Genetic Variability and Population Structure of Camelus from Kazakhstan Inferred from 17 STR Markers
Abstract
1. Introduction
2. Materials and Methods
2.1. Sampling Information
2.2. Laboratory Protocol
2.3. Genetic Structure Analysis
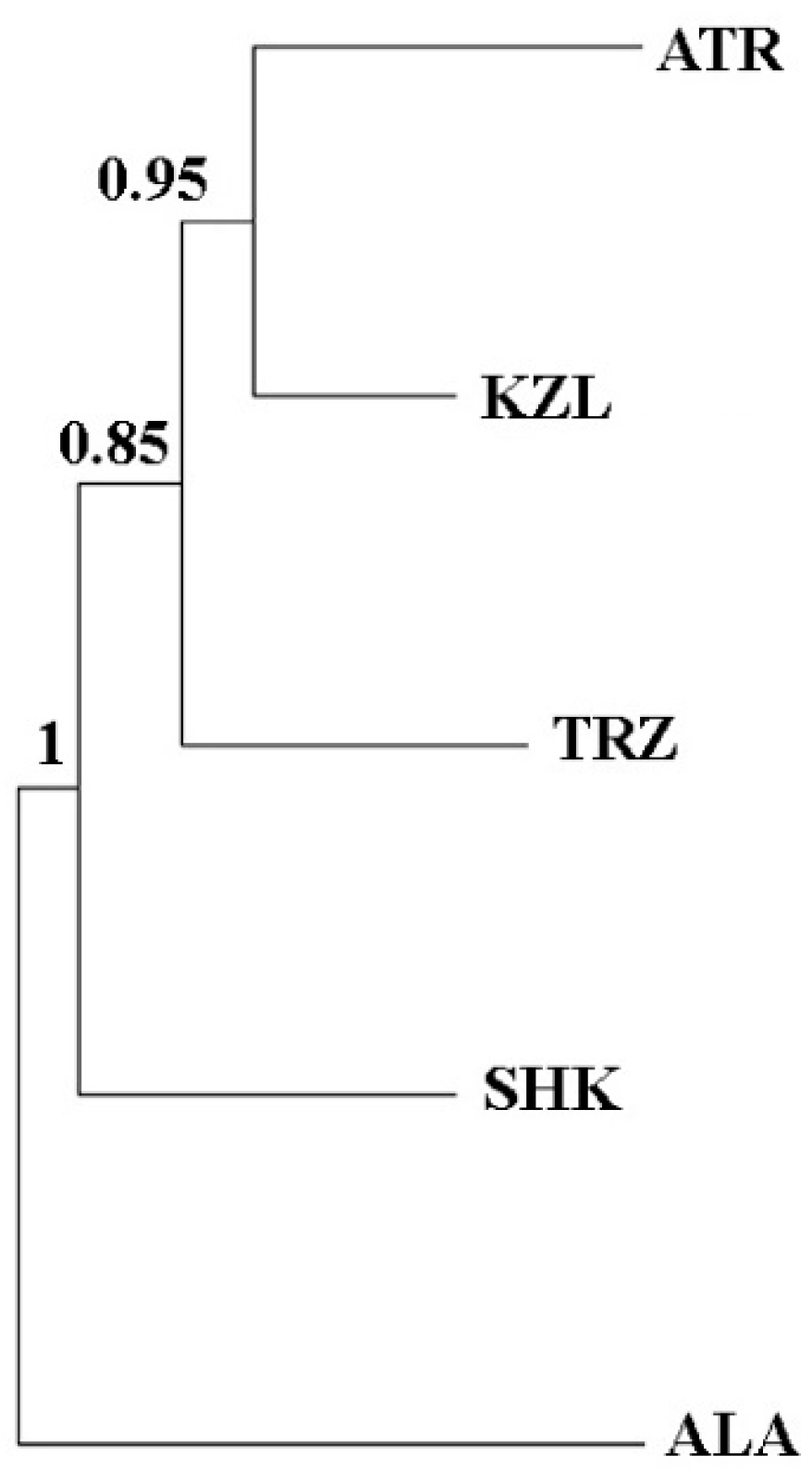
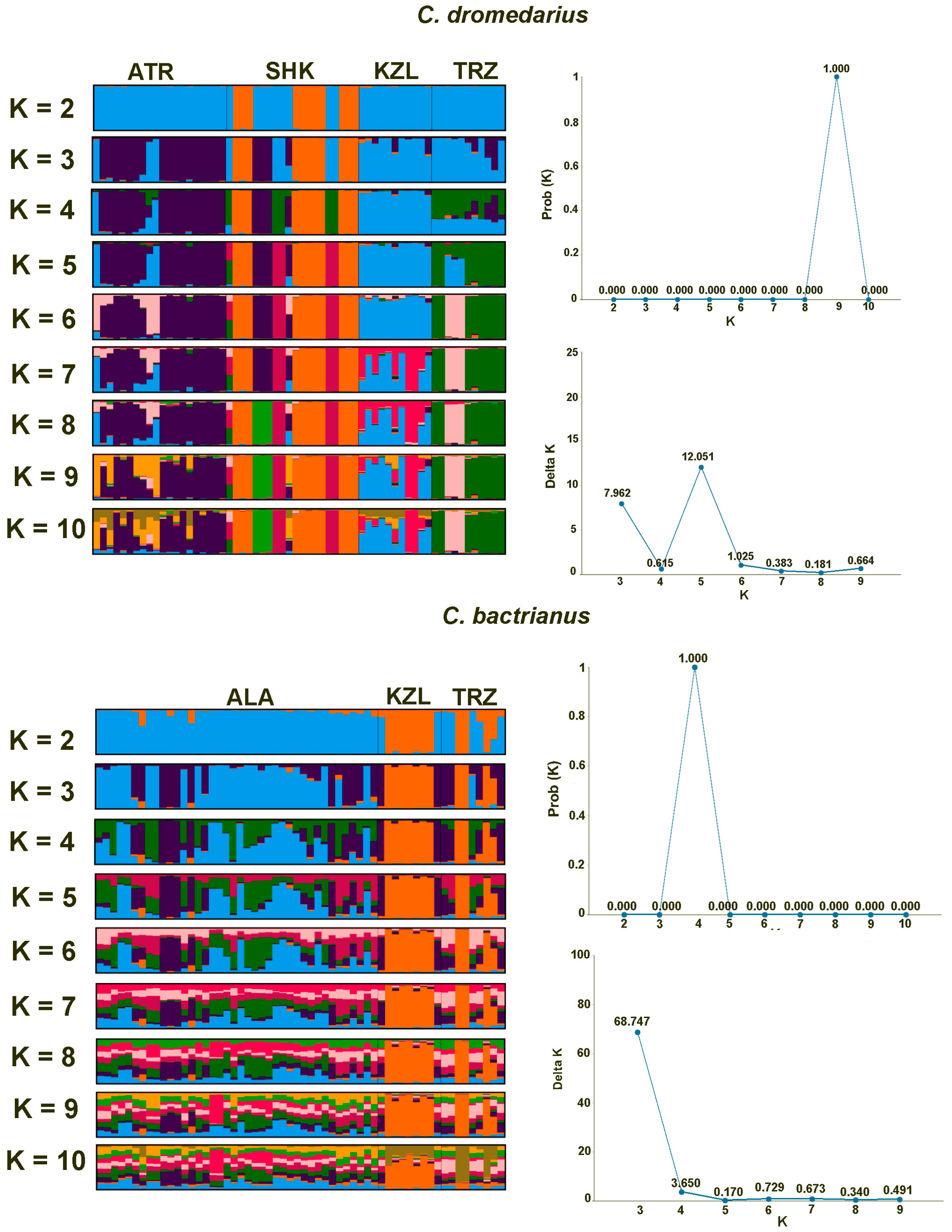
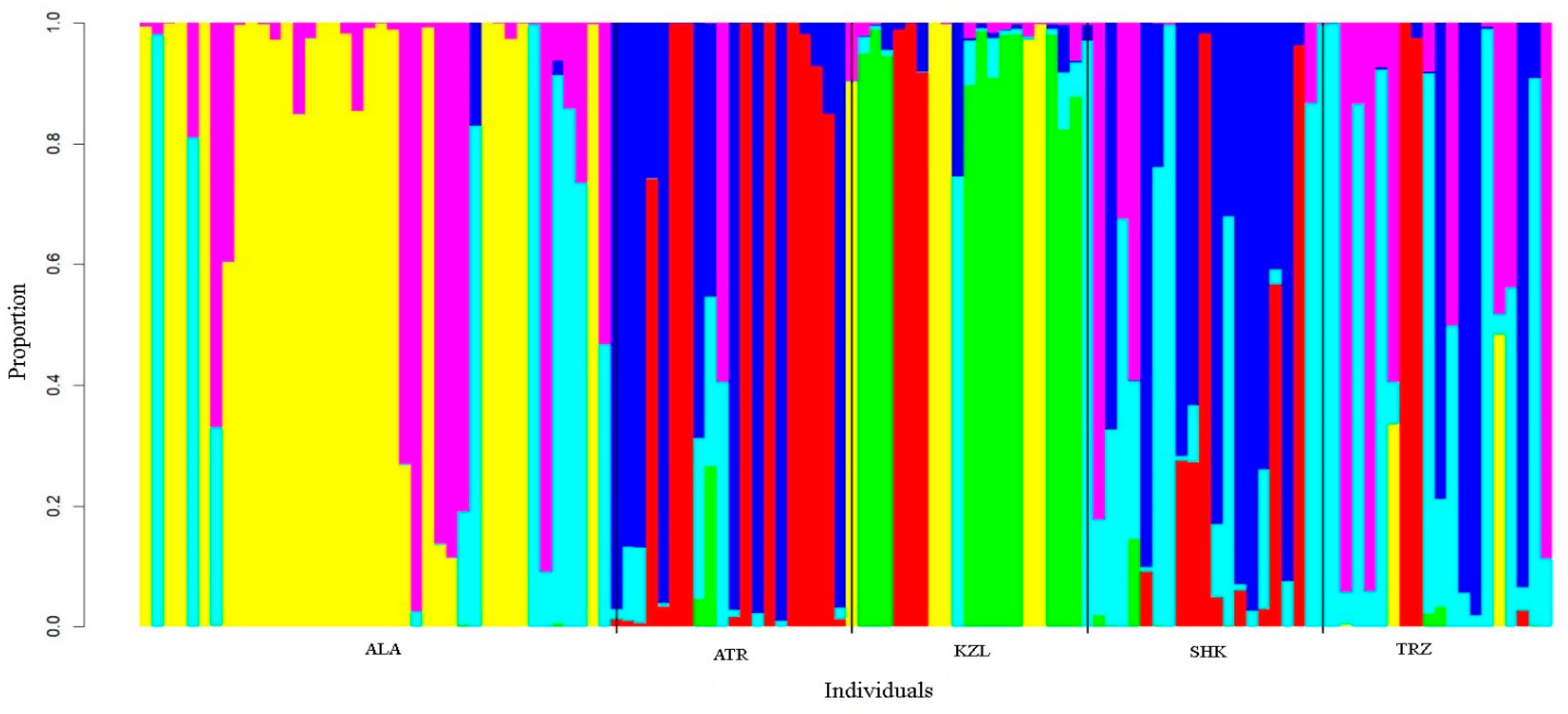
3. Results
3.1. Genetic Polymorphisms and Diversity
3.1.1. Species-Level Genetic Diversity
3.1.2. Regional Population-Level Genetic Diversity
3.2. Genetic Structure and Differentiation
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Available online: https://ielc.libguides.com/sdzg/factsheets/extantcamels/taxonomy?utm_source (accessed on 3 January 2024).
- Mirzaei, F. Production and trade of camel products in some Middle East countries. J. Agric. Econ. Dev. 2012, 1, 153–160. [Google Scholar]
- Vyas, S.; Sharma, N.; Sheikh, F.D.; Singh, S.; Sena, D.S.; Bissa, U.K. Reproductive status of Camelus bactrianus during early breeding season in India. Asian Pac. J. Reprod. 2015, 3, 61–64. [Google Scholar] [CrossRef]
- Faye, B.; Konuspayeva, G. The Encounter between Bactrian and Dromedary Camels in Central Asia. In Camels in Asia. In Camels in Asia and North-Africa—Interdisciplinary Prospectives on Their Past and Present Significance; Knoll, E.-M., Burger, P., Eds.; Austrian Academy of Sciences Press: Vienna, Austria, 2012; pp. 27–33, 248–250. [Google Scholar]
- Dioli, M. Dromedary (Camelus dromedarius) and bactrian camel (Camelus bactrianus) crossbreeding husbandry practices in Turkey and Kazakhstan: An in-depth review. Pastor. Res. Policy Pract. 2020, 10, 6. [Google Scholar] [CrossRef]
- Baimukanov, D.A.; Yuldashbaev, Y.A.; Iskhan, K.Z.H.; Demin, V.A. Concept for the development of productive and pedigree camel breeding in the Republic of Kazakhstan for 2021–2030. Agrar. Sci. 2020, 340, 52–60. (In Russian) [Google Scholar] [CrossRef]
- Imamura, K.; Salmurzauli, R.; Iklasov, M.K.; Baibayssov, A.; Matsui, K.; Nurtazin, S.T. The distribution of the two domestic camel species in Kazakhstan caused by the demand of industrial stockbreeding. J. Arid Land Stud. 2017, 26, 233–236. [Google Scholar] [CrossRef]
- Akhmetsadykova, S.N.; Konuspayeva, G.; Akhmetsadykov, N. Camel breeding in Kazakhstan and future perspectives. Anim. Front. 2022, 12, 71–77. [Google Scholar] [CrossRef]
- Eldala. Available online: https://eldala.kz/novosti/zhivotnovodstvo/12959-pogolove-loshadey-v-kazakhstane-vyroslo-na-12-za-god (accessed on 15 January 2024).
- Silbermayr, K.; Orozco-terWengel, P.; Charruau, P.; Enkhbileg, D.; Walzer, C.; Vogl, C.; Schwarzenberger, F.; Kaczensky, P.; Burger, P.A. High Mitochondrial Differentiation Levels Between Wild and Domestic Bactrian Camels: A Basis for Rapid Detection of Maternal Hybridization. Anim. Genet. 2010, 41, 315–318. [Google Scholar] [CrossRef]
- Chuluunbat, B.; Charruau, P.; Silbermayr, K.; Khorloojav, T.; Burger, P. Genetic diversity and population structure of Mongolian domestic Bactrian camels (Camelus Bactrianus). Anim. Genet. 2014, 45, 550–558. [Google Scholar] [CrossRef]
- Ming, L.; Yi, L.; Sa, R.; Wang, Z.X.; Wang, Z.; Ji, R. Genetic diversity and phylogeographic structure of Bactrian camels shown by mitochondrial sequence variations. Anim. Genet. 2017, 48, 217–220. [Google Scholar] [CrossRef]
- Mahmoud, A.H.; Abu-Tarbush, F.M.; Alshaik, M.; Aljumaah, R.; Saleh, A. Genetic diversity and population genetic structure of six dromedary camel (Camelus dromedarius) populations in Saudi Arabia. Saudi J. Biol. Sci. 2020, 27, 1384–1389. [Google Scholar] [CrossRef]
- Ming, L.; Yuan, L.; Yi, L. Whole genome sequencing of 128 camels across Asia reveals origin and migration of domestic Bactrian camels. Commun. Biol. 2020, 3, 1. [Google Scholar] [CrossRef] [PubMed]
- Wu, H.; Guang, X.; Al-Fageeh, M.B.; Cao, J.; Pan, S.; Zhou, H.; Zhang, L.; Abutarboush, M.H.; Xing, Y.; Xie, Z.; et al. Camelid Genomes Reveal Evolution and Adaptation to Desert Environments. Nat. Commun. 2014, 5, 5188. [Google Scholar] [CrossRef]
- Elmira, A.; Nuradin, A.; Svitojus, A.; Galymzhan, А. Genetic typing of South Kazakhstan populations’ dairy camels using DNA technology. Anim. Biotechnol. 2019, 31, 547–554. [Google Scholar] [CrossRef] [PubMed]
- Dossybayev, K.; Ualiyeva, D.; Amandykova, M.; Kapasuly, T.; Mussayeva, A.; Orazymbetova, Z.; Shaltenbay, G.; Bekmanov, B. Genetic differentiation of Camelus bactrianus from Kazakhstan. In Proceedings of the ISAG 2023 39th International Society for Animal Genetics Conference, Cape Town, South Africa, 2–7 July 2023; pp. 24–25. [Google Scholar]
- Amandykova, M.; Dossybayev, K.; Mussayeva, A.; Bekmanov, B.; Saitou, N. Comparative Analysis of the Polymorphism of the Casein Genes in Camels Bred in Kazakhstan. Diversity 2022, 14, 285. [Google Scholar] [CrossRef]
- Amandykova, M.; Dossybayev, K.; Mussayeva, A.; Zhunusbayeva, Z.; Bekmanov, B. A Study of the Genetic Structure of Hybrid Camels in Kazakhstan. Genes 2023, 14, 1373. [Google Scholar] [CrossRef] [PubMed]
- Hoffmann, I.; Marsan, P.A.; Barker, J.S.F.; Cothran, E.G.; Hanotte, O.; Lenstra, J.A.; Milan, D.; Weigend, S.; Simianer, H. New MoDAD marker sets to be used in diversity studies for the major farm animal species: Recommendations of a joint ISAG/FAO working group. In Proceedings of the 29th International Conference of Animal Genetics, ISAG2004/TOKYO, Tokyo, Japan, 11–16 September 2004. [Google Scholar]
- Penedo, M.C.T.; Caetano, A.R.; Cordova, K.I. Six microsatellite markers for South American camelids. Animal Genetics 1999, 30, 399. [Google Scholar] [CrossRef]
- Obreque, V.; Mancilla, R.; García-Huidobro, J.; Cothran, E.G.; Hinrichsen, P. Thirteen new dinucleotide microsatellites in Alpaca. Anim. Genet. 1999, 30, 397–398. [Google Scholar] [CrossRef]
- Lang, K.D.; Wang, Y.; Plante, Y. Fifteen Polymorphic Dinucleotide Microsatellites in Llamas and Alpacas. Anim. Genet. 1996, 27, 293. [Google Scholar] [CrossRef]
- Mariasegaram, M.; Pullenayegum, S.; Jahabar, A.M.; Shah, R.S.; Penedo, M.C.; Wernery, U.; Sasse, J. Isolation and characterization of eight microsatellite markers in Camelus dromedarius and cross-species amplification in C. bactrianus and Lama pacos. Anim Genet. 2002, 33, 385–387. [Google Scholar] [CrossRef]
- Evdotchenko, D.; Han, Y.; Bartenschlager, H.; Preuss, S.; Geldermann, H. New polymorphic microsatellite loci for different camel species. Mol. Ecol. Notes 2003, 3, 431–434. [Google Scholar] [CrossRef]
- Bolstein, D.; White, R.L.; Skolnick, M.; Davis, R.W. Construction of a genetic linkage map in man using restriction fragment length polymorphisms. Am. J. Hum. Genet. 1980, 32, 314–331. [Google Scholar]
- Liu, K.; Muse, S.V. Powermarker: Integrated analysis environment for genetic maker data. Bioinformatics 2005, 21, 2128–2129. [Google Scholar] [CrossRef] [PubMed]
- Weir, B.S. Genetic Data Analysis II; Sinauer Associates: Sunderland, MA, USA, 1996. [Google Scholar]
- Weir, B.S.; Cockerham, C.C. Estimating F-Statistics for the Analysis of Population Structure. Evolution 1984, 38, 1358–1370. [Google Scholar] [CrossRef] [PubMed]
- Goudet, J. FSTAT, a Program to Estimate and Test Gene Diversities and Fixation Indices (Version 2.9.3). 2001. Available online: https://www2.unil.ch/popgen/softwares/fstat.htm (accessed on 23 September 2024).
- Peakall, R.; Smouse, P.E. GenAlEx 6: Genetic analysis in Excel: Population genetic software for teaching and research. Mol. Ecol. Notes 6 2006, 6, 288–295. [Google Scholar] [CrossRef]
- Raymond, M.; Rousset, F. Genepop (version 1.2), population genetics software for exact tests and ecumenicism. J. Hered. 1995, 86, 248–249. [Google Scholar] [CrossRef]
- Rousset, F. Genepop’007: A complete reimplementation of the Genepop software for Windows and Linux. Mol. Ecol. Resour. 2008, 8, 103–106. [Google Scholar] [CrossRef]
- Saitou, N.; Nei, M. The Neighbor-Joining Method: A New Method for Reconstructing Phylogenetic Trees. Mol. Biol. Evol. 1987, 4, 406–425. [Google Scholar]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef]
- Felsenstein, J. Confidence Limits on Phylogenies: An Approach Using the Bootstrap. Evolution 1985, 39, 783–791. [Google Scholar] [CrossRef]
- Belkhir, K. GENETIX 4.05, Logiciel Sous Windows TM Pour la Génétique des Populations. 2004, p. 9. Available online: http://www.genetix.univ-montp2.fr/genetix/genetix.htm (accessed on 15 September 2024).
- Pritchard, J.K.; Stephens, M.; Donnelly, P. Inference of population structure using multilocus genotype data. Genetics 2000, 155, 945–959. [Google Scholar] [CrossRef]
- Evanno, G.; Regnaut, S.; Goudet, J. Detecting the number of clusters of individuals using the software STRUCTURE: A simulation study. Mol Ecol. 2005, 14, 2611–2620. [Google Scholar] [CrossRef] [PubMed]
- Earl, D.A.; Von Holdt, B.M. Structure Harvester: A website and program for visualizing STRUCTURE output and implementing the Evanno method. Conserv. Genet. Resour. 2012, 4, 359–361. [Google Scholar] [CrossRef]
- Van Rossum, G.; Drake, F.L. Python 3 Reference Manual; CreateSpace: Scotts Valley, CA, USA, 2009; ISBN 1441412697. [Google Scholar]
- Jakobsson, M.; Rosenberg Noah, A. CLUMPP: A cluster matching and permutation program for dealing with label switching and multimodality in analysis of population structure. Bioinformatics 2007, 23, 1801–1806. [Google Scholar] [CrossRef]
- Rosenberg, N.A. Distruct: A program for the graphical display of population structure. Mol. Ecol. Notes 2004, 4, 137–138. [Google Scholar] [CrossRef]
- Anderson, E.C.; Thompson, E.A. A Model-Based Method for Identifying Species Hybrids Using Multilocus Genetic Data. Genetics 2002, 160, 1217–1229. [Google Scholar] [CrossRef]
- Chen, B.; Cole, J.W.; Grond-Ginsbach, C. Departure from Hardy–Weinberg Equilibrium and Genotyping Error. Front. Genet. 2017, 8, 167. [Google Scholar] [CrossRef]
- Lachance, J. Detecting Selection-Induced Departures from Hardy–Weinberg Proportions. Genet. Sel. Evol. 2009, 41, 15. [Google Scholar] [CrossRef]
- Nolte, M.; Kotzé, A.; Van der Bank, F.H.; Grobler, J.P. Microsatellite Markers Reveal Low Genetic Differentiation Among Southern African Camelus dromedarius Populations. S. Afr. J. Anim. Sci. 2005, 35, 152–161. [Google Scholar] [CrossRef][Green Version]
- Orazymbetova, Z.; Ualiyeva, D.; Dossybayev, K.; Torekhanov, A.; Sydykov, D.; Mussayeva, A.; Baktybayev, G. Genetic Diversity of Kazakhstani Equus caballus (Linnaeus, 1758) Horse Breeds Inferred from Microsatellite Markers. Vet. Sci. 2023, 10, 598. [Google Scholar] [CrossRef]
- Yakubu, A.; Okpeku, M.; Shoyombo, A.J.; Onasanya, G.O.; Dahloum, L.; Çelik, S.; Oladepo, A. Exploiting Morphobiometric and Genomic Variability of African Indigenous Camel Populations—A Review. Front. Genet. 2022, 13, 1021615. [Google Scholar] [CrossRef]
- Al-Swailem, S.; Shehata, M.M.; Al-Busadah, K.A.; Fallatah, M.H.; Ejaz, A. Evaluation of the genetic variability of microsatellite markers in Saudi Arabian camels. J. Food. Agric. Environ. 2009, 7, 636–639. [Google Scholar]
- Piro, M.; Mabsoute, F.E.; El Khattaby, N.; Laghouaouta, H.; Boujenane, I. Genetic variability of dromedary camel populations based on microsatellite markers. Animal 2020, 14, 2452–2462. [Google Scholar] [CrossRef]
- Patel, A.C.; Jisha, T.K.; Disha, U.; Rakesh, P. Molecular characterization of camel breeds of Gujarat using microsatellite markers. Livest. Sci. 2015, 181, 85–88. [Google Scholar] [CrossRef]
- Eltanany, M.; Elfaroug Sidahmed, O.; Distl, O. Assessment of genetic diversity and differentiation of two major camel ecotypes (Camelus dromedarius) in Sudan using microsatellite markers. Arch. Anim. Breed. 2015, 58, 269–275. [Google Scholar] [CrossRef]
- Spencer, P.; Woolnough, A. Assessment and Genetic Characterisation of Australian Camels Using Microsatellite Polymorphisms. Livest. Sci. 2010, 129, 241–245. [Google Scholar] [CrossRef]
- Ould Ahmed, M.; Ben Salem, F.; Bedhiaf, S.; Rekik, B.; Djemali, M. Genetic diversity in Tunisian dromedary (Camelus dromedarius) populations using microsatellite markers. Livest. Sci. 2010, 132, 182–185. [Google Scholar] [CrossRef]
- Hedayat-Evrigh, N.; Khalkhali-Evrigh, R.; Vahedi, V.; Pourasad, K.; Sharifi, R.S. Genetic Diversity and Population Structure of Old-World Camelids (Camelus dromedarius and Camelus bactrianus) in Iran Using Mitochondrial DNA. Kafkas Üniv. Vet. Fak. Derg. 2018, 24, 467–471. [Google Scholar] [CrossRef]
- Hussain, T.; Hussain, F.; Wajid, A.; Ellahi Babar, M.; Musthafa, M.M.; Marikar, F.M.M.T. Microsatellite marker based genetic diversity in Mareecha and Barela breeds of dromedary camel from Pakistan. Vet. Stanica 2021, 52, 307–314. [Google Scholar] [CrossRef]
- Konuspayeva, G.; Faye, B.; Bengoumi, M. Mineral Status in Camel Milk: A Critical Review. Anim. Front. 2022, 12, 52–60. [Google Scholar] [CrossRef]
- Raziq, A.; Younas, M.; Khan, M.S.; Iqbal, A. Milk Production Potential as Affected by Parity and Age in the Kohi Dromedary Camel. J. Camel Pract. Res. 2010, 17, 195–198. [Google Scholar]
- Köhler-Rollefson, I. The camel in Rajasthan: Agricultural diversification and pastoral risk management. J. Arid. Environ. 2001, 49, 453–465. [Google Scholar]
- Burger, P.A. The history of Old World camelids in the light of molecular genetics. Trop. Anim. Health Prod. 2016, 48, 905–913. [Google Scholar] [CrossRef]
- Mburu, D.N.; Ochieng, J.W.; Kuria, S.G.; Jianlin, H.; Kaufmann, B.; Rege, J.E.; Hanotte, O. Genetic diversity and relationships of indigenous Kenyan camel (Camelus dromedarius) populations: Implications for their classification. Anim. Genet. 2003, 34, 26–32. [Google Scholar] [CrossRef] [PubMed]
- Kadim, I.T.; Mahgoub, O.; Al-Marzooqi, W.; Al-Zadjali, S.; Annamalai, K.; Mansour, M.H. Camel meat production and quality: A review. Meat Sci. 2014, 98, 289–297. [Google Scholar] [CrossRef]
- Hedrick, P.W. Genetic Variability in a Conservation Context. Nat. Rev. Genet. 2005, 6, 459–470. [Google Scholar] [CrossRef]
- Charlesworth, D.; Charlesworth, B. Inbreeding Depression and Its Evolutionary Consequences. Annu. Rev. Ecol. Syst. 1987, 18, 237–268. [Google Scholar] [CrossRef]
- Mahmoud, A.H.; AlShaikh, M.A.; Aljummah, R.S.; Mohammed, O.B. Genetic variability of camel (Camelus dromedarius) populations in Saudi Arabia based on microsatellites analysis. Afr. J. Biotechnol. 2012, 11, 11173–11180. [Google Scholar] [CrossRef]
- Mahmoud, A.H.; AlShaikh, M.A.; Aljummah, R.S.; Mohammed, O.B. Genetic characterization of Majaheem camel population in Saudi Arabia based on microsatellite markers. Res. J. Biotechnol. 2013, 8, 26–30. [Google Scholar]
- Xiaohong, H.; Xiuli, H.; Weijun, G.; Kechuan, T.; Wenbin, Z.; Yuehui, M. Genetic variability and relationship of 10 Bactrian camel populations revealed by microsatellite markers. Biodivers. Sci. 2012, 20, 199–206. [Google Scholar] [CrossRef]
- Shaltenbay, G.; Kapassuly, T.; Dossybayev, K. Analysis of the genetic variability of camel populations in Kazakhstan using ISSR-PCR markers. BIO Web Conf. 2024, 100, 03015. [Google Scholar] [CrossRef]


| Location, Farm | (Sample Size), Taxon |
|---|---|
| (ATR) Atyrau, WKZ | (n = 20), C. dromedarius |
| (KZL) Kyzylorda, SWKZ | (n = 10), C. dromedarius; (n = 10), C. bactrianus |
| (SHK) Shymkent, SKZ | (n = 20), C. dromedarius |
| (TRZ) Taraz, SKZ | (n = 10), C. dromedarius; (n = 10), C. bactrianus |
| (ALA) Almaty, SEKZ | (n = 20), C. bactrianus |
| Locus | Na | Ne | I | Ho | He | uHe | FIS | FIT | Fst | Nm | PIC | HWE |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CVRL05 | 8.400 | 5.035 | 1.787 | 0.747 | 0.791 | 0.811 | 0.055 | 0.120 | 0.068 | 3.418 | 0.824 | ns |
| LCA66 | 10.600 | 5.781 | 2.025 | 0.677 | 0.826 | 0.848 | 0.181 | 0.262 | 0.099 | 2.272 | 0.917 | * |
| VOLP08 | 7.800 | 3.867 | 1.561 | 0.614 | 0.721 | 0.741 | 0.148 | 0.275 | 0.149 | 1.425 | 0.824 | ns |
| VOLP03 | 13.000 | 9.063 | 2.304 | 0.603 | 0.871 | 0.900 | 0.308 | 0.359 | 0.073 | 3.168 | 0.941 | *** |
| CMS15 | 9.000 | 5.179 | 1.798 | 0.649 | 0.775 | 0.798 | 0.163 | 0.256 | 0.111 | 1.999 | 0.855 | ** |
| YWLL09 | 4.200 | 2.622 | 1.112 | 0.472 | 0.601 | 0.617 | 0.214 | 0.298 | 0.106 | 2.106 | 0.590 | ns |
| VOLP32 | 4.400 | 3.363 | 1.265 | 0.650 | 0.686 | 0.704 | 0.053 | 0.114 | 0.065 | 3.593 | 0.736 | ns |
| YWLL38 | 6.600 | 3.526 | 1.490 | 0.710 | 0.713 | 0.731 | 0.004 | 0.059 | 0.055 | 4.256 | 0.716 | ns |
| CVRL07 | 6.400 | 3.442 | 1.417 | 0.430 | 0.690 | 0.708 | 0.377 | 0.456 | 0.127 | 1.722 | 0.787 | *** |
| YWLL44 | 7.200 | 4.165 | 1.599 | 0.759 | 0.745 | 0.764 | −0.019 | 0.048 | 0.066 | 3.520 | 0.763 | ns |
| CMS50 | 12.800 | 8.121 | 2.248 | 0.800 | 0.860 | 0.882 | 0.070 | 0.136 | 0.071 | 3.288 | 0.923 | ns |
| LCA63 | 5.200 | 3.759 | 1.424 | 0.770 | 0.724 | 0.742 | −0.064 | 0.013 | 0.073 | 3.194 | 0.760 | ns |
| VOLP67 | 15.200 | 10.132 | 2.468 | 0.809 | 0.895 | 0.918 | 0.095 | 0.141 | 0.051 | 4.697 | 0.940 | *** |
| YWLL08 | 12.600 | 6.905 | 2.162 | 0.860 | 0.846 | 0.867 | −0.017 | 0.082 | 0.097 | 2.322 | 0.933 | ns |
| VOLP10 | 12.600 | 6.005 | 2.112 | 0.808 | 0.822 | 0.844 | 0.017 | 0.105 | 0.090 | 2.536 | 0.903 | * |
| CMS13 | 8.600 | 5.158 | 1.843 | 0.790 | 0.802 | 0.822 | 0.015 | 0.079 | 0.065 | 3.570 | 0.840 | ns |
| CVRL06 | 5.000 | 3.402 | 1.315 | 0.450 | 0.658 | 0.675 | 0.316 | 0.458 | 0.207 | 0.957 | 0.804 | *** |
| Mean | 8.800 | 5.266 | 1.761 | 0.682 | 0.766 | 0.787 | 0.113 | 0.192 | 0.093 | 2.826 | 0.827 | ns |
| Source of Variation | d.f. | SS | MS | Est. Var. | % of Var. |
|---|---|---|---|---|---|
| Five regional populations of camels | |||||
| Among population | 4 | 128.580 | 32.145 | 0.608 | 8% |
| Among individuals | 95 | 743.075 | 7.822 | 1.076 | 15% |
| Within individuals | 100 | 567.000 | 5.670 | 5.670 | 77% |
| Total | 199 | 1438.655 | 7.354 | 100% | |
| FST = 0.083 (p ≥ 0.001); FIS = 0.159 (p ≥ 0.001); FIT = 0.229 (p ≥ 0.001) | |||||
| Two species of camels | |||||
| Among species | 1 | 27.777 | 27.777 | 0.203 | 3% |
| Among individuals | 98 | 844.338 | 8.616 | 1.475 | 20% |
| Within individuals | 100 | 566.500 | 5.665 | 5.665 | 77% |
| Total | 199 | 1438.615 | 7.344 | 100% | |
| FST = 0.028 (p ≥ 0.001); FIS = 0.207 (p ≥ 0.001); FIT = 0.229 (p ≥ 0.001) | |||||
| Pop | ALA | ATR | SHK | KZL | TRZ |
|---|---|---|---|---|---|
| ALA | 0.088 | 0.075 | 0.064 | 0.065 | |
| ATR | 0.854 | 0.059 | 0.041 | 0.050 | |
| SHK | 0.615 | 0.466 | 0.052 | 0.062 | |
| KZL | 0.555 | 0.339 | 0.429 | 0.044 | |
| TRZ | 0.574 | 0.433 | 0.553 | 0.385 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shaltenbay, G.; Ualiyeva, D.; Kapassuly, T.; Kozhakhmet, A.; Orazymbetova, Z.; Kulboldin, T.; Yergali, K.; Amandykova, M.; Bekmanov, B.; Dossybayev, K. Genetic Variability and Population Structure of Camelus from Kazakhstan Inferred from 17 STR Markers. Diversity 2025, 17, 459. https://doi.org/10.3390/d17070459
Shaltenbay G, Ualiyeva D, Kapassuly T, Kozhakhmet A, Orazymbetova Z, Kulboldin T, Yergali K, Amandykova M, Bekmanov B, Dossybayev K. Genetic Variability and Population Structure of Camelus from Kazakhstan Inferred from 17 STR Markers. Diversity. 2025; 17(7):459. https://doi.org/10.3390/d17070459
Chicago/Turabian StyleShaltenbay, Gulfairuz, Daniya Ualiyeva, Tilek Kapassuly, Altynay Kozhakhmet, Zarina Orazymbetova, Temirlan Kulboldin, Kanagat Yergali, Makpal Amandykova, Bakhytzhan Bekmanov, and Kairat Dossybayev. 2025. "Genetic Variability and Population Structure of Camelus from Kazakhstan Inferred from 17 STR Markers" Diversity 17, no. 7: 459. https://doi.org/10.3390/d17070459
APA StyleShaltenbay, G., Ualiyeva, D., Kapassuly, T., Kozhakhmet, A., Orazymbetova, Z., Kulboldin, T., Yergali, K., Amandykova, M., Bekmanov, B., & Dossybayev, K. (2025). Genetic Variability and Population Structure of Camelus from Kazakhstan Inferred from 17 STR Markers. Diversity, 17(7), 459. https://doi.org/10.3390/d17070459







