Genetic Testing of a High-End ‘Angel Skin’ Precious Coral Necklace Identifies a Species New to the Precious Coral Trade and Potentially New to Science
Abstract
1. Introduction
2. Materials and Methods
2.1. Description of the ‘Angel Skin’ Coral Necklace
2.2. Visual Inspection and Molecular Fingerprinting
2.3. Initial Genetic Testing Using the Coral-ID Method
2.4. Comparing the DNA of an ‘Angel Skin’ Coral Bead with an Extended Pleurocorallium Reference Data Set
2.5. Comparison of the DNA Sequences of the ‘Angel Skin’ Coral Necklace with Additional Samples
3. Results
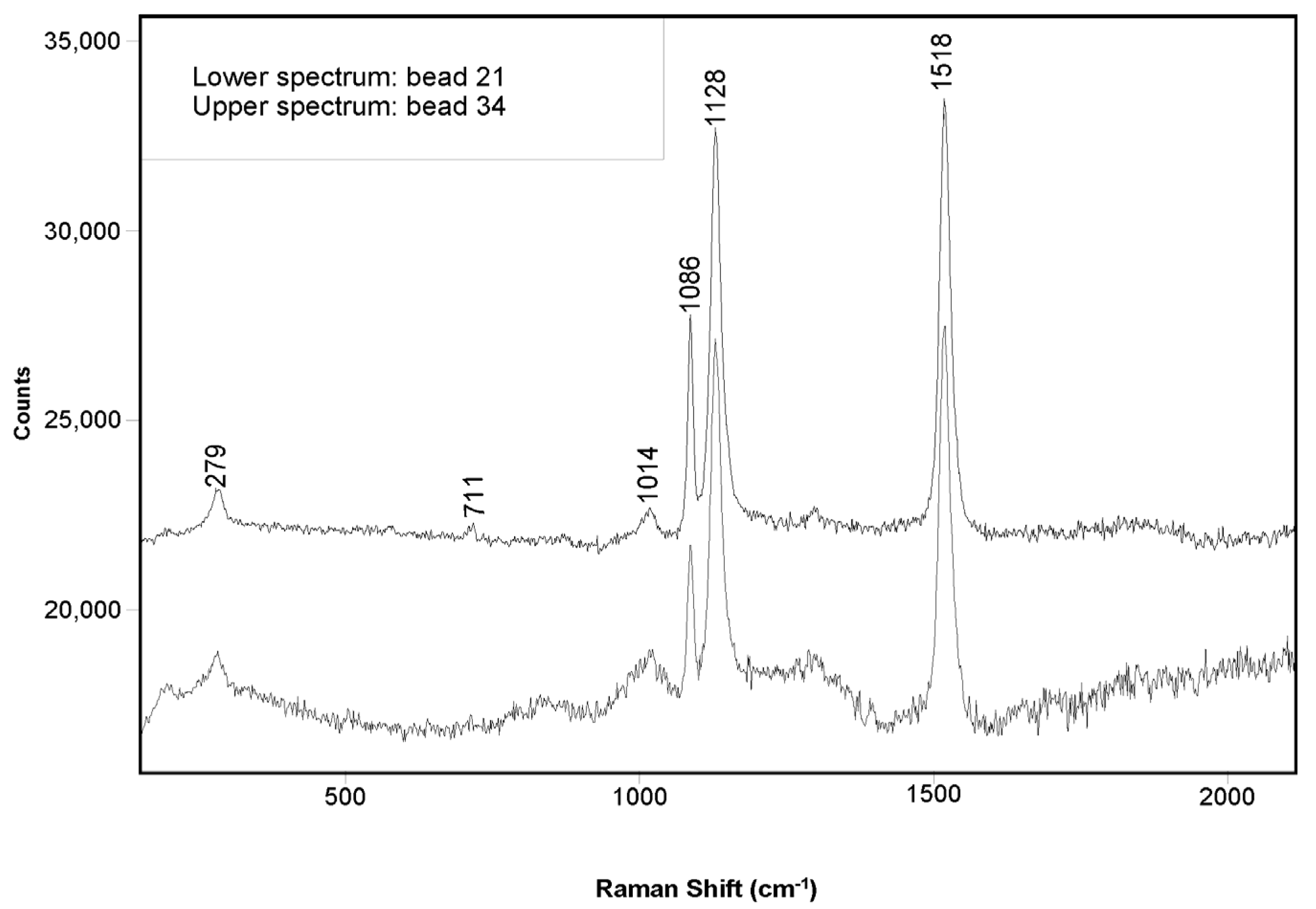
3.1. Visual Inspection and Molecular Fingerprinting
3.2. Initial Genetic Testing Using the Coral-ID Method
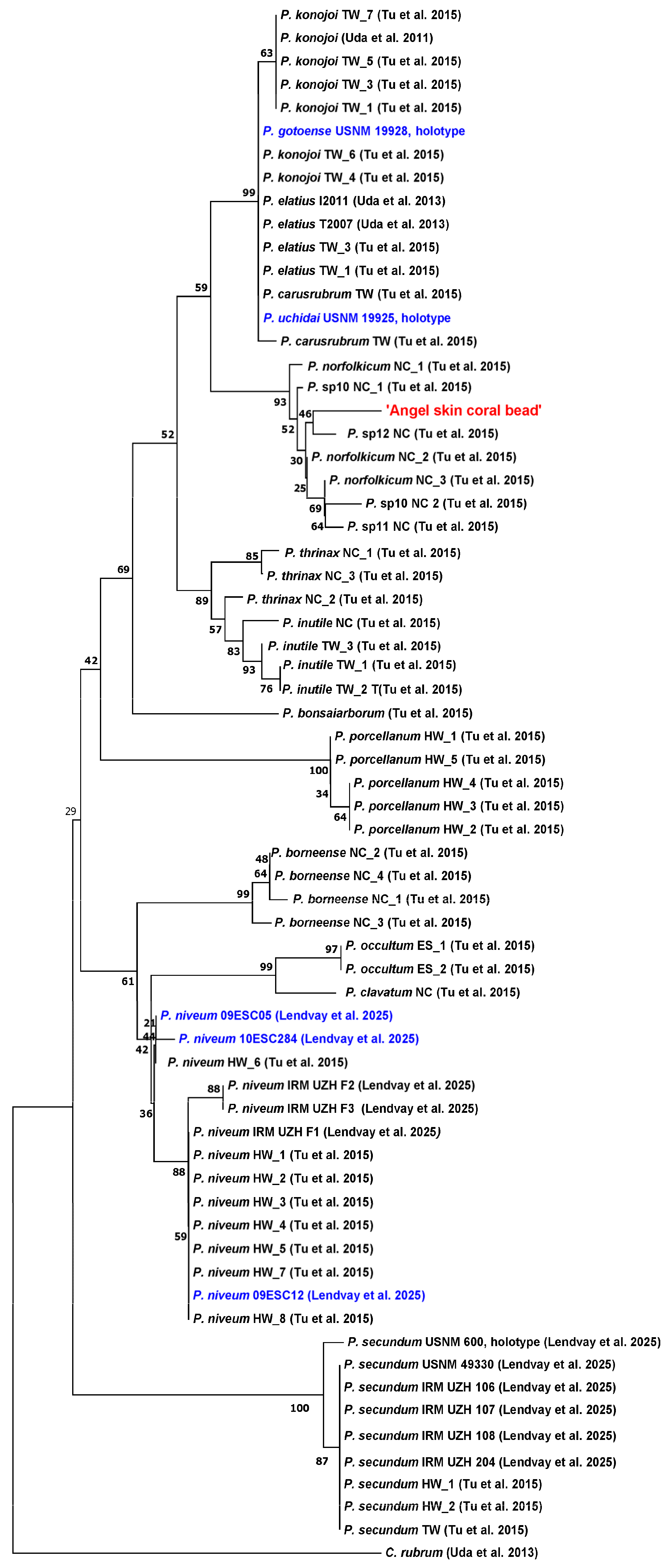
3.3. Comparing the DNA of an ‘Angel Skin’ Coral Bead with an Extended Pleurocorallium Reference Data Set
| Species | Species Author | Distribution Area | No. of Reference Samples | Base-Pair Differences from the ‘Angel Skin’ Coral Necklace | |
|---|---|---|---|---|---|
| IGS-mtMutS-LR | Only LR | ||||
| P. bonsaiarborum | Tu, Dai, and Jeng 2016 | South-West Pacific | 1 | 106 | 4 |
| P. borneense | Bayer 1950 | North-West Pacific | 4 | 122–124 | 4 |
| P. carusrubrum | Tu, Dai, and Jeng 2012 | North-West Pacific | 1 | 85 | 4 |
| P. clavatum | Tu, Dai, and Jeng 2016 | North-West Pacific | 1 | 136 | 5 |
| P. elatius | Ridley 1882 | North-West Pacific | 5 | 82–84 | 4 |
| P. gotoense | Nonaka et al. 2012 | North-West Pacific | 1 | 83 | 4 |
| P. inutile | Kishinouye 1902 | North-West Pacific | 4 | 275–313 | 3–4 |
| P. johnsoni | Gray 1860 | North-East Atlantic | n.a. | n.a. | |
| P. konojoi | Kishinouye 1903 | North-West Pacific | 7 | 82–84 | 4 |
| P. niveum | Bayer 1956 | Central North Pacific | 14 | 114–118 | 1–4 |
| P. norfolkicum | Tu, Dai, and Jeng 2016 | South-West Pacific | 3 | 6–7 | 1–2 |
| P. occultum | Tu, Altuna, and Jeng 2015 | North-East Atlantic | 2 | 127 | 4 |
| P. porcellanum | Pasternak 1981 | Central North Pacific | 5 | 135–137 | 5 |
| P. pusillum | Kishinouye 1903 | North-West Pacific | n.a. | n.a. | |
| P. secundum | Dana 1846 | Central and North-West Pacific | 9 | 129 | 7 |
| P. thrinax | Bayer and Stefani, 1996 | South-West Pacific | 3 | 274–281 | 3–4 |
| P. uchidai | Nonaka et al. 2012 | North-West Pacific | 1 | 83 | 4 |
| P. sp10 | sensu Tu, Dai, and Jeng 2015 | South-West Pacific | 2 | 8–16 | 1–2 |
| P. sp11 | sensu Tu, Dai, and Jeng 2015 | South-West Pacific | 1 | 18 | 2 |
| P. sp12 | sensu Tu, Dai, and Jeng 2015 | South-West Pacific | 1 | 18 | 1 |
| Corallium sp1 | sensu Ardila et al. 2012 | South-West Pacific | 4 | n.a. | 5 |
| P. sp1 | sensu Ardila et al. 2012 | South-West Pacific | 3 | n.a. | 2–4 |
| P. sp2 | sensu Ardila et al. 2012 | South-West Pacific | 1 | n.a. | 4 |
3.4. Comparison of the ‘Angel Skin’ Coral Necklace’s DNA with Additional Samples
4. Discussion
4.1. Taxonomic and Geographic Origin Assignment of the ‘Angel Skin’ Coral Necklace
4.2. Taxonomic Inferences of the Newly Analyzed Taxa
4.3. General Considerations for the International Coral Trade
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- CIBJO. CIBJO Guides. Precious Coral; Coral Commission of the World Jewellery Confederation: Milano, Italy, 2024. [Google Scholar]
- Tu, T.-H.; Dai, C.-F.; Jeng, M.-S. Precious corals (Octocorallia: Coralliidae) from the Northern West Pacific region with descriptions of two new species. Zootaxa 2012, 3395, 1–17. [Google Scholar] [CrossRef]
- Lendvay, B.; Cartier, L.E.; Costantini, F.; Iwasaki, N.; Everett, M.V.; Krzemnicki, M.S.; Kratzer, A.; Morf, N.V. Coral-ID: A forensically validated genetic test to identify precious coral material and its application to objects seized from illegal traffic. Forensic Sci. Int. Genet. 2022, 58, 102663. [Google Scholar] [CrossRef] [PubMed]
- de Carvalho, R.G. Understanding Precious Coral Jewelry. In: Obsessed by Pearls, Assael. 2020. Available online: https://assael.com/blog/understanding-precious-corals (accessed on 4 May 2025).
- Branstrator, B. 5 Things to Know About … Coral. National Jeweler. 2021. Available online: https://nationaljeweler.com/articles/9973-5-things-to-know-about-coral (accessed on 4 May 2025).
- O’Steen, D. Defining Exceptional—The Beauty & Luxury of Angel Skin Coral. In: Obsessed by Pearls, Assael. 2018. Available online: https://assael.com/blog/defining-exceptional-the-beauty-luxury-of-angel-skin-coral (accessed on 4 May 2025).
- CIBJO. The Coral Book; Coral Commission of The World Jewellery Confederation: Milano, Italy, 2020. [Google Scholar]
- Cooper, E.W.T.; Torntore, S.J.; Leung, A.S.M.; Shadbolt, T.; Dawe, C. Guide to the Identification of Precious and Semi-Precious Corals: In Commercial Trade; TRAFFIC North America and WWF: Vancouver, BC, Canada, 2011. [Google Scholar]
- Lendvay, B.; Morf, N.V.; Cartier, L.E.; Krzemnicki, M.S.; Nonaka, M. Trace-DNA from a century-old holotype specimen resolves taxonomic uncertainties: The case of the Hawaiian pink precious coral (Pleurocorallium secundum), a CITES-listed species used in jewelry. Coral Reefs 2025, in press. [Google Scholar]
- Bersani, D.; Lottici, P.P. Applications of Raman spectroscopy to gemology. Anal. Bioanal. Chem. 2010, 397, 2631–2646. [Google Scholar] [CrossRef]
- Bergamonti, L.; Bersani, D.; Csermely, D.; Lottici, P.P. The nature of the pigments in corals and pearls: A contribution from Raman spectroscopy. Spectrosc Let 2011, 44, 453–458. [Google Scholar] [CrossRef]
- Macchia, M.; Resta, V.; Quarta, G.; Calcagnile, L. Precious coral non-destructive characterization by Raman and XRF spectroscopy. X-Ray Spectrom 2016, 45, 281–287. [Google Scholar] [CrossRef]
- Karampelas, S.; Fritsch, E.; Rondeau, B.; Andouche, A.; Métivier, B. Identification of the endangered pink-to-red Stylaster corals by Raman spectroscopy. Gems Gemol. 2009, 45, 48–52. [Google Scholar] [CrossRef]
- Smith, C.P.; McClure, S.F.; Eaton-Magaña, S.; Kondo, D.M. Pink-to-red coral: A guide to determining origin of color. Gems Gemol 2007, 43, 4–15. [Google Scholar] [CrossRef]
- Lendvay, B.; Cartier, L.E.; Gysi, M.; Meyer, J.B.; Krzemnicki, M.S.; Kratzer, A.; Morf, N.V. DNA fingerprinting: An effective tool for taxonomic identification of precious corals in jewelry. Sci. Rep. 2020, 10, 8287. [Google Scholar] [CrossRef]
- Tu, T.-H.; Dai, C.-F.; Jeng, M.-S. Phylogeny and systematics of deep-sea precious corals (Anthozoa: Octocorallia: Coralliidae). Mol. Phylogenet. Evol. 2015, 84, 173–184. [Google Scholar] [CrossRef]
- Nonaka, M.; Muzik, K.; Iwasaki, N. Descriptions of two new species and designation of three neotypes of Japanese Coralliidae from recently discovered specimens that were collected by Kishinouye, and the introduction of a statistical approach to sclerite abundance and size. Zootaxa 2012, 3428, 1–67. [Google Scholar] [CrossRef]
- Kishinouye, K. Kosango. Dobutsugaku Zasshi 1903, 15, 372. [Google Scholar]
- Nonaka, M.; Hayashibara, T. A report on Coralliidae (Cnidaria: Octocorallia) specimens collected from the Emperor Seamounts with descriptions of three new species. Species Divers 2021, 26, 297–342. [Google Scholar] [CrossRef]
- Takata, K.; Iwase, F.; Iguchi, A.; Yuasa, H.; Taninaka, H.; Iwasaki, N.; Uda, K.; Suzuki, T.; Nonaka, M.; Kikuchi, T.; et al. Genome-wide SNP data revealed notable spatial genetic structure in the deep-sea precious coral Corallium japonicum. Front. Mar. Sci. 2021, 8, 667481. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef]
- Ardila, N.E.; Giribet, G.; Sánchez, J.A. A time-calibrated molecular phylogeny of the precious corals: Reconciling discrepancies in the taxonomic classification and insights into their evolutionary history. BMC Evol. Biol. 2012, 12, 246. [Google Scholar] [CrossRef]
- Linacre, A.; Gusmão, L.; Hecht, W.; Hellmann, A.P.; Mayr, W.R.; Parson, W.; Prinz, M.; Schneider, P.M.; Morling, N. ISFG: Recommendations regarding the use of non-human (animal) DNA in forensic genetic investigations. Forensic. Sci. Int. Genet. 2011, 5, 501–505. [Google Scholar] [CrossRef] [PubMed]
- Shearer, T.L.; Van Oppen, M.J.H.; Romano, S.L.; Wörheide, G. Slow mitochondrial DNA sequence evolution in the Anthozoa (Cnidaria). Mol. Ecol. 2002, 11, 2475–2487. [Google Scholar] [CrossRef]
- Takata, K.; Taninaka, H.; Nonaka, M.; Iwase, F.; Kikuchi, T.; Suyama, Y.; Nagai, S.; Yasuda, N. Multiplexed ISSR genotyping by sequencing distinguishes two precious coral species (Anthozoa: Octocorallia: Coralliidae) that share a mitochondrial haplotype. PeerJ 2019, 7, e7769. [Google Scholar] [CrossRef]
- Iwasaki, N. Asian regional report on the biology, fishery and trade of precious and semi-precious corals. In Global Report on the Biology, Fishery Trade of Precious Corals; FAO: Rome, Italy, 2019; pp. 191–254. [Google Scholar]
- Shiraishi, H. Seeing Red. Precious Coral Trade in East Asia; TRAFFIC: Tokyo, Japan, 2018. [Google Scholar]
- Bayer, F.M. Descriptions and redescriptions of the Hawaiian Octocorals collected by the US Fish Commission steamer “Albatross” (2. Gorgonacea: Scleraxonia). Pac. Sci. 1956, 10, 67–95. [Google Scholar]
- Tu, T.-H.; Dai, C.-F.; Jeng, M.-S. Taxonomic revision of Coralliidae with descriptions of new species from New Caledonia and the Hawaiian Archipelago. Mar. Biol. Res. 2016, 12, 1003–1038. [Google Scholar] [CrossRef]
- Tu, T.-H.; Altuna, A.; Jeng, M.-S. Coralliidae (Anthozoa: Octocorallia) from the INDEMARES 2010 expedition to North and Northwest Spain (Northeast Atlantic), with delimitation of a new species using both morphological and molecular approaches. Zootaxa 2015, 3926, 301–328. [Google Scholar] [CrossRef] [PubMed]
- CIBJO. Coral Guide for Customs. Classification & Identification of Coral Materials; The World Jewellery Confederation: Milano, Italy, 2017. [Google Scholar]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lendvay, B.; Cartier, L.E.; Sato, A.; Krzemnicki, M.S.; Nonaka, M.; Yasuda, N.; Takata, K.; Hayashibara, T.; Morf, N.V.; Iwasaki, N. Genetic Testing of a High-End ‘Angel Skin’ Precious Coral Necklace Identifies a Species New to the Precious Coral Trade and Potentially New to Science. Diversity 2025, 17, 395. https://doi.org/10.3390/d17060395
Lendvay B, Cartier LE, Sato A, Krzemnicki MS, Nonaka M, Yasuda N, Takata K, Hayashibara T, Morf NV, Iwasaki N. Genetic Testing of a High-End ‘Angel Skin’ Precious Coral Necklace Identifies a Species New to the Precious Coral Trade and Potentially New to Science. Diversity. 2025; 17(6):395. https://doi.org/10.3390/d17060395
Chicago/Turabian StyleLendvay, Bertalan, Laurent E. Cartier, Akitsugu Sato, Michael S. Krzemnicki, Masanori Nonaka, Nina Yasuda, Kenji Takata, Takeshi Hayashibara, Nadja V. Morf, and Nozomu Iwasaki. 2025. "Genetic Testing of a High-End ‘Angel Skin’ Precious Coral Necklace Identifies a Species New to the Precious Coral Trade and Potentially New to Science" Diversity 17, no. 6: 395. https://doi.org/10.3390/d17060395
APA StyleLendvay, B., Cartier, L. E., Sato, A., Krzemnicki, M. S., Nonaka, M., Yasuda, N., Takata, K., Hayashibara, T., Morf, N. V., & Iwasaki, N. (2025). Genetic Testing of a High-End ‘Angel Skin’ Precious Coral Necklace Identifies a Species New to the Precious Coral Trade and Potentially New to Science. Diversity, 17(6), 395. https://doi.org/10.3390/d17060395






