | 45 | 14 | 16 | 0.2147 | 0.8788 | −1.2687 |
| OLA22 | P | 27 | 11 | 11 | 0.1959 | 0.886 | −0.9923 |
| TAUI12 | P | 27 | 12 | 10 | 0.2228 | 0.9088 | −0.4084 |
| TAUI16 | P | 29 | 8 | 7 | 0.1669 | 0.8498 | −0.1386 |
| EVEN ATLANTIC/ARCTIC | A | 96 | 13 | 12 | 0.1846 | 0.8498 | −0.5223 |
| EVEN PACIFIC | P | 128 | 23 | 21 | 0.2017 | 0.8761 | −1.3291 |
| ALL EVEN | 224 | 26 | 23 | 0.1942 | 0.8646 | −1.3118 | |
| ALL | 635 | 31 | 29 | 0.1817 | 0.8812 | −1.4119 | |
| Lineage | Even-Year Spawning Lineage | Odd-Year Spawning Lineage | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Ocean Basin * | A | P | P | P | P | A | A | A | A | A | A | A | P | P | P | ||
| Haplotype | GenBank Acc. Number (1018 bp) | GenBank Acc. Number (1141 bp) | UMBA18 | OLA16 | OLA22 | TAUI12 | TAUI16 | SCOT17 | KOLA19 | VARZ23 | UMBA21 | KER17 | MEZ23 | PYAS17 | TAUI11 | TAUI17 | OLA09 |
| A1 | MT328254 | PQ817431 | 25 | 10 | 5 | 5 | 7 | 1 | 4 | 2 | - | 2 | 1 | 6 | 12 | 5 | 7 |
| A1-8 | MT328262 | PQ817432 | 2 | 1 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| A1-9 | MT328263 | PQ817433 | - | - | 1 | 1 | - | - | - | - | - | - | - | - | - | - | - |
| A1-13 | MT328267 | PQ817434 | 2 | 1 | - | - | - | 13 | 3 | 12 | 4 | 6 | 3 | 3 | - | - | - |
| A2 | MT328278 | PQ817435 | 13 | 8 | 3 | 1 | 4 | - | - | - | - | - | - | - | - | - | 1 |
| A2-1 | MT328279 | PQ817436 | - | - | 1 | - | - | - | - | - | - | - | - | - | - | - | - |
| A2-5 | MT328283 | - | - | 1 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| A3 | MT328285 | PQ817437 | 8 | 3 | - | 1 | 1 | - | - | - | - | - | - | - | - | - | - |
| A3-1 | MT328286 | PQ817438 | - | 1 | - | - | - | - | - | - | - | - | - | - | 1 | - | - |
| A4 | MT328290 | PQ817439 | - | 1 | - | - | - | - | 1 | - | - | - | - | - | 4 | - | 2 |
| A4-1 | MT328291 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1 | - | - |
| A5 | MT328292 | PQ817440 | 1 | 2 | - | 1 | 1 | - | - | 1 | - | - | - | - | 2 | 4 | 3 |
| A5-3 | MT328295 | - | - | 1 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| A6 | MT328297 | PQ817441 | 1 | - | 1 | 3 | 1 | - | - | - | - | - | - | - | - | - | - |
| A6-2 | MT328299 | - | - | 1 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| B1 | MT328300 | PQ817442 | 7 | - | 3 | 5 | 5 | 13 | 8 | 13 | 12 | 15 | 9 | 12 | 5 | 10 | 21 |
| B1-1 | MT328301 | PQ817443 | - | - | 1 | 1 | - | - | - | - | - | - | - | - | - | 1 | - |
| B1-13 | MT328313 | PQ817445 | - | - | 1 | - | - | - | - | - | - | - | - | - | - | - | - |
| B1-15 | MT328315 | PQ817446 | - | - | - | - | - | 8 | 7 | 14 | 3 | 6 | 7 | 8 | 1 | - | - |
| B1-33 | MT328333 | - | - | - | - | 1 | - | - | - | - | - | - | - | - | - | - | - |
| B1-42 | MT328342 | - | - | 1 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| B1-45 | MT328345 | PQ817447 | 2 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| B1-59 | MT328359 | PQ817448 | 4 | - | - | - | - | 2 | 2 | - | 1 | 5 | 1 | 2 | - | - | - |
| B2 | MT328360 | PQ817449 | 22 | 8 | 7 | 4 | 7 | - | - | - | - | - | - | - | 1 | 1 | 4 |
| B2-9 | MT328369 | PQ817450 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1 |
| B3 | MT328373 | PQ817451 | 6 | 6 | 3 | 3 | 3 | - | - | - | - | - | - | - | - | - | - |
| B3-1 | MT328374 | PQ817452 | - | - | - | 1 | - | - | - | - | - | - | - | - | - | - | - |
| B3-9 | - | PQ817453 | - | - | 1 | - | - | - | - | - | - | - | - | - | - | - | - |
| B4 | MT328382 | PQ817454 | - | - | - | - | - | 11 | 5 | 8 | 10 | 16 | 4 | 10 | 14 | 7 | 18 |
| B5 | MT328389 | PQ817455 | - | - | - | - | - | - | - | - | - | - | - | - | 4 | 4 | 3 |
| B6 | MT328390 | PQ817456 | 3 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| Lineage | Odd-Year Spawning Lineage | Even-Year Spawning Lineage | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Ocean Basin * | P | P | P | A | A | A | A | A | A | A | P | P | P | P | A |
| TAUI11 | TAUI17 | OLA09 | SCOT17 | KOLA19 | VARZ23 | UMBA21 | KER17 | MEZ23 | PYAS17 | TAUI12 | TAUI16 | OLA16 | OLA22 | UMBA18 | |
| TAUI11 | NS | * | *** | *** | *** | *** | *** | *** | *** | *** | *** | *** | *** | *** | |
| TAUI17 | 0.009 | NS | *** | *** | *** | *** | *** | *** | *** | *** | *** | *** | *** | *** | |
| OLA09 | 0.021 | 0.000 | *** | *** | *** | *** | *** | *** | *** | *** | *** | *** | *** | *** | |
| SCOT17 | 0.066 * | 0.069 * | 0.065 * | NS | NS | NS | NS | NS | NS | *** | *** | *** | *** | *** | |
| KOLA19 | 0.051 * | 0.049 * | 0.048 * | 0.000 | NS | NS | NS | NS | NS | *** | *** | *** | *** | *** | |
| VARZ23 | 0.087 * | 0.084 * | 0.091 * | 0.000 | 0.000 | NS | * | NS | NS | *** | *** | *** | *** | *** | |
| UMBA21 | 0.064 * | 0.049 * | 0.016 | 0.013 | 0.014 | 0.043 | NS | NS | NS | *** | *** | *** | *** | *** | |
| KER17 | 0.068 * | 0.056 * | 0.03 * | 0.016 | 0.007 | 0.047 * | 0.000 | NS | NS | *** | *** | *** | *** | *** | |
| MEZ23 | 0.091 * | 0.07 * | 0.064 * | 0.007 | 0.000 | 0.000 | 0.012 | 0.01 | NS | *** | *** | *** | *** | *** | |
| PYAS17 | 0.053 * | 0.045 * | 0.029 * | 0.007 | 0.000 | 0.014 | 0.000 | 0.000 | 0.000 | *** | *** | *** | *** | *** | |
| TAUI12 | 0.072 * | 0.036 * | 0.097 * | 0.127 * | 0.096 * | 0.13 * | 0.157 * | 0.147 * | 0.131 * | 0.124 * | NS | * | NS | * | |
| TAUI16 | 0.075 * | 0.056 * | 0.102 * | 0.137 * | 0.113 * | 0.139 * | 0.183 * | 0.17 * | 0.158 * | 0.141 * | 0.000 | NS | NS | NS | |
| OLA16 | 0.095 * | 0.098 * | 0.16 * | 0.173 * | 0.154 * | 0.172 * | 0.223 * | 0.219 * | 0.197 * | 0.187 * | 0.011 | 0.000 | NS | NS | |
| OLA22 | 0.073 * | 0.043 * | 0.084 * | 0.124 * | 0.096 * | 0.127 * | 0.153 * | 0.147 * | 0.128 * | 0.121 * | 0.000 | 0.000 | 0.021 | NS | |
| UMBA18 | 0.084 * | 0.079 * | 0.117 * | 0.136 * | 0.114 * | 0.139 * | 0.175 * | 0.169 * | 0.153 * | 0.139 * | 0.009 | 0.000 | 0.005 | 0.000 | |
| (a) Among all samples (4 groups: EVEN-A, EVEN-P, ODD-A, ODD-P). | ||||
| Source of variation | d.f. | Sum of squares | Variance composition (%) | Fixation indexes |
| Among groups | 3 | 47.221 | 0.09824 (10.27%) | FCT: 0.10274 *** |
| Among populations within groups | 11 | 11.233 | 0.00444 (0.46%) | FSC: 0.00518 ns |
| Within populations | 620 | 529.163 | 0.85349 (99.26%) | FST: 0.10739 *** |
| Total | 634 | 587.617 | 0.95618 | |
| (b) Among odd-year spawning populations and groups (2 groups: ODD-A, ODD-P). | ||||
| Source of variation | d.f. | Sum of squares | Variance composition | Fixation indexes |
| Among groups | 1 | 9.222 | 0.04471 (5.39%) | FCT: 0.05394 ** |
| Among populations within groups | 8 | 8.141 | 0.00597 (0.72%) | FSC: 0.00761 ns |
| Within populations | 401 | 312.115 | 0.77834 (93.89%) | FST: 0.06113 *** |
| Total | 410 | 329.478 | 0.82902 | |
| (c) Among even-year spawning populations and groups (2 groups: EVEN-A, EVEN-P). | ||||
| Source of variation | d.f. | Sum of squares | Variance composition | Fixation indexes |
| Among groups | 1 | 0.829 | −0.00228 (−0.13%) | FCT: −0.00230 ns |
| Among populations within groups | 3 | 3.092 | 0.00126 (0.13%) | FSC: 0.00127 ns |
| Within populations | 219 | 217.048 | 0.99109 (100.10%) | FST: −0.00103 ns |
| Total | 223 | 220.968 | 0.99007 | |
| (d) Among Atlantic populations and groups (2 groups: ATL-ODD, ATL-EVEN). | ||||
| Source of variation | d.f. | Sum of squares | Variance composition | Fixation indexes |
| Among groups | 1 | 22.415 | 0.15004 (15.78%) | FCT: 0.15776 ns |
| Among populations within groups | 6 | 5.593 | 0.00348 (0.37%) | FSC: 0.00434 ns |
| Within populations | 362 | 288.704 | 0.79752 (83.86%) | FST: 0.16142 *** |
| Total | 369 | 316.712 | 0.95104 | |
| (e) Among Pacific populations and groups (2 groups: PAC-ODD, PAC-EVEN). | ||||
| Source of variation | d.f. | Sum of squares | Variance composition | Fixation indexes |
| Among groups | 1 | 12.94 | 0.08907 (8.68%) | FCT: 0.08678 * |
| Among populations within groups | 5 | 5.64 | 0.00536 (0.52%) | FSC: 0.00572 ns |
| Within populations | 258 | 240.459 | 0.93201 (90.80%) | FST: 0.09200 *** |
| Total | 264 | 259.038 | 1.02644 | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zelenina, D.A.; Soshnina, V.A.; Gordeev, I.I.; Alekseev, M.Y.; Zadelenov, V.A.; Mugue, N.S. The Splendour and Misery of European Pink Salmon, Oncorhynchus gorbuscha: Abundant Odd Lineage vs. Depressed Even Lineage—Insights from cytb Gene Analysis. Diversity 2025, 17, 563. https://doi.org/10.3390/d17080563
Zelenina DA, Soshnina VA, Gordeev II, Alekseev MY, Zadelenov VA, Mugue NS. The Splendour and Misery of European Pink Salmon, Oncorhynchus gorbuscha: Abundant Odd Lineage vs. Depressed Even Lineage—Insights from cytb Gene Analysis. Diversity. 2025; 17(8):563. https://doi.org/10.3390/d17080563
Chicago/Turabian StyleZelenina, Daria A., Valeria A. Soshnina, Ilya I. Gordeev, Maksim Yu. Alekseev, Vladimir A. Zadelenov, and Nikolai S. Mugue. 2025. "The Splendour and Misery of European Pink Salmon, Oncorhynchus gorbuscha: Abundant Odd Lineage vs. Depressed Even Lineage—Insights from cytb Gene Analysis" Diversity 17, no. 8: 563. https://doi.org/10.3390/d17080563
APA StyleZelenina, D. A., Soshnina, V. A., Gordeev, I. I., Alekseev, M. Y., Zadelenov, V. A., & Mugue, N. S. (2025). The Splendour and Misery of European Pink Salmon, Oncorhynchus gorbuscha: Abundant Odd Lineage vs. Depressed Even Lineage—Insights from cytb Gene Analysis. Diversity, 17(8), 563. https://doi.org/10.3390/d17080563








