Description of a New Species of Hainania Koller (Teleostei, Cypriniformes, Xenocyprididae) from Guangdong Province, Southern China †
Abstract
1. Introduction
2. Materials and Methods
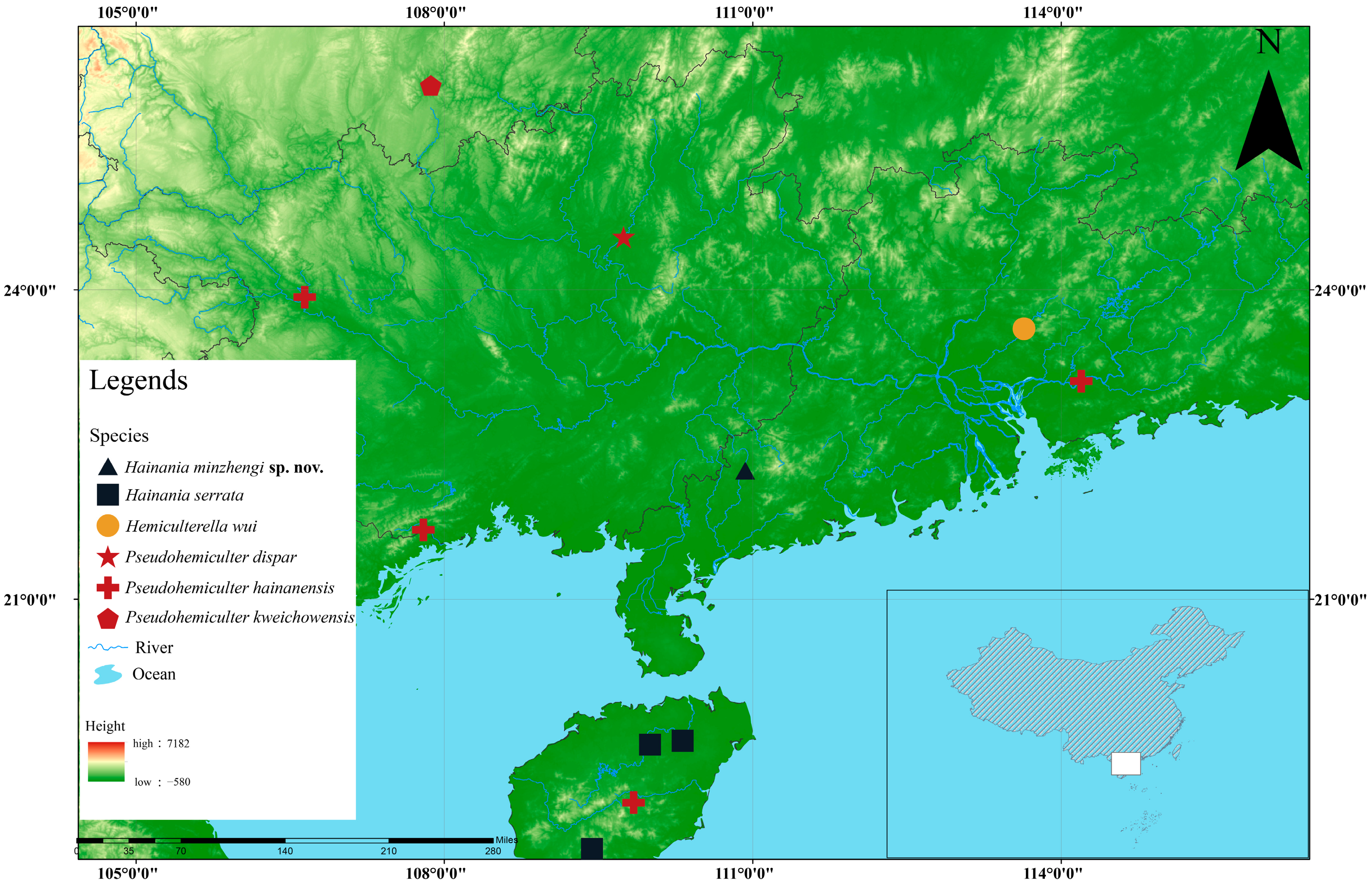
2.1. Sample Collection and Taxon Sampling
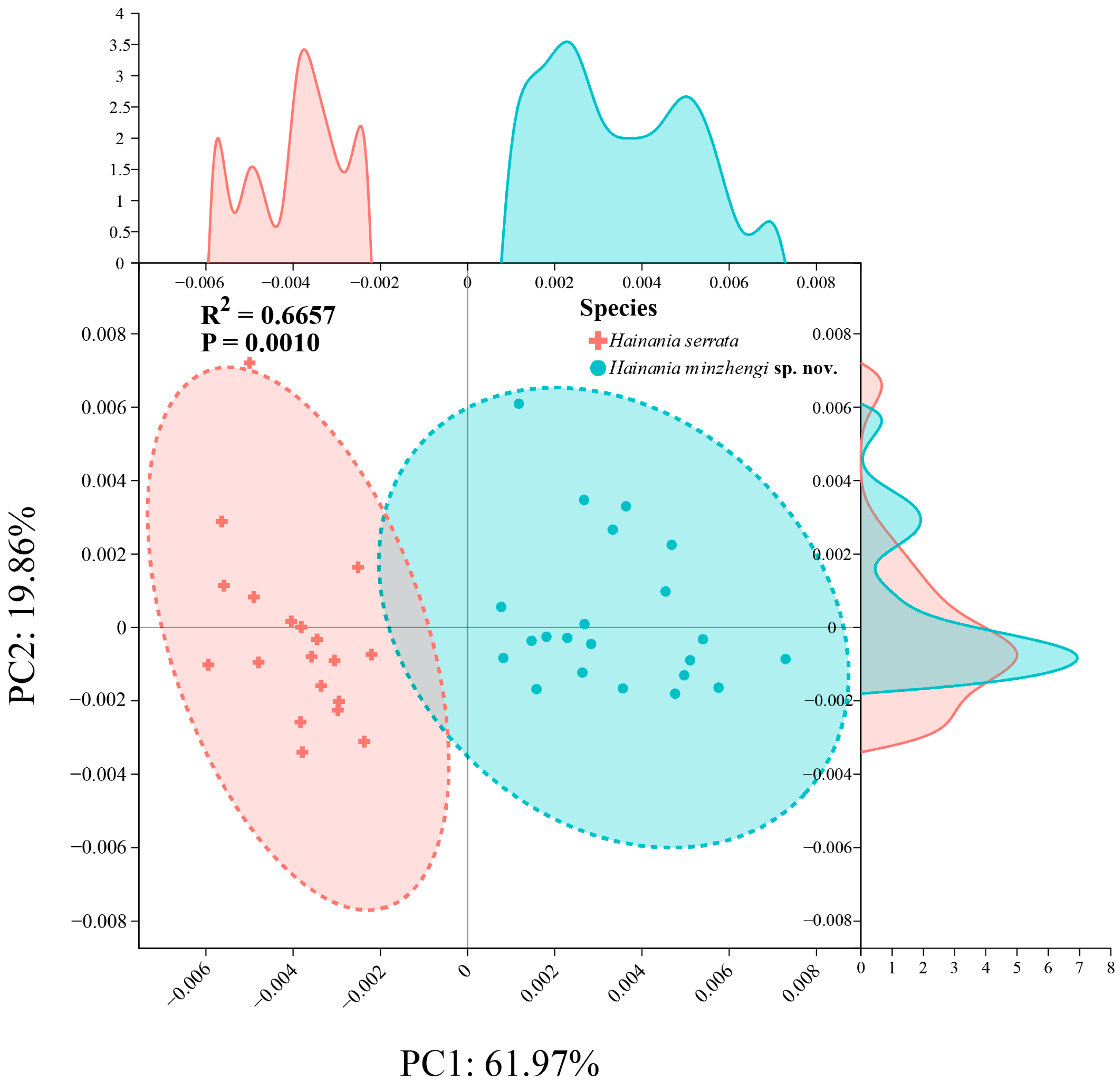
2.2. Morphometric Data Acquiring
2.3. DNA Extraction, Amplification, Sequencing, and Phylogenetic Analysis
3. Results
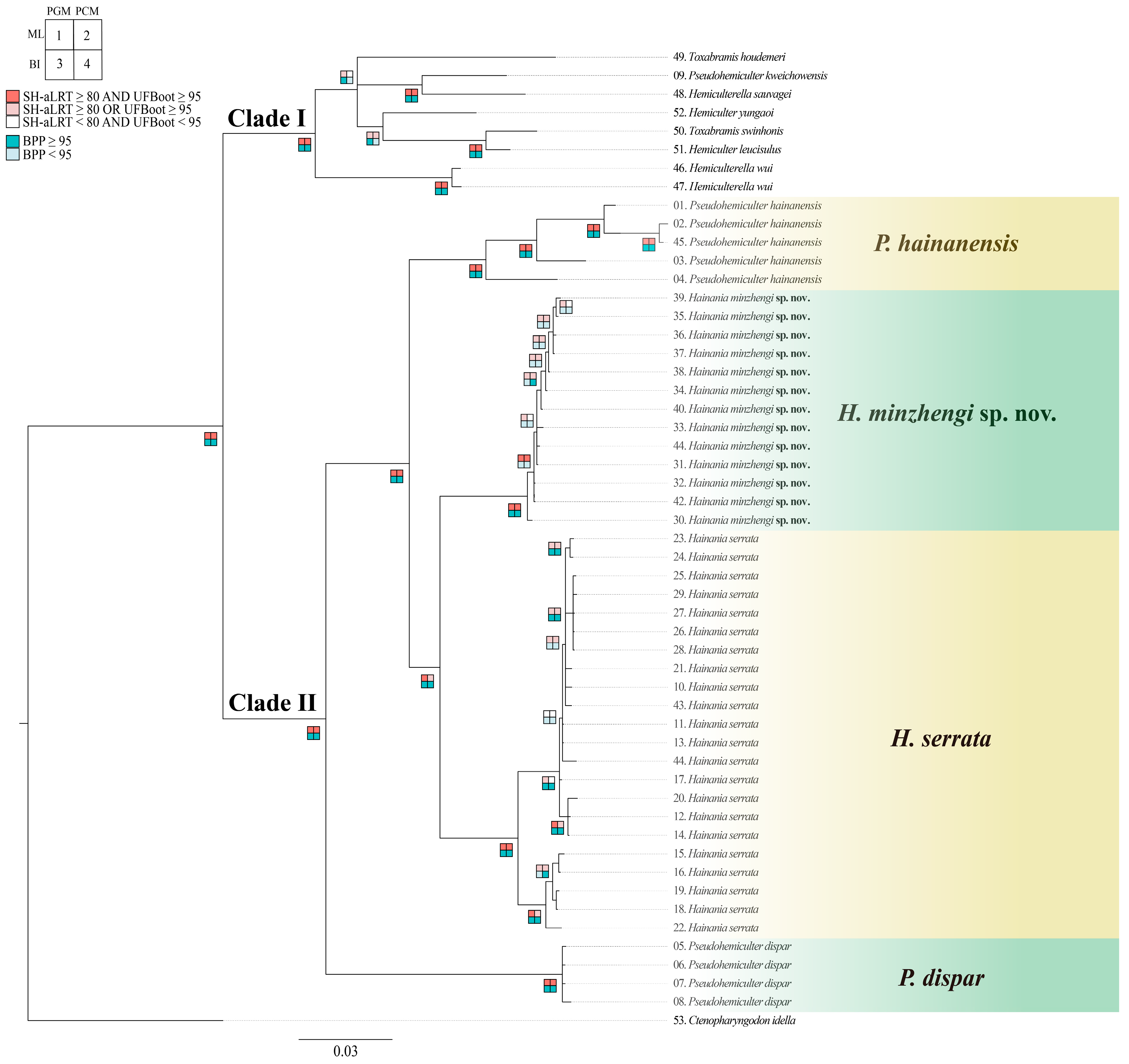
3.1. Phylogenetic Analysis
3.2. Systematics
3.3. Key to Chinese Species of Hainania and Pseudohemiculter
| Hainania |
| Pseudohemiculter |
| Ha. mingzhengi sp. nov. |
| Ha. serrata |
| P. kweichowensis |
| 4 |
| P. dispar |
| P. hainanensis |
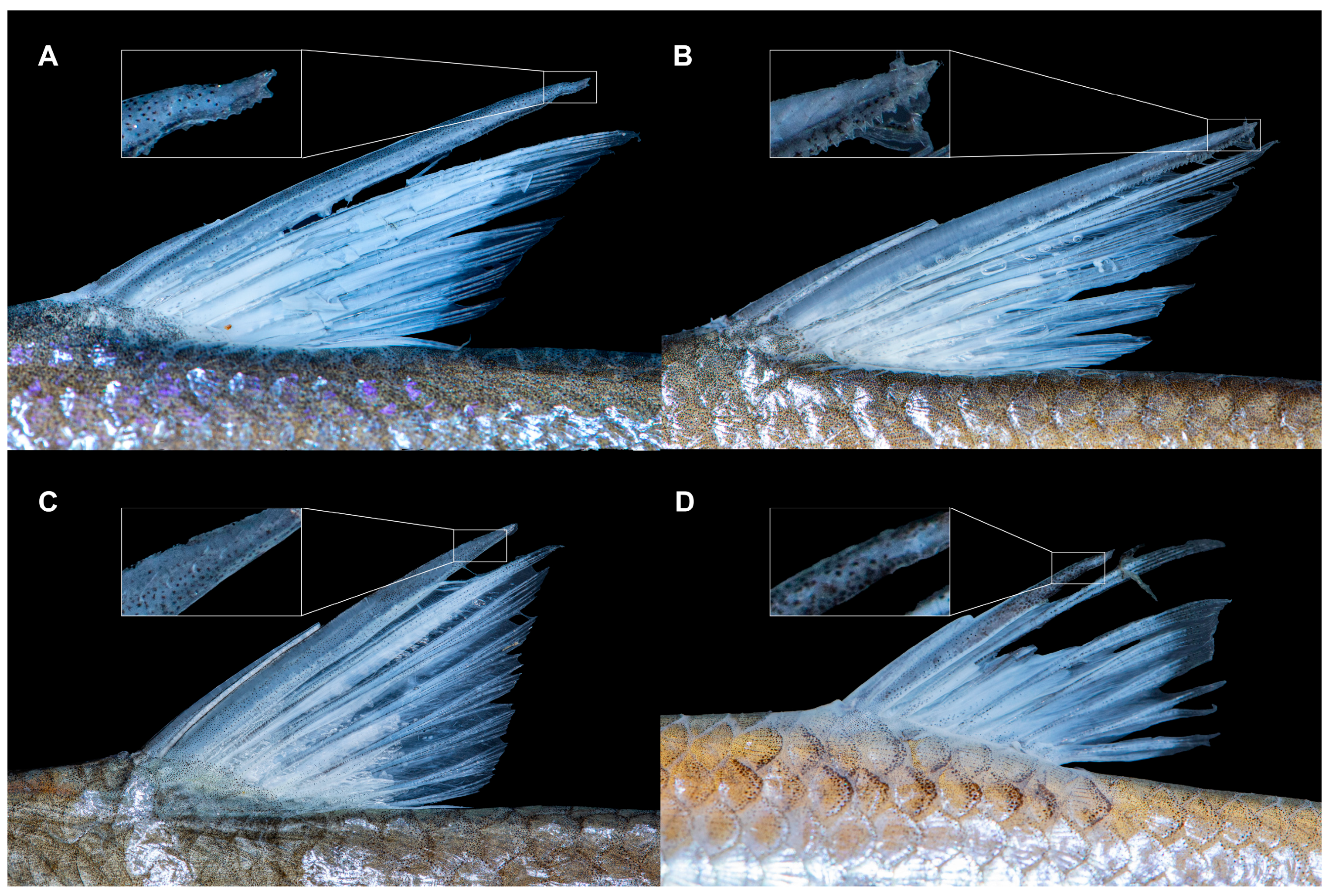
3.4. Taxonomic Description
4. Discussion
5. Comparative Material
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Koller, O. Fische von der Insel Hai-Nan. Ann. Naturhist. Mus. Wien 1927, 41, 25–49. (In German) [Google Scholar]
- Nichols, J.T.; Pope, C.H. The Fishes of Hainan. Bulletin of the AMNH; v. 54, Article 2; American Museum of Natural History Library: New York, NY, USA, 1927; pp. 321–394, Pl. 26. [Google Scholar]
- Myers, G.S. On the fishes described by Koller from Hainan in 1926 and 1927. Lingnan Sci. J. 1931, 10, 255–262. [Google Scholar]
- Bănărescu, P.M. Further studies on the systematics of Cultrinae with reidentification of 44 type specimens (Pisces, Cyprinidae). Rev. Roum. Biol. Série Zool. 1971, 16, 9–20. [Google Scholar]
- Wu, X.W. The Cyprinid Fishes of China; Shanghai Science and Tecnology Press: Shanghai, China, 1964; Volume 1. [Google Scholar]
- Chen, Y.Y. (Ed.) Fauna Sinica. Osteichthyes. Cypriniformes II; Science Press: Beijing, China, 1998. [Google Scholar]
- Yue, P.Q.; Chen, Y.Y. [Chief Compilers] China Red Data Book of Endangered Animals; Pisces; Science Press: Beijing, China; Hong Kong, China; New York, NY, USA, 1998; (In Chinese and English). [Google Scholar]
- Dai, Y.; Yang, J. Phylogeny and Zoogeography of the Cyprinid Hemicultrine Group (Cyprinidae: Cultrinae). Zool. Stud. 2003, 42, 73–92. [Google Scholar]
- Kottelat, M. The fishes of the inland waters of southeast Asia: A catalogue and core bibliography of the fishes known to occur in freshwaters, mangroves and estuaries. Raffles Bull. Zool. 2013, 27, 1–663. [Google Scholar]
- Tang, K.L.; Agnew, M.K.; Hirt, M.V.; Lumbantobing, D.N.; Sado, T.; Teoh, V.H.; Yang, L.; Bart, H.L.; Harris, P.M.; He, S.; et al. Limits and phylogenetic relationships of East Asian fishes in the subfamily Oxygastrinae (Teleostei: Cypriniformes: Cyprinidae). Zootaxa 2013, 3681, 101–135. [Google Scholar] [CrossRef]
- Cheng, P.; Yu, D.; Tang, Q.; Yang, J.; Chen, Y.; Liu, H. Macro-evolutionary patterns of East Asian opsariichthyin-xenocyprinin-cultrin fishes related to the formation of river and river-lake environments under monsoon climate. Water Biol. Secur. 2022, 1, 100036. [Google Scholar] [CrossRef]
- Deng, Q.; Li, M.; Yu, D.; Chen, L.; Li, W.; Cai, X.; Liu, H. Molecular phylogenetic analysis of the East Asian hemicultrine fishes (Teleostei: Cyprinidae: Xenocypridinae), with suggestions to their generic classification and redescription of the recently described species Hemiculter yungaoi Vasil’eva et al. 2022. J. Fish Biol. 2024, 105, 239–253. [Google Scholar] [CrossRef] [PubMed]
- Pan, J.H.; Zhong, L.; Zheng, C.Y.; Wu, H.L.; Liu, J.H. (Eds.) The Freshwater Fishes of Guangdong Province. Guangdong Science and Technology Press: Guangzhou, China, 1991. (In Chinese) [Google Scholar]
- Mai, D.Y. Identification of the Fresh-Water Fishes of North Viet Nam; Ha Noi, Scientific & Technology, Publisher: Hanoi, Vietnam, 1978. (In Vietnamese) [Google Scholar]
- Hammer, Ø.; Harper, D.A.; Ryan, P.D. PAST: Paleontological statistical software package for education and data analysis. Palaeontol. Electron. 2001, 4, 1–9. [Google Scholar]
- Katoh, K.; Standley, D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef]
- Larsson, A. AliView: A fast and lightweight alignment viewer and editor for large datasets. Bioinformatics 2014, 30, 3276–3278. [Google Scholar] [CrossRef]
- Brown, J.W.; Walker, J.F.; Smith, S.A. Phyx: Phylogenetic tools for unix. Bioinformatics 2017, 33, 1886–1888. [Google Scholar] [CrossRef]
- Nguyen, L.T.; Schmidt, H.A.; von Haeseler, A.; Minh, B.Q. IQ-TREE: A fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 2015, 32, 268–274. [Google Scholar] [CrossRef]
- Kalyaanamoorthy, S.; Minh, B.Q.; Wong, T.K.F.; von Haeseler, A.; Jermiin, L.S. ModelFinder: Fast model selection for accurate phylogenetic estimates. Nat. Methods 2017, 14, 587–589. [Google Scholar] [CrossRef]
- Minh, B.Q.; Nguyen, M.A.; von Haeseler, A. Ultrafast approximation for phylogenetic bootstrap. Mol. Biol. Evol. 2013, 30, 1188–1195. [Google Scholar] [CrossRef]
- Hoang, D.T.; Chernomor, O.; von Haeseler, A.; Minh, B.Q.; Vinh, L.S. UFBoot2: Improving the Ultrafast Bootstrap Approximation. Mol. Biol. Evol. 2018, 35, 518–522. [Google Scholar] [CrossRef]
- Guindon, S.; Dufayard, J.F.; Lefort, V.; Anisimova, M.; Hordijk, W.; Gascuel, O. New algorithms and methods to estimate maximum-likelihood phylogenies: Assessing the performance of PhyML 3.0. Syst. Biol. 2010, 59, 307–321. [Google Scholar] [CrossRef]
- Ronquist, F.; Teslenko, M.; van der Mark, P.; Ayres, D.L.; Darling, A.; Höhna, S.; Larget, B.; Liu, L.; Suchard, M.A.; Huelsenbeck, J.P. MrBayes 3.2: Efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 2012, 61, 539–542. [Google Scholar] [CrossRef] [PubMed]
- Lanfear, R.; Frandsen, P.B.; Wright, A.M.; Senfeld, T.; Calcott, B. PartitionFinder 2: New Methods for Selecting Partitioned Models of Evolution for Molecular and Morphological Phylogenetic Analyses. Mol. Biol. Evol. 2017, 34, 772–773. [Google Scholar] [CrossRef] [PubMed]
- Rambaut, A. Figtree Ver 1.4.4. Institute of Evolutionary Biology; University of Edinburgh: Edinburgh, UK, 2018. [Google Scholar]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular Evolutionary Genetics Analysis Version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef] [PubMed]
- Kimura, M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 1980, 16, 111–120. [Google Scholar] [CrossRef]
- Zhang, R.; Zhu, T.; Yu, F. The New Mitochondrial Genome of Hemiculterella wui (Cypriniformes, Xenocyprididae): Sequence, Structure, and Phylogenetic Analyses. Genes 2023, 14, 2110. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.L.; Hsu, K.C.; Luo, J.; Wang, C.; Chan, B.P.; Li, J.; Kuo, P.; Lin, H. Genetic diversity and population history of Tanichthys albonubes (Teleostei: Cyprinidae): Implications for conservation. Aquat. Conserv.-Mar. Freshw. Ecosyst. 2018, 28, 422–434. [Google Scholar] [CrossRef]
- Zhang, W.J.; Wang, J.J.; Li, C.; Chen, J.Q.; Li, W.; Jiang, S.Y.; Hsu, K.C.; Zhao, M.; Lin, H.D.; Zhao, J. Spatial genetic structure of Opsariichthys hainanensis in South China. Mitochondrial DNA Part A DNA Mapp. Seq. Anal. 2020, 31, 98–107. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Li, C.; Chen, J.; Wang, J.; Jin, J.; Jiang, S.; Yan, L.; Lin, H.D.; Zhao, J. Phylogeographic structure of the dwarf snakehead (Channa gachua) around Gulf of Tonkin: Historical biogeography and pronounced effects of sea-level changes. Ecol. Evol. 2021, 11, 12583–12595. [Google Scholar] [CrossRef] [PubMed]
- Zhou, M.; Xia, J.; Li, C. Divergence of the freshwater sleeper, Neodontobutis hainanensis (Chen, 1985) (Teleostei, Odontobutidae), in the Pearl River basin and on Hainan Island of southern China. ZooKeys 2024, 1197, 183–196. [Google Scholar] [CrossRef]
- Zhu, H. On the biogeographical origin of Hainan Island in China. Plant Sci. J. 2020, 38, 839–843. [Google Scholar] [CrossRef]





| Number | Species | Locality | Voucher ID | Cyt b | COI |
|---|---|---|---|---|---|
| 1 | Pseudohemiculter hainanensis | Boluo County, Huizhou City, Guangdong Province, China | IHBHZ202407001 | PV164763 | PV165721 |
| 2 | Pseudohemiculter hainanensis | Youjiang District, Baise City, Guangxi Zhuang Autonomous Region, China | IHBBS202301001 | PV164764 | PV165722 |
| 3 | Pseudohemiculter hainanensis | Qiongzhong Li and Miao Autonomous County, Hainan Province, China | IHBQZ202308001 | PV164765 | PV165723 |
| 4 | Pseudohemiculter hainanensis | Naliang Town, Fangcheng District, Fangchenggang City, Guangxi Zhuang Autonomous Region, China | IHBFCG202405001 | PV164766 | PV165724 |
| 5 | Pseudohemiculter dispar | Jiangkou Town, Luzhai County, Liuzhou City, Guangxi Zhuang Autonomous Region, China. | IHBLZ202410001 | PV164767 | PV165725 |
| 6 | Pseudohemiculter dispar | Jiangkou Town, Luzhai County, Liuzhou City, Guangxi Zhuang Autonomous Region, China. | IHBLZ202410002 | PV164768 | PV165726 |
| 7 | Pseudohemiculter dispar | Jiangkou Town, Luzhai County, Liuzhou City, Guangxi Zhuang Autonomous Region, China. | IHBLZ202410003 | PV164769 | PV165727 |
| 8 | Pseudohemiculter dispar | Jiangkou Town, Luzhai County, Liuzhou City, Guangxi Zhuang Autonomous Region, China. | IHBLZ202410004 | PV164770 | PV165728 |
| 9 | Pseudohemiculter kweichowensis | Sandu Shui Autonomous County, Qiannan Buyi and Miao Autonomous Prefecture, Guizhou Province, China | IHBQN202307001 | PV164771 | PV165729 |
| 10 | Hainania serrata | Dingan County, Hainan Province, China | IHBHK202308002 | PV164772 | PV165730 |
| 11 | Hainania serrata | Dingan County, Hainan Province, China | IHBHK202308004 | PV164773 | PV165731 |
| 12 | Hainania serrata | Dingan County, Hainan Province, China | IHBHK202308005 | PV164774 | PV165732 |
| 13 | Hainania serrata | Dingan County, Hainan Province, China | IHBHK202308006 | PV164775 | PV165733 |
| 14 | Hainania serrata | Dingan County, Hainan Province, China | IHBHK202308008 | PV164776 | PV165734 |
| 15 | Hainania serrata | Dingan County, Hainan Province, China | IHBHK202308010 | PV164777 | PV165735 |
| 16 | Hainania serrata | Dingan County, Hainan Province, China | IHBHK202308011 | PV164778 | PV165736 |
| 17 | Hainania serrata | Dingan County, Hainan Province, China | IHBHK202308013 | PV164779 | PV165737 |
| 18 | Hainania serrata | Dingan County, Hainan Province, China | IHBHK202308014 | PX024142 | PV991209 |
| 19 | Hainania serrata | Chengmai County, Hainan Province, China | IHBHK202308015 | PX024143 | PV991208 |
| 20 | Hainania serrata | Chengmai County, Hainan Province, China | IHBHK202308017 | PX024144 | PV991210 |
| 21 | Hainania serrata | Chengmai County, Hainan Province, China | IHBHK202308018 | PX024145 | PV991202 |
| 22 | Hainania serrata | Chengmai County, Hainan Province, China | IHBHK202308019 | PX024146 | PV991211 |
| 23 | Hainania serrata | Chengmai County, Hainan Province, China | IHBHK202308020 | PX024147 | PV991200 |
| 24 | Hainania serrata | Chengmai County, Hainan Province, China | IHBHK202308021 | PX024148 | PV991207 |
| 25 | Hainania serrata | Jiyang District, Sanya City, Hainan Province, China | IHBSY202505001 | PX024149 | PV991201 |
| 26 | Hainania serrata | Jiyang District, Sanya City, Hainan Province, China | IHBSY202505002 | PX024150 | PV991205 |
| 27 | Hainania serrata | Jiyang District, Sanya City, Hainan Province, China | IHBSY202505003 | PX024151 | PV991204 |
| 28 | Hainania serrata | Jiyang District, Sanya City, Hainan Province, China | IHBSY202505004 | PX024152 | PV991206 |
| 29 | Hainania serrata | Jiyang District, Sanya City, Hainan Province, China | IHBSY202505005 | PX024153 | PV991203 |
| 30 | Hainania minzhengi sp. nov. | Shuikou Town, Xinyi City, Maoming City, Guangdong Province, China | IHBMM202407002 | PV164780 | PV165738 |
| 31 | Hainania minzhengi sp. nov. | Shuikou Town, Xinyi City, Maoming City, Guangdong Province, China | IHBMM202407004 | PV164781 | PV165739 |
| 32 | Hainania minzhengi sp. nov. | Shuikou Town, Xinyi City, Maoming City, Guangdong Province, China | IHBMM202407005 | PX024154 | PV991196 |
| 33 | Hainania minzhengi sp. nov. | Shuikou Town, Xinyi City, Maoming City, Guangdong Province, China | IHBMM202407006 | PX024155 | PV991197 |
| 34 | Hainania minzhengi sp. nov. | Shuikou Town, Xinyi City, Maoming City, Guangdong Province, China | IHBMM202407007 | PX024156 | PV991198 |
| 35 | Hainania minzhengi sp. nov. | Shuikou Town, Xinyi City, Maoming City, Guangdong Province, China | IHBMM202407008 | PV164782 | PV165740 |
| 36 | Hainania minzhengi sp. nov. | Shuikou Town, Xinyi City, Maoming City, Guangdong Province, China | IHBMM202407009 | PX024157 | PV991199 |
| 37 | Hainania minzhengi sp. nov. | Shuikou Town, Xinyi City, Maoming City, Guangdong Province, China | IHBMM202407010 | PV164783 | PV165741 |
| 38 | Hainania minzhengi sp. nov. | Shuikou Town, Xinyi City, Maoming City, Guangdong Province, China | IHBMM202407011 | PX024158 | PV991192 |
| 39 | Hainania minzhengi sp. nov. | Shuikou Town, Xinyi City, Maoming City, Guangdong Province, China | IHBMM202407013 | PV164784 | PV165742 |
| 40 | Hainania minzhengi sp. nov. | Shuikou Town, Xinyi City, Maoming City, Guangdong Province, China | ASIZB 248490 | PX024159 | PV991193 |
| 41 | Hainania minzhengi sp. nov. | Shuikou Town, Xinyi City, Maoming City, Guangdong Province, China | ASIZB 248491 | PX024160 | PV991194 |
| 42 | Hainania minzhengi sp. nov. | Shuikou Town, Xinyi City, Maoming City, Guangdong Province, China | ASIZB 248492 | PX024161 | PV991195 |
| 43 | Hainania serrata | Qiongzhong Li and Miao Autonomous County, Hainan Province, China | / | OP251698.1 | OP251568.1 |
| 44 | Hainania serrata | Qiongzhong Li and Miao Autonomous County, Hainan Province, China | / | OP251699.1 | OP251569.1 |
| 45 | Pseudohemiculter hainanensis | Market of Laibin City, Guangxi Zhuang Autonomous Region, China | / | NC_065693.1 | NC_065693.1 |
| 46 | Hemiculterella wui | Conghua District, Guangzhou City, Guangdong Province, China | IHBGZ202408001 | PV164785 | PV165743 |
| 47 | Hemiculterella wui | / | / | NC_084163.1 | NC_084163.1 |
| 48 | Hemiculterella sauvagei | Xin Village, Cuiping Distrct, Yibin City, Sichuan Province, China | / | NC_026693.1 | NC_026693.1 |
| 49 | Toxabramis houdemeri | Fusui County, Chongzuo City, Guangxi Zhuang Autonomous Region, China | / | NC_029348.1 | NC_029348.1 |
| 50 | Toxabramis swinhonis | Dianshan Lake, Qingpu District, Shanghai City, China | / | NC_029249.1 | NC_029249.1 |
| 51 | Hemiculter leucisulus | / | / | NC_022929.1 | NC_022929.1 |
| 52 | Hemiculter yungaoi | Shapo Reservoir, Longhua District, Haikou City, Hainan Province, China | / | OP251707.1 | OP251579.1 |
| 53 | Ctenopharyngodon idella | / | / | NC_010288.1 | NC_010288.1 |
| P. hainanensis | P. dispar | P. kweichowensis | Ha. serrata | Ha. minzhengi sp. nov. | |
|---|---|---|---|---|---|
| P. hainanensis | 0.0544 | ||||
| P. dispar | 0.1023 | 0.0040 | |||
| P. kweichowensis | 0.1217 | 0.1249 | / | ||
| Ha. serrata | 0.0677 | 0.1042 | 0.1195 | 0.0185 | |
| Ha. minzhengi sp. nov. | 0.0673 | 0.0969 | 0.1206 | 0.0483 | 0.0046 |
| Characteristics | Hainania minzhengi (n = 19) | Hainania serrata (n = 22) | Pseudohemiculter dispar (n = 3) | Pseudohemiculter hainanensis (n = 3) | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Holotype | Range | Mean | S.D. | Range | Mean | S.D. | Range | Mean | S.D. | Range | Mean | S.D. | |
| Lateral line scales | 54 | 52–54 | 53 | / | 55–56 | 55 | / | 50–52 | 52 | / | 48–52 | 49 | / |
| Dorsal fin rays | III, 7 | III, 7 | III, 7 | / | III, 7 | III, 7 | / | III, 7 | III, 7 | / | III, 7 | III, 7 | / |
| Anal-fin rays | III, 14 | III, 13–14 | III, 13 | / | III, 15 | III, 15 | / | III, 16 | III, 16 | / | III, 13 | III, 13 | / |
| Pectoral fin rays | I, 13 | I, 13–14 | I, 13 | / | I, 14 | I, 14 | / | I, 14 | I, 14 | / | I, 13 | I, 13 | / |
| Pelvic fin rays | I, 7 | I, 6–7 | I, 7 | / | I, 6 | I, 6 | / | I, 8 | I, 8 | / | I, 7 | I, 7 | / |
| Caudal fin rays | 2 + 16 | 2 + 16 | 2 + 16 | / | 2 + 16 | 2 + 16 | / | 2 + 16 | 2 + 16 | / | 2 + 16 | 2 + 16 | / |
| Standard length (SL, mm) | 84.64 | 69.8–110.9 | 90.7 | 11.5 | 64.5–117.4 | 94.9 | 16.9 | 123.4–149.1 | 139.7 | 14.1 | 66.9–99.5 | 79.8 | 17.3 |
| In percentage of SL (%) | |||||||||||||
| Head length (HL) | 24.3 | 20.6−24.5 | 23.4 | 0.9 | 21.9−26.4 | 24.1 | 1.2 | 21.7–24.2 | 22.8 | 1.2 | 16.8–24.5 | 21.8 | 4.3 |
| Body depth | 23.4 | 14.6−26.5 | 23.2 | 2.4 | 19.4−25.5 | 23.0 | 1.6 | 21.8–25.4 | 24.2 | 2.0 | 25.4–26.3 | 25.7 | 0.5 |
| Predorsal length | 51.6 | 41.7−51.9 | 49.5 | 2.2 | 46.2−52.3 | 50.3 | 1.5 | 49.0–52.3 | 50.9 | 1.7 | 51.9–53.1 | 52.5 | 0.6 |
| Prepelvic length | 48.5 | 35.3−48.4 | 45.9 | 2.8 | 43.8−48.0 | 46.2 | 1.3 | 45.7–48.8 | 47.3 | 1.6 | 48–50 | 49.1 | 1.0 |
| Preanal length | 69.6 | 60.2−71.0 | 67 | 2.3 | 65.2−70.9 | 67.6 | 1.5 | 70.5–73.7 | 72.0 | 1.7 | 67.9–69.5 | 68.6 | 0.8 |
| Dorsal-fin height | 19.5 | 10.6−24.1 | 20.9 | 2.8 | 16.1−25.5 | 21.7 | 2.4 | 17.6–17.7 | 17.7 | 0.1 | 18–18.6 | 18.5 | 0.3 |
| Dorsal-fin base length | 10.5 | 6.5−10.5 | 8.1 | 1.1 | 7.4−10.7 | 9.0 | 0.9 | 8.7–10.5 | 9.6 | 0.9 | 9.4–11.5 | 10.4 | 1.0 |
| Anal-fin height | 12.2 | 7.7−15.5 | 10.3 | 1.5 | 8.1−11.4 | 9.8 | 1.1 | 9.2–10.2 | 9.6 | 0.6 | 11–11.8 | 11.4 | 0.4 |
| Anal-fin base length | 17.7 | 13.6−19.0 | 16.7 | 1.2 | 15.1−18.5 | 16.6 | 1.1 | 15.0–17.6 | 16.3 | 1.3 | 16.9–17.2 | 17.1 | 0.1 |
| Pelvic-fin length | 16.2 | 12.3−16.9 | 14.5 | 1.1 | 19.2−23.8 | 21.5 | 1.3 | 14.1–15.7 | 15.0 | 0.8 | 14–14.9 | 14.4 | 0.5 |
| Pectoral-fin length | 22.0 | 18.7−22.9 | 20.8 | 1.1 | 13.1−18.3 | 15.8 | 1.2 | 18.2–21.1 | 20.1 | 1.6 | 19.8–20.9 | 20.2 | 0.6 |
| Caudal-fin length | 22.5 | 18.9−28.5 | 23.6 | 2.6 | 8.4−10.3 | 9.1 | 0.5 | 21.4–25.5 | 23.5 | 2.0 | 18–23.2 | 20.9 | 2.6 |
| Caudal-peduncle length (CPL) | 15.4 | 13.5−19.1 | 16.4 | 1.3 | 19.2−25.1 | 22.6 | 1.5 | 14.6–16.4 | 15.3 | 1.0 | 14.7–16.9 | 15.7 | 1.2 |
| Caudal-peduncle depth (CPD) | 9.6 | 8.0−10.5 | 9.5 | 0.5 | 18.0−23.4 | 21.6 | 1.5 | 8.8–9 | 8.9 | 0.1 | 8.3–9.1 | 8.8 | 0.4 |
| P–V distance | 24.2 | 21.1−27.4 | 24.1 | 1.4 | 19.2−23.8 | 21.5 | 1.3 | 22.9–25.4 | 24.5 | 1.4 | 23.1–27.2 | 24.7 | 2.2 |
| V–A distance | 21.3 | 19.7−24.2 | 21.5 | 1.2 | 13.1−18.3 | 15.8 | 1.2 | 22.7–24.8 | 23.6 | 1.1 | 18.9–20.6 | 19.9 | 0.8 |
| In percentage of HL (%) | |||||||||||||
| Head depth | 59.4 | 56.5−75.4 | 61.8 | 4.1 | 55.4−63.7 | 59.2 | 2.2 | 61.2–70.6 | 64.5 | 5.4 | 72.5–102.6 | 83.0 | 17.1 |
| Head width | 41.0 | 35.5−44.6 | 40.3 | 2.1 | 37.8−42.7 | 40.0 | 1.5 | 42.6–44.8 | 43.7 | 1.1 | 52–78.7 | 62.0 | 14.6 |
| Snout length | 27.6 | 25.0−29.0 | 26.8 | 1.0 | 26.8−32.9 | 30.2 | 1.5 | 30.7–33.3 | 32.4 | 33.3 | 26.6–40.7 | 32.2 | 7.5 |
| Eye diameter | 33.1 | 30.4−32.9 | 31.1 | 0.6 | 24.6−29.7 | 27.6 | 1.3 | 26.3–29.7 | 28.2 | 1.7 | 29.9–52.2 | 38.8 | 11.8 |
| Interorbital width | 32.6 | 26.2−35.4 | 29.9 | 2.4 | 24.6−33.9 | 27.5 | 2.2 | 27.7–31.2 | 29.5 | 1.8 | 32.7–53.9 | 40.4 | 11.8 |
| Snout length/eye diameter (%) | 83.4 | 80.0−93.0 | 86.2 | 3.2 | 101.0−121.5 | 109.3 | 5.5 | 111.2–117.0 | 115.0 | 3.3 | 77.9–89 | 84.0 | 5.6 |
| CPD/CPL (%) | 62.3 | 48.9–63.1 | 58.4 | 3.8 | 47.9−69.4 | 58.5 | 5.6 | 54.8–60.8 | 58.5 | 3.2 | 53.2–61.6 | 56.6 | 4.4 |
| Morphometric Measurements | PC1 | PC2 |
|---|---|---|
| HL/SL | 0.1057 | −0.2265 |
| In percentage of HL | ||
| Head depth | −0.1359 | 0.3471 |
| Head width | −0.0563 | 0.1947 |
| Snout length | 0.3620 | 0.4390 |
| Eye diameter | −0.3858 | 0.1265 |
| Interorbital distance | −0.3553 | 0.7028 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lei, H.; Gong, Z.; Li, X. Description of a New Species of Hainania Koller (Teleostei, Cypriniformes, Xenocyprididae) from Guangdong Province, Southern China. Diversity 2025, 17, 549. https://doi.org/10.3390/d17080549
Lei H, Gong Z, Li X. Description of a New Species of Hainania Koller (Teleostei, Cypriniformes, Xenocyprididae) from Guangdong Province, Southern China. Diversity. 2025; 17(8):549. https://doi.org/10.3390/d17080549
Chicago/Turabian StyleLei, Haotian, Ziyu Gong, and Xuankun Li. 2025. "Description of a New Species of Hainania Koller (Teleostei, Cypriniformes, Xenocyprididae) from Guangdong Province, Southern China" Diversity 17, no. 8: 549. https://doi.org/10.3390/d17080549
APA StyleLei, H., Gong, Z., & Li, X. (2025). Description of a New Species of Hainania Koller (Teleostei, Cypriniformes, Xenocyprididae) from Guangdong Province, Southern China. Diversity, 17(8), 549. https://doi.org/10.3390/d17080549






