Dynamics of Phyto- and Bacterioplankton in Southern Baikal and Irkutsk Reservoir During the Open Water Period of 2023 According to Metabarcoding Data
Abstract
1. Introduction
2. Materials and Methods
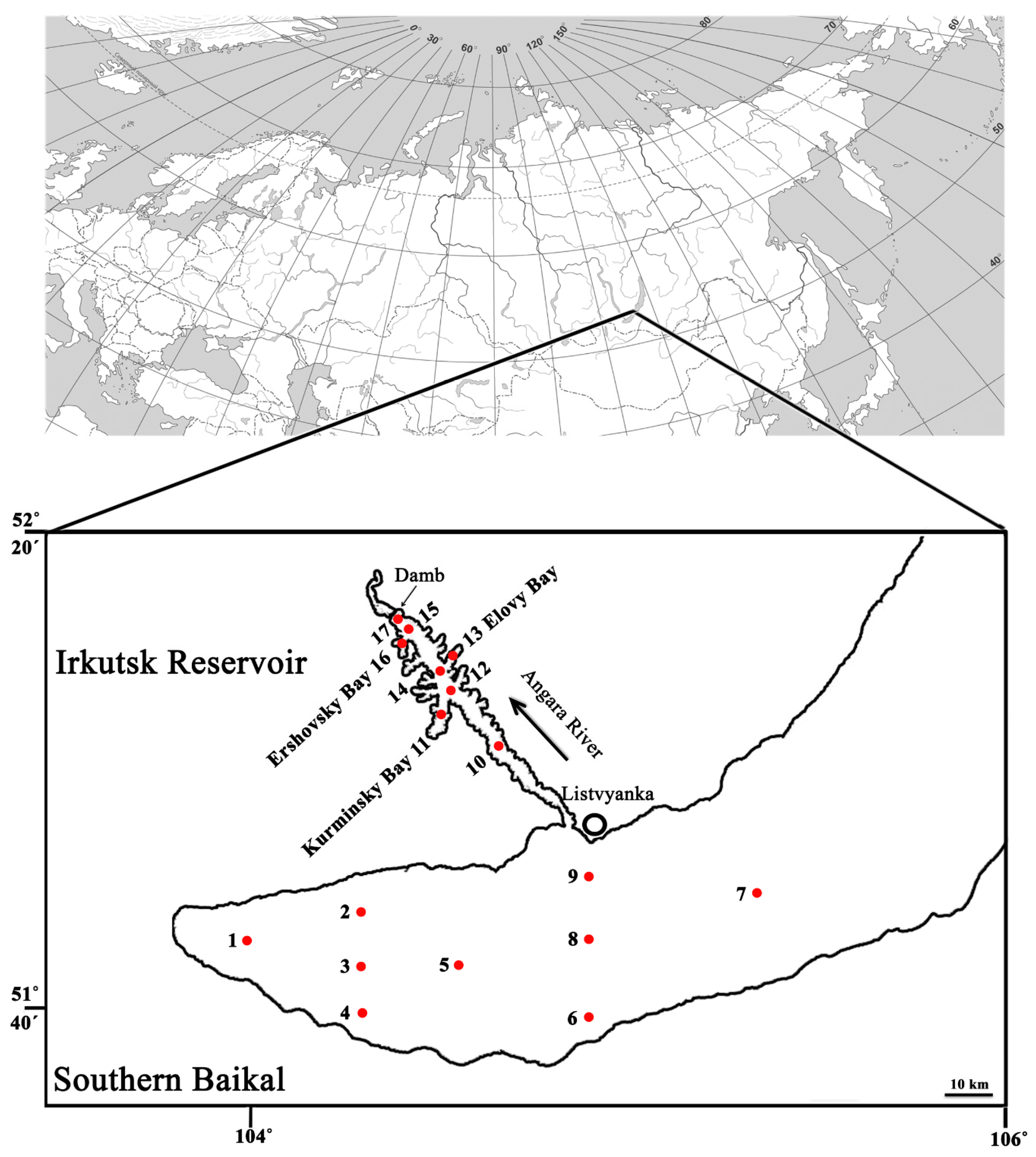
2.1. Sampling and Environmental Parameters
2.2. Amplicon Library Preparation, Sequencing, Raw Data Processing, and Quality Control
2.3. Bioinformatics, Statistical Analyses, and Data Visualization
3. Results
3.1. Difference of Environmental Factors and General Diversity Metrics Observed for Bacterioplankton and Microeukaryotic Communities
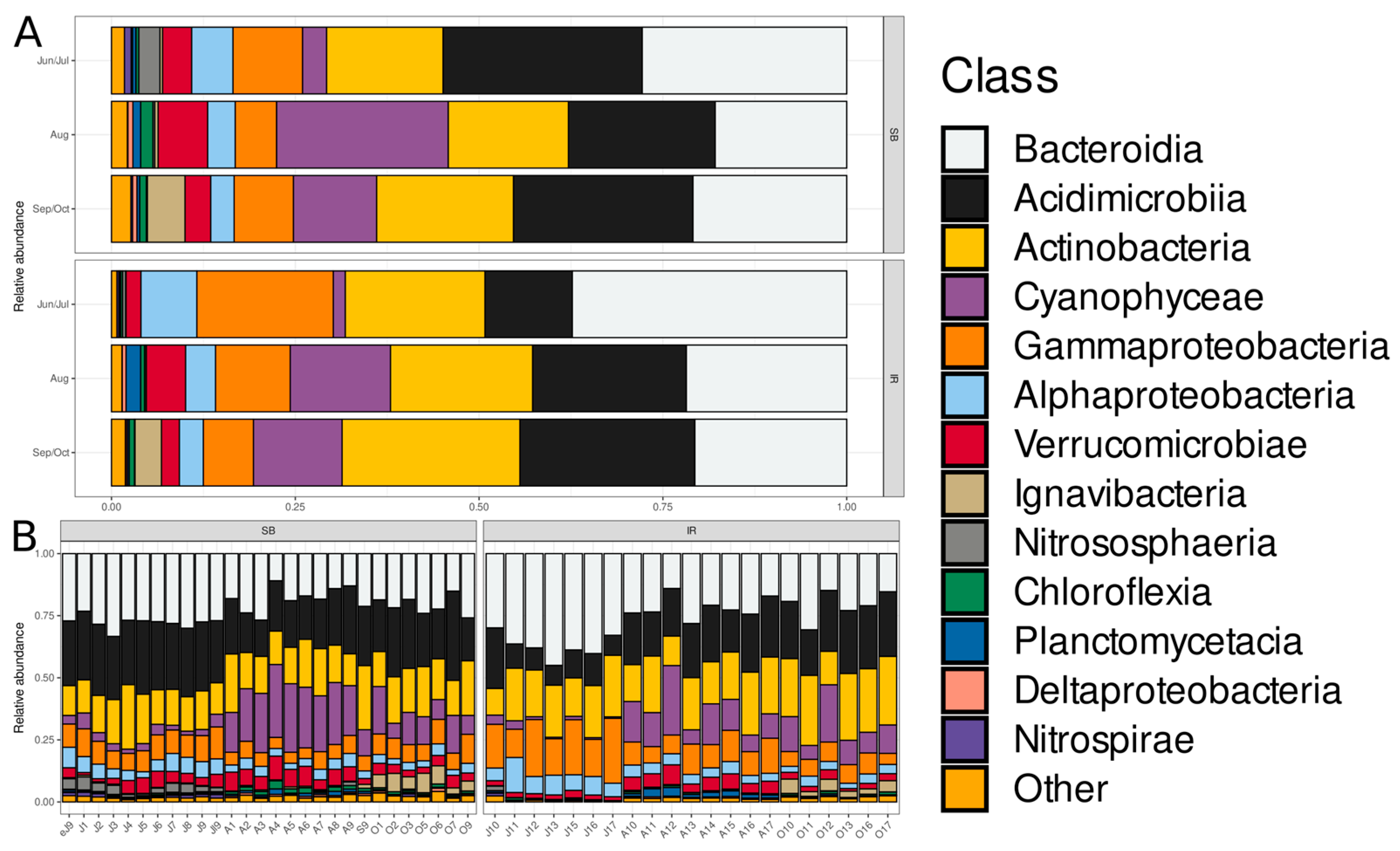
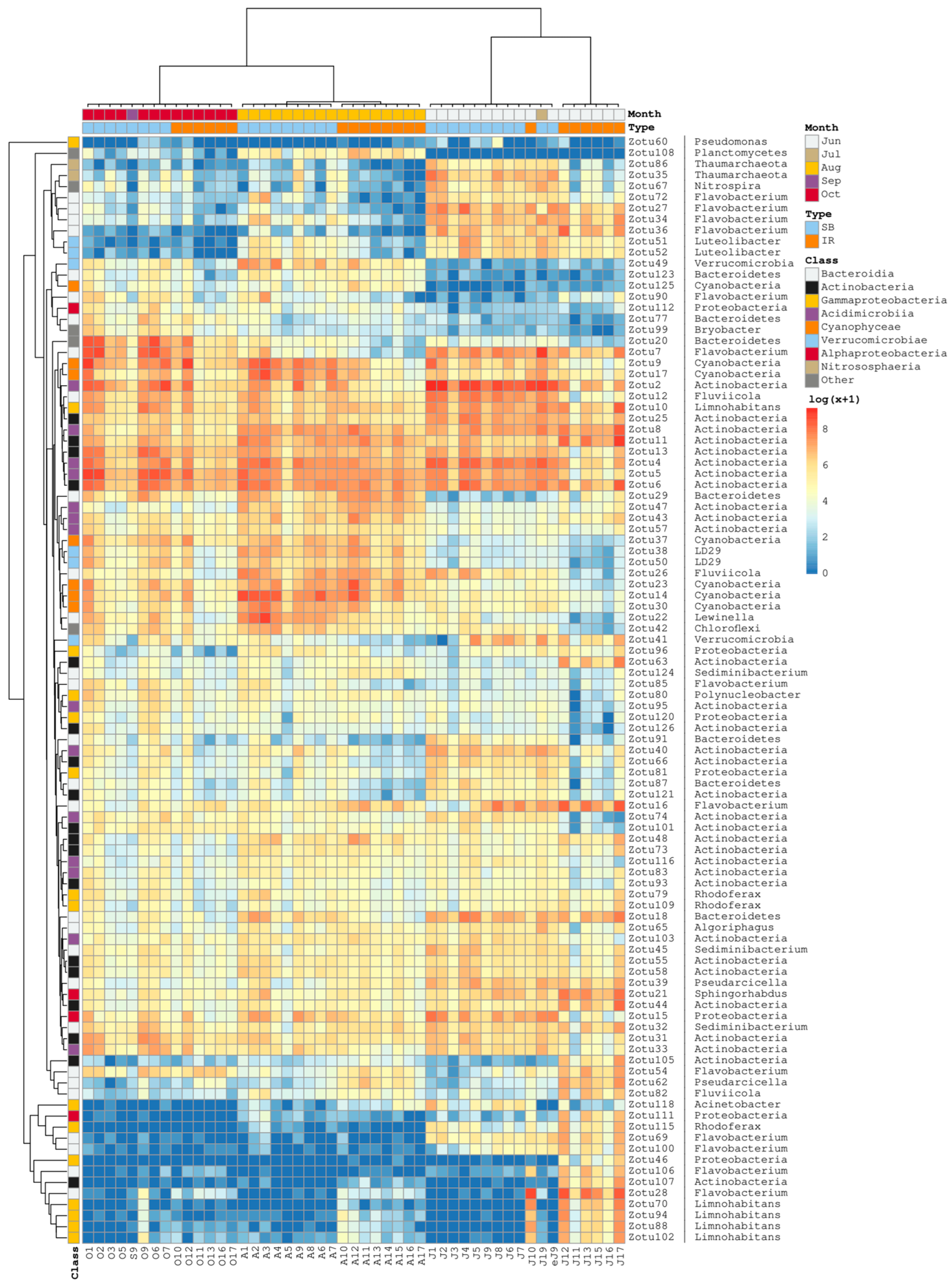
3.2. Taxonomic Composition and Dynamics of Bacterioplankton Communities
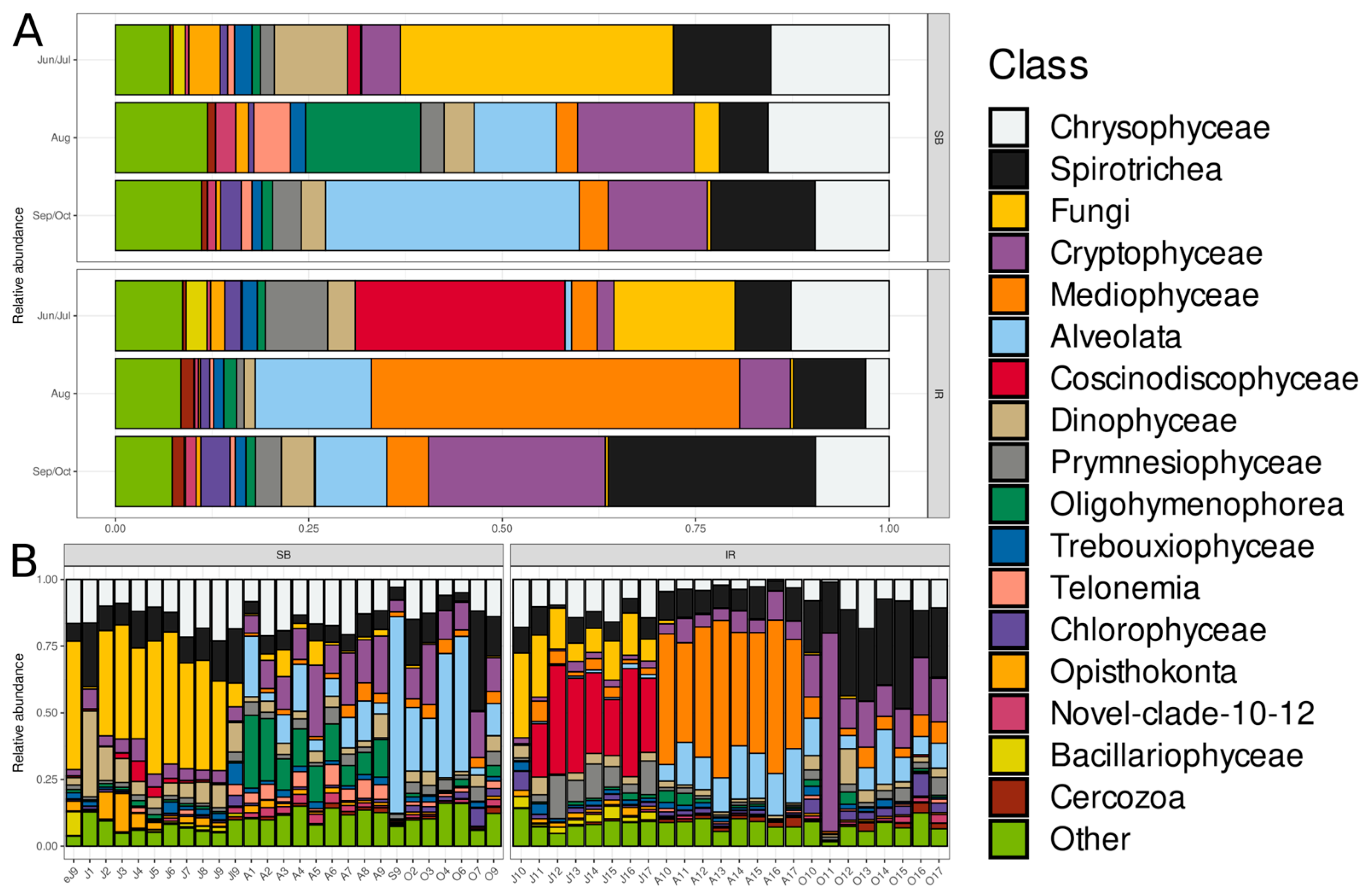
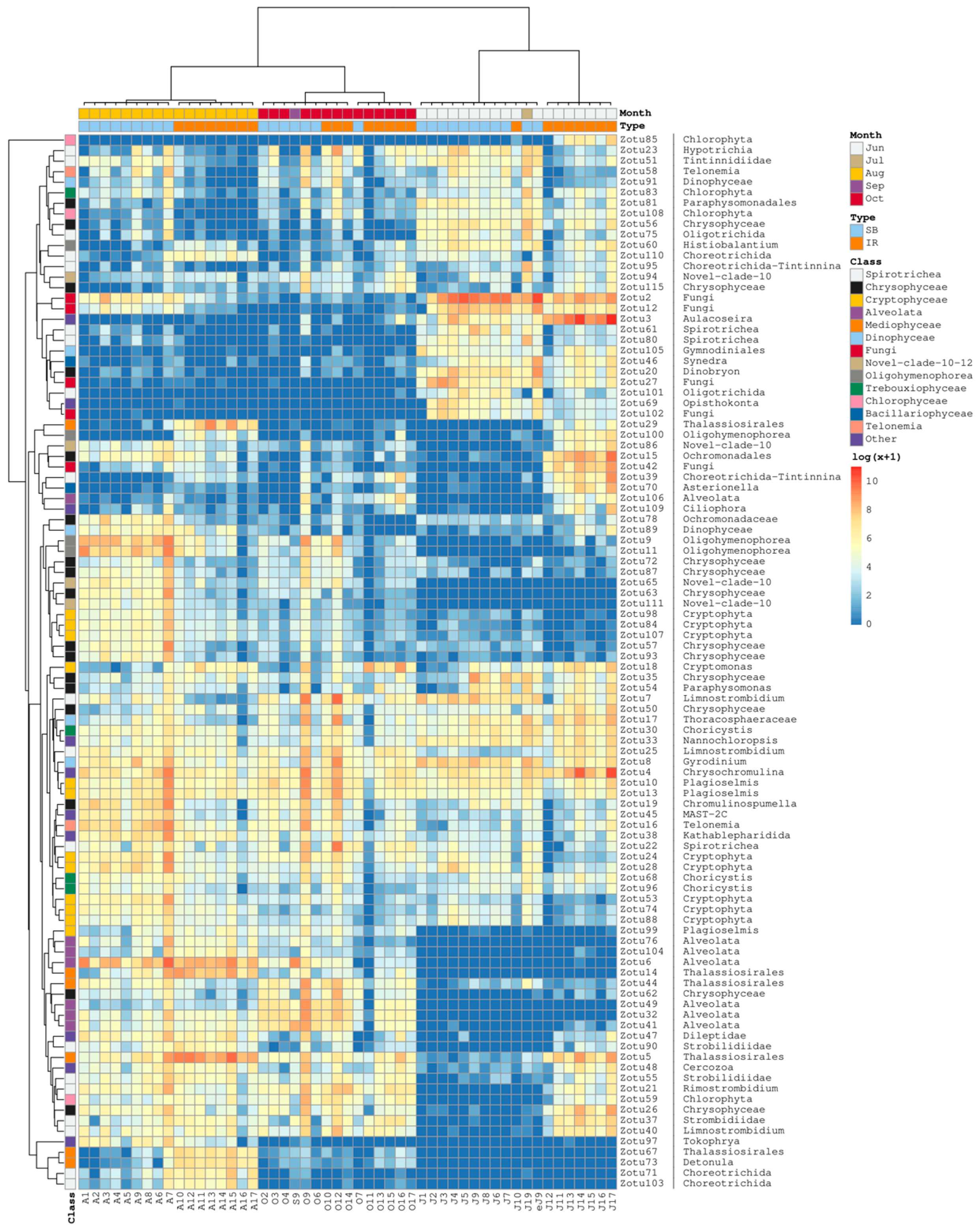
3.3. Taxonomic Composition and Dynamics of Microeukaryotic Communities
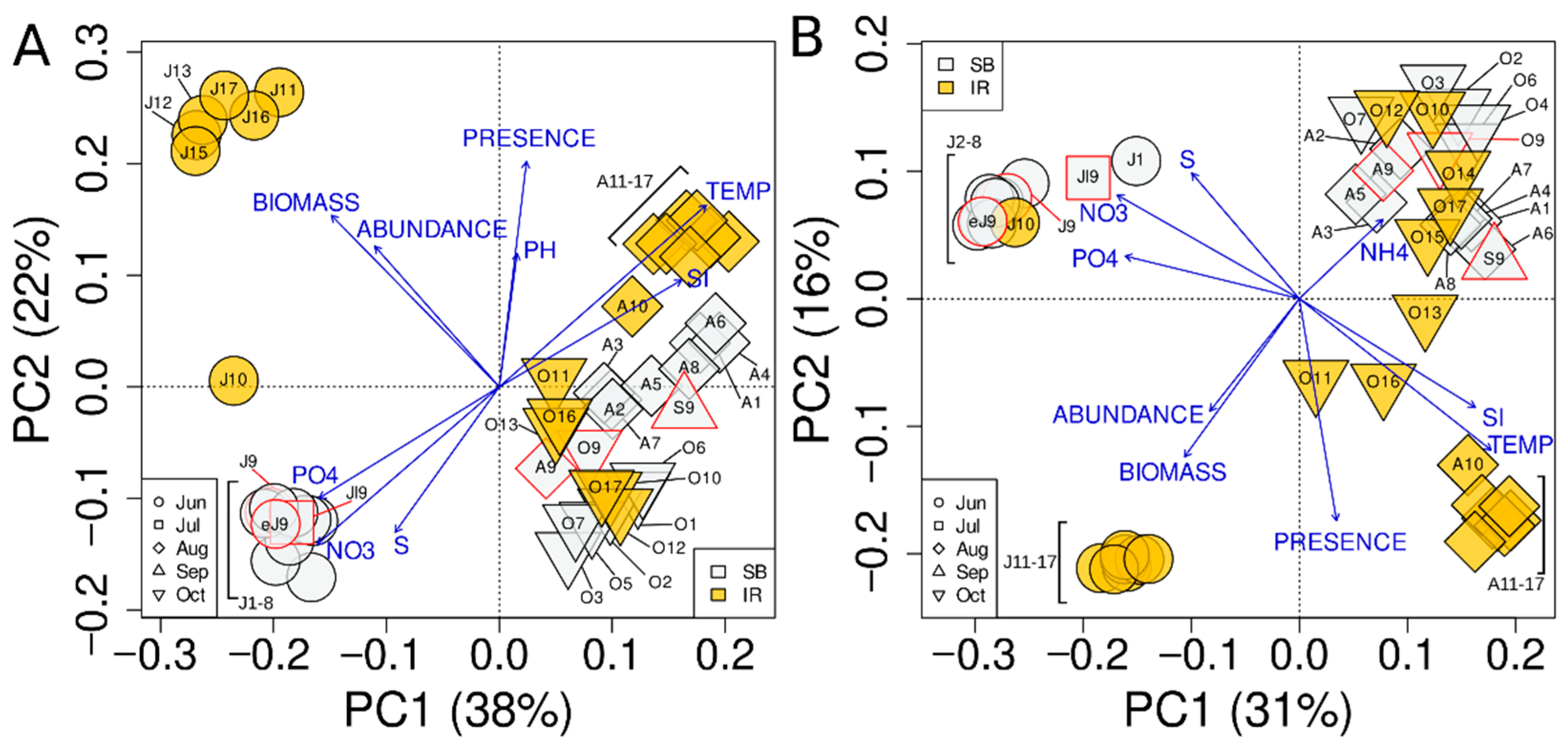
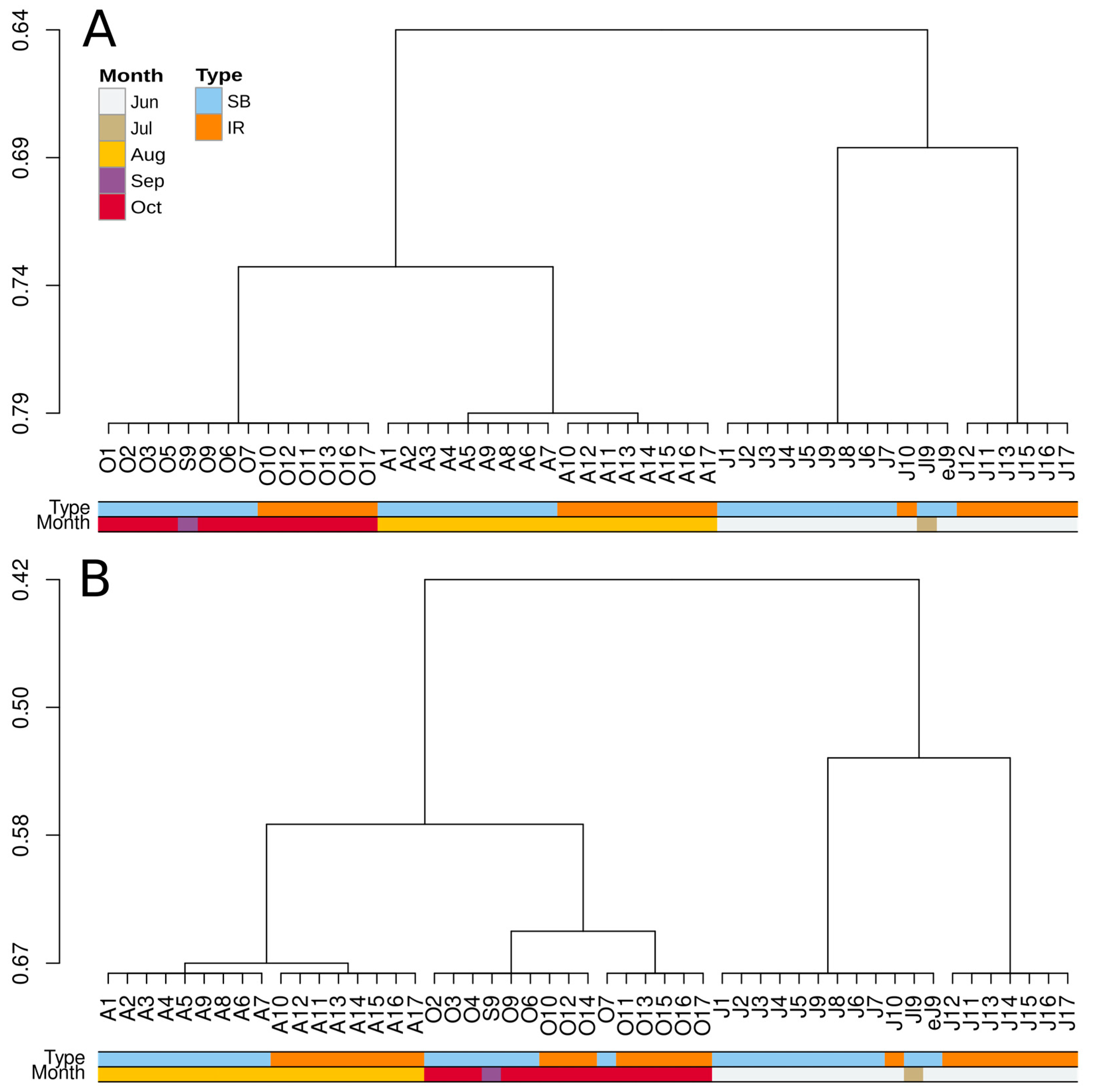
3.4. Ordination and Clustering of the Community Profiles
3.5. Bacterioplankton ASV Sequence Matches Against SSU rRNA Reference Databases in Terms of Environmental, Ecological, and Geographical Context
3.6. Microeukaryotic ASV Sequence Matches Against SSU rRNA Reference Databases in Terms of Environmental, Ecological, and Geographical Context
3.7. Dynamics of Cyanobacterial Abundance
3.8. Dynamics of Diatom Abundance Across Different Classes
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| ANOVA | analysis of variance |
| ASV | amplicon sequencing variant |
| FDR | false-discovery rate |
| HTS | high-throughput sequencing |
| IR | Irkutsk Reservoir |
| OTU | operational taxonomic unit |
| PCA | principal component analysis |
| RDA | redundancy analysis |
| SB | Southern Baikal |
References
- Banerji, A.; Bagley, M.; Elk, M.; Pilgrim, E.; Marinson, J.; Domingo, J.S. Spatial and temporal dynamics of a freshwater eukaryotic plankton community revealed via 18S rRNA gene metabarcoding. Hydrobiologia 2018, 818, 71–86. [Google Scholar] [CrossRef] [PubMed]
- Choi, T.-J.; Malik, A.; An, H.-E.; Kim, J.-I.; Do, T.D.; Kim, C.-B. Seasonal diversity of microeukaryotes in the Han River, Korea through 18S rRNA gene metabarcoding. Evol. Bioinform. Online 2022, 18, 1–10. [Google Scholar] [CrossRef]
- David, G.M.; Moreira, D.; Reboul, G.; Annenkova, N.V.; Galindo, L.J.; Bertolino, P.; López-Archilla, A.I.; Jardillier, L.; López-García, P. Environmental drivers of plankton protist communities along latitudinal and vertical gradients in the oldest and deepest freshwater lake. Environ. Microbiol. 2021, 23, 1436–1451. [Google Scholar] [CrossRef]
- Lin, Y.; Wenjun, Z.W.; Zhang, X.; Zhou, X.; He, L.; Lv, J.; Zhao, Z. Environmental DNA metabarcoding revealed the impacts of anthropogenic activities on phytoplankton diversity in Dianchi Lake and its three inflow rivers. Ecol. Evol. 2023, 13, e10088. [Google Scholar] [CrossRef]
- Santoferrara, L.; Burki, F.; Filker, S.; Logares, R.; Dunthorn, M.; Mcmanus, G.B. Perspectives from Ten Years of Protist Studies by High-Throughput Metabarcoding. J. Eukaryot. Microbiol. 2020, 67, 612–622. [Google Scholar] [CrossRef]
- Martin, J.L.; Santi, I.; Pitta, P.; John, U.; Gypens, N. Towards quantitative metabarcoding of eukaryotic plankton: An approach to improve 18S rRNA gene copy number bias. MBMG 2022, 6, 245–259. [Google Scholar] [CrossRef]
- Latz, M.A.C.; Grujcic, V.; Brugel, S.; Lycken, J.; Jphn, U.; Karlson, B.; Andersson, A.; Andersson, A.F. Short- and long- read metabarcoding of the eukaryotic rRNA operon: Evaluation of primers and comparison to shotgun metagenomics sequencing. Mol. Ecol. Resour. 2022, 22, 2304–2318. [Google Scholar] [CrossRef] [PubMed]
- Pierella Karlusich, J.J.; Lombard, F.; Irisson, J.-O.; Bowler, C.; Foster, R.A. Coupling Imaging and Omics in Plankton Surveys: State-of-the-Art, Challenges, and Future Directions. Front. Mar. Sci. 2022, 9, 878803. [Google Scholar] [CrossRef]
- Vaulot, D.; Sim, C.W.H.; Ong, D.; Teo, B.; Biwer, C.; Jamy, M.; Lopes Dos Santos, A. metaPR2: A database of eukaryotic 18S rRNA metabarcodes with an emphasis on protists. Mol. Ecol. Resour. 2022, 22, 3188–3201. [Google Scholar] [CrossRef]
- Skopina, M.Y.; Kuznetzova, E.V.; Tikhonenkov, D.V.; Zhang, H.; Gerasimova, E.A.; Tsvetkov, A.I.; Kosolapov, D.B. Distribution, community structure and metabolic potential of bacterioplankton in a small boreal lake: Microscopy-based survey and 16S rRNA-based DNA metabarcoding. Limnologica 2024, 109, 126213. [Google Scholar] [CrossRef]
- Romero, P.E.; Calla-Quispe, E.; Castillo Vilcahuaman, C.; Yokoo, M.; Fuentes-Rivera, H.L.; Ramirez, J.L.; Ampuero, A.; Ibáñez, A.J.; Wong, P. From the Andes to the desert: 16S rRNA metabarcoding characterization of aquatic bacterial communities in the Rimac River, the main source of water for Lima, Peru. PLoS ONE 2021, 16, e0250401. [Google Scholar] [CrossRef] [PubMed]
- Mikhailov, I.S.; Galachyants, Y.P.; Bukin, Y.S.; Petrova, D.P.; Bashenkhaeva, M.V.; Sakirko, M.V.; Blinov, V.V.; Titova, L.A.; Zakharova, Y.R.; Likhoshway, Y.V. Seasonal Succession and Coherence Among Bacteria and Microeukaryotes in Lake Baikal. Microb. Ecol. 2022, 84, 404–422. [Google Scholar] [CrossRef] [PubMed]
- Umanskaya, M.V.; Gorbunov, M.Y.; Bykova, S.V.; Tarasova, N.G. Diversity and Transformation of the Freshwater Planktonic Protist Community Along the Estuarine Tributary Zone of the Large Plainland Reservoir: 18S RRNA Gene Metabarkoding. Biol. Bull. 2023, 50, 707–723. [Google Scholar] [CrossRef]
- Umanskaya, V.; Gorbunov, M.Y.; Krasnova, Y.S.; Tarasova, N.G. Microscopic examination and 16S metabarcoding for comparative analysis of cyanobacteria community structure in a plainland reservoir. Biosphere 2023, 15, 246–260. (In Russian) [Google Scholar]
- Umanskaya, M.V.; Gorbunov, M.Y. Phylogenetic structure of bacterioplankton in water bodies of the Kuibyshev Reservoir basin during the period of mass development of cyanobacteria. Microbiology 2024, 93, 876–890. [Google Scholar] [CrossRef]
- Yasnitskiy, V.N. Plankton of Lake Baikal in the area of Kultuk and Slyudyanka. Izv. ESB RGS 1924, 47, 147–152. (In Russian) [Google Scholar]
- Kozhova, O.M. Phytoplankton of the Irkutsk reservoir. In Biology of the Irkutsk Reservoir; Nauka: Moscow, Russia, 1964; pp. 41–114. (In Russian) [Google Scholar]
- Vorobyova, S.S. Phytoplankton of Angara Reservoirs; Nauka Siberian Publishing Company of the RAS: Novosibirsk, Russia, 1995; 126p. (In Russian) [Google Scholar]
- Firsova, A.D.; Galachyants, Y.; Bessudova, A.Y.; Titova, L.A.; Sakirko, M.V.; Marchenkov, A.M.; Hilkhanova, D.V.; Nalimova, M.A.; Buzevich, V.V.; Mikhailov, I.S.; et al. Environmental factors affecting distribution and diversity of phytoplankton in the Irkutsk Reservoir ecosystem in June 2023. Diversity 2023, 15, 1070. [Google Scholar] [CrossRef]
- Firsova, A.; Galachyants, Y.; Bessudova, A.; Mikhailov, I.; Titova, L.; Marchenkov, A.; Hilkhanova, D.; Nalimova, M.; Buzevich, V.; Likhoshway, Y. Summer phytoplankton species composition and abundance in the southern basin of Lake Baikal and Irkutsk Reservoir. LFWB 2023, 6, 204–228. [Google Scholar] [CrossRef]
- Firsova, A.; Galachyants, Y.; Bessudova, A.; Hilkhanova, D.; Titova, L.; Nalimova, M.; Buzevich, V.; Marchenkov, A.; Sakirko, M.; Likhoshway, Y. The Influence of Waters of Lake Baikal on the Spatiotemporal Dynamics of Phytoplankton in the Irkutsk Reservoir. Water 2024, 16, 3284. [Google Scholar] [CrossRef]
- Rusch, D.B.; Halpern, A.L.; Sutton, G.; Heidelberg, K.B.; Williamson, S.; Yooseph, S.; Wu, D.; Eisen, J.A.; Hoffman, J.M.; Remington, K.; et al. The Sorcerer II Global Ocean Sampling Expedition: Northwest Atlantic through Eastern Tropical Pacific. PLoS Biol. 2007, 5, e77. [Google Scholar] [CrossRef]
- Bukin, Y.S.; Mikhailov, I.S.; Petrova, D.P.; Galachyants, Y.P.; Zakharova, Y.R.; Likhoshway, Y.V. The effect of metabarcoding 18S rRNA region choice on diversity of microeukaryotes including phytoplankton. World J. Microbiol. Biotechnol. 2023, 39, 1–18. [Google Scholar] [CrossRef]
- Baker, G.C.; Smith, J.J.; Cowan, D.A. Review and re-analysis of domain-specific 16S primers. J. Microbiol. Methods 2003, 55, 541–555. [Google Scholar] [CrossRef]
- Bradley, I.M.; Pinto, A.J.; Guest, J.S. Design and evaluation of Illumina MiSeq-compatible, 18S rRNA gene-specific primers for improved characterization of mixed phototrophic communities. Appl. Environ. Microbiol. 2016, 82, 5878–5891. [Google Scholar] [CrossRef]
- Edgar, R.C. Search and clustering orders of magnitude faster than BLAST. Bioinformatics 2010, 26, 2460–2461. [Google Scholar] [CrossRef] [PubMed]
- Rognes, T.; Flouri, T.; Nichols, B.; Quince, C.; Mahe, F. VSEARCH: A versatile open source tool for metagenomics. PeerJ 2016, 4, e2584. [Google Scholar] [CrossRef]
- Schloss, P.D.; Westcott, S.L.; Ryabin, T.; Hall, J.R.; Hartmann, M.; Hollister, E.B.; Lesniewski, R.A.; Oakley, B.B.; Parks, D.H.; Robinson, C.J. Introducing mothur: Open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 2009, 75, 7537–7541. [Google Scholar] [CrossRef] [PubMed]
- Guillou, L.; Bachar, D.; Audic, S.; Bass, D.; Berney, C.; Bittner, L.; Boutte, C.; Burgaud, G.; de Vargas, C.; Decelle, J.; et al. The Protist Ribosomal Reference database (PR2): A catalog of unicellular eukaryote Small Sub-Unit rRNA sequences with curated taxonomy. Nucleic Acids Res. 2013, 41, D597–D604. [Google Scholar] [CrossRef]
- Pruesse, E.; Peplies, J.; Glöckner, F.O. SINA: Accurate high-throughput multiple sequence alignment of ribosomal RNA genes. Bioinformatics 2012, 28, 1823–1829. [Google Scholar] [CrossRef]
- Oksanen, J.; Simpson, G.L.; Blanchet, G.F.; Kindt, R.; Legendre, P.; Minchin, P.R.; O’Hara, R.B.; Solymos, P.; Stevens, M.H.H.; Szoecs, E.; et al. Vegan: Community Ecology Package. R Package Version 2.5–6. 2022. Available online: https://CRAN.R-project.org/package=vegan (accessed on 24 November 2022).
- McMurdie, P.J.; Holmes, S. phyloseq: An R package for reproducible interactive analysis and graphics of microbiome census data. PLoS ONE 2013, 8, e61217. [Google Scholar] [CrossRef]
- Kolde, R. _Pheatmap: Pretty Heatmaps_. R Package Version 1.0.12. 2019. Available online: https://CRAN.R-project.org/package=pheatmap (accessed on 16 May 2025).
- Bodenhofer, U.; Kothmeier, A.; Hochreiter, S. APCluster: An R package for affinity propagation clustering. Bioinformatics 2011, 27, 2463–2464. [Google Scholar] [CrossRef]
- Andersson, A.; Zhao, L.; Brugel, S.; Figueroa, D.; Huseby, S. Metabarcoding vs Microscopy: Comparison of methods to monitor phytoplankton communities. ACS EST Water 2023, 3, 2671–2680. [Google Scholar] [CrossRef]
- Martens, N.; Russnak, V.; Woudhouse, J.; Grossart, H.-P.; Schaum, E. Metabarcoding reveals potentially mixotrophic flagellates and picophytoplankton as key groups of phytoplankton in the Elbe estuary. Environ. Res. 2024, 252, 119126. [Google Scholar] [CrossRef]
- Bessudova, A.; Galachyants, Y.; Firsova, A.; Hilkhanova, D.; Nalimova, M.; Marchenkov, A.; Mikhailov, I.; Sakirko, M.; Likhoshway, Y. Changes in diversity of silica-scaled Chrysophytes during lake–river–reservoir transition (Baikal–Angara–Irkutsk Reservoir). Life 2023, 13, 2052. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.-S.; Yun, H.-S.; Lee, J.-H.; Lee, K.-H.; Chpi, J.-S.; Won, D.H.; Kim, Y.J.; Kim, H.-S.; Yoon, H.-S. Comparison of Metabarcoding and Microscopy Methodologies to Analyze Diatom Communities in Five Estuaries Along the Southern Coast of the Korean Peninsula. Microb. Ecol. 2024, 87, 95. [Google Scholar] [CrossRef]
- Urbach, E.K.; Vergin, L.; Young, L.; Morse, A.; Larson, G.L.; Giovannoni, S.J. Unusual bacterioplankton community structure in ultra-oligotrophic Crater Lake. Limnol. Oceanogr. 2001, 46, 557–572. [Google Scholar] [CrossRef]
- Kaluzhnaya, O.V.; Itskovich, V.B.; McCormack, G.P. Phylogenetic diversity of bacteria associated with the endemic freshwater sponge Lubomirskia baicalensis. World J. Microbiol. Biotechnol. 2011, 27, 1955–1959. [Google Scholar] [CrossRef]
- Clingenpeel, S.; Macur, R.E.; Kan, J.; Inskeep, W.P.; Lovalvo, D.; Varley, J.; Mathur, E.; Nealson, K.; Gorby, Y.; Jiang, H.; et al. Yellowstone Lake: High-energy geochemistry and rich bacterial diversity. Environ. Microbiol. 2011, 13, 2172–2185. [Google Scholar] [CrossRef]
- Allgaier, M.; Grossart, H.P. Diversity and seasonal dynamics of Actinobacteria populations in four lakes in northeastern Germany. Appl. Environ. Microbiol. 2006, 72, 3489–3497. [Google Scholar] [CrossRef]
- Warnecke, F.; Amann, R.; Pernthaler, J. Actinobacterial 16S rRNA genes from freshwater habitats cluster in four distinct lineages. Environ. Microbiol. 2004, 6, 242–253. [Google Scholar] [CrossRef]
- Brinkmeyer, R.; Knittel, K.; Jürgens, J.; Weyland, H.; Amann, R.; Helmke, E. Diversity and structure of bacterial communities in Arctic versus Antarctic pack ice. Appl. Environ. Microbiol. 2003, 69, 6610–6619. [Google Scholar] [CrossRef]
- Van den Wyngaert, S.; Salcher, M.M.; Pernthaler, J.; Zeder, M.; Posch, T. Quantitative dominance of seasonally persistent filamentous cyanobacteria (Planktothrix rubescens) in the microbial assemblages of a temperate lake. Limnol. Oceanog. 2011, 56, 97–109. [Google Scholar] [CrossRef]
- Cheng, S.M.; Foght, J.M. Cultivation-independent and -dependent characterization of Bacteria resident beneath John Evans Glacier. FEMS Microbiol. Ecol. 2007, 59, 318–330. [Google Scholar] [CrossRef]
- Crump, B.C.; Kling, G.W.; Bahr, M.; Hobbie, J.E. Bacterioplankton community shifts in an arctic lake correlate with seasonal changes in organic matter source. Appl. Environ. Microbiol. 2003, 69, 2253–2268. [Google Scholar] [CrossRef] [PubMed]
- Newton, R.J.; Jones, S.E.; Helmus, M.R.; McMahon, K.D. Phylogenetic ecology of the freshwater Actinobacteria acI lineage. Appl. Environ. Microbiol. 2007, 73, 7169–7176. [Google Scholar] [CrossRef] [PubMed]
- Xing, P.; Hahn, M.W.; Wu, Q.L. Low taxon richness of bacterioplankton in high-altitude lakes of the eastern tibetan plateau, with a predominance of Bacteroidetes and Synechococcus spp. Appl. Environ. Microbiol. 2009, 75, 7017–7025. [Google Scholar] [CrossRef]
- Van Trappen, S.; Mergaert, J.; Van Eygen, S.; Dawyndt, P.; Cnockaert, M.C.; Swings, J. Diversity of 746 heterotrophic bacteria isolated from microbial mats from ten Antarctic lakes. Syst. Appl. Microbiol. 2002, 25, 603–610. [Google Scholar] [CrossRef]
- Zwart, G.; Hiorns, W.D.; Methé, B.A.; van Agterveld, M.P.; Huismans, R.; Nold, S.C.; Zehr, J.P.; Laanbroek, H.J. Nearly identical 16S rRNA sequences recovered from lakes in North America and Europe indicate the existence of clades of globally distributed freshwater bacteria. Syst. Appl. Microbiol. 1998, 21, 546–556. [Google Scholar] [CrossRef]
- Urbach, E.; Scanlan, D.J.; Distel, D.L.; Waterbury, J.B.; Chisholm, S.W. Rapid diversification of marine picophytoplankton with dissimilar light-harvesting structures inferred from sequences of Prochlorococcus and Synechococcus (Cyanobacteria). J. Mol. Evol. 1998, 46, 188–201. [Google Scholar] [CrossRef]
- Wolf, A.; Wiese, J.; Jost, G.; Witzel, K.P. Wide geographic distribution of bacteriophages that lyse the same indigenous freshwater isolate (Sphingomonas sp. strain B18). Appl. Environ. Microbiol. 2003, 69, 2395–2398. [Google Scholar] [CrossRef]
- Pernthaler, J.; Zöllner, E.; Warnecke, F.; Jürgens, K. Bloom of filamentous bacteria in a mesotrophic lake: Identity and potential controlling mechanism. Appl. Environ. Microbiol. 2004, 70, 6272–6281. [Google Scholar] [CrossRef]
- Simpson, J.M.; Santo Domingo, J.W.; Reasoner, D.J. Assessment of equine fecal contamination: The search for alternative bacterial source-tracking targets. FEMS Microbiol. Ecol. 2004, 47, 65–75. [Google Scholar] [CrossRef] [PubMed]
- Zwart, G.J.M.; Crump, B.C.; Agterveld, M.; Hagen, F.; Han, S.K. Typical freshwater bacteria: An analysis of available 16S rRNA gene sequences from plankton of lakes and rivers. Aquat. Microb. Ecol. 2002, 28, 141–155. [Google Scholar] [CrossRef]
- Blunt, S.M.; Sackett, J.D.; Rosen, M.R.; Benotti, M.J.; Trenholm, R.A.; Vanderford, B.J.; Hedlund, B.P.; Moser, D.P. Association between degradation of pharmaceuticals and endocrine-disrupting compounds and microbial communities along a treated wastewater effluent gradient in Lake Mead Sci. Total Environ. 2017, 622, 1640–1648. [Google Scholar] [CrossRef] [PubMed]
- Crosbie, N.D.; Pöckl, M.; Weisse, T. Dispersal and phylogenetic diversity of nonmarine picocyanobacteria, inferred from 16S rRNA gene and cpcBA-intergenic spacer sequence analyses. Appl. Environ. Microbiol. 2023, 69, 5716–5721. [Google Scholar] [CrossRef]
- Simek, K.; Pernthaler, J.; Weinbauer, M.G.; Hornák, K.; Dolan, J.R.; Nedoma, J.; Masín, M.; Amann, R. Changes in bacterial community composition and dynamics and viral mortality rates associated with enhanced flagellate grazing in a mesoeutrophic reservoir. Appl. Environ. Microbiol. 2001, 67, 2723–2733. [Google Scholar] [CrossRef]
- Stein, L.Y.; Jones, G.; Alexander, B.; Elmund, K.; Wright-Jones, C.; Nealson, K.H. Intriguing microbial diversity associated with metal-rich particles from a freshwater reservoir. FEMS Microbiol. Ecol. 2002, 42, 431–440. [Google Scholar] [CrossRef]
- Tamaki, H.; Hanada, S.; Kamagata, Y.; Nakamura, K.; Nomura, N.; Nakano, K.; Matsumura, M. Flavobacterium limicola sp. nov., a psychrophilic, organic-polymer-degrading bacterium isolated from freshwater sediments. Int. J. Syst. Evol. Microbiol. 2003, 53, 519–526. [Google Scholar] [CrossRef]
- Eder, W.; Schmidt, M.; Koch, M.; Garbe-Schönberg, D.; Huber, R. Prokaryotic phylogenetic diversity and corresponding geochemical data of the brine-seawater interface of the Shaban Deep, Red Sea. Environ. Microbiol. 2002, 4, 758–763. [Google Scholar] [CrossRef] [PubMed]
- Zwart, G.; Huismans, R.; van Agterveld, M.P.; Van de Peer, Y.; De Rijk, P.; Eenhoorn, H.; Muyzer, G.; van Hannen, E.J.; Gons, H.J.; Laanbroek, H.J. Divergent members of the bacterial division Verrucomicrobiales in a temperate freshwater lake. FEMS Microbiol. Ecol. 1998, 25, 159–169. [Google Scholar] [CrossRef]
- Balmonte, J.P.; Arnosti, C.; Underwood, S.; McKee, B.A.; Teske, A. Riverine Bacterial Communities Reveal Environmental Disturbance Signatures within the Betaproteobacteria and Verrucomicrobia. Front. Microbiol. 2016, 7, 1441. [Google Scholar] [CrossRef]
- Reardon, C.L.; Cummings, D.E.; Petzke, L.M.; Kinsall, B.L.; Watson, D.B.; Peyton, B.M.; Geesey, G.G. Composition and diversity of microbial communities recovered from surrogate minerals incubated in an acidic uranium-contaminated aquifer. Appl. Environ. Microbiol. 2004, 70, 6037–6046. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Jeon, C.O.; Park, W.; Padmanabhan, P.; DeRito, C.; Snape, J.R.; Madsen, E.L. Discovery of a bacterium, with distinctive dioxygenase, that is responsible for in situ biodegradation in contaminated sediment. Proc. Natl. Acad. Sci. USA 2003, 100, 13591–13596. [Google Scholar] [CrossRef] [PubMed]
- Nelson, D.M.; Ohene-Adjei, S.; Hu, F.S.; Cann, I.K.; Mackie, R.I. Bacterial diversity and distribution in the holocene sediments of a northern temperate lake. Microb. Ecol. 2007, 54, 252–263. [Google Scholar] [CrossRef]
- Matsubayashi, M.; Shimada, Y.; Li, Y.Y.; Harada, H.; Kubota, K. Phylogenetic diversity and in situ detection of eukaryotes in anaerobic sludge digesters. PLoS ONE 2017, 12, e0172888. [Google Scholar] [CrossRef]
- Lefèvre, E.; Bardot, C.; Noël, C.; Carrias, J.F.; Viscogliosi, E.; Amblard, C.; Sime-Ngando, T. Unveiling fungal zooflagellates as members of freshwater picoeukaryotes: Evidence from a molecular diversity study in a deep meromictic lake. Environ. Microbiol. 2007, 9, 61–71. [Google Scholar] [CrossRef]
- Medlin, L.K.; Kaczmarska, I. Evolution of the diatoms: V. Morphological and cytological support for the major clades and a taxonomic revision. Phycologia 2006, 43, 245–270. [Google Scholar] [CrossRef]
- Edvardsen, B.; Eikrem, W.; Throndsen, J.; Sáez, A.G.; Probert, I.; Medlin, L.K. Ribosomal DNA phylogenies and a morphological revision provide the basis for a revised taxonomy of the Prymnesiales (Haptophyta). Eur. J. Psychol. 2011, 46, 202–228. [Google Scholar] [CrossRef]
- Ki, J.S. Comparative molecular analysis of freshwater centric diatoms with particular emphasis on the nuclear ribosomal DNA of Stephanodiscus (Bacillariophyceae). Algae 2009, 24, 129–138. [Google Scholar] [CrossRef]
- Mukherjee, I.; Hodoki, Y.; Nakano, S. Kinetoplastid flagellates overlooked by universal primers dominate in the oxygenated hypolimnion of Lake Biwa, Japan. FEMS Microbiol. Ecol. 2015, 91, fiv083. [Google Scholar] [CrossRef]
- Rossi, A.; Boscaro, V.; Carducci, D.; Serra, V.; Modeo, L.; Verni, F.; Fokin, S.I.; Petroni, G. Ciliate communities and hidden biodiversity in freshwater biotopes of the Pistoia province (Tuscany, Italy). Eur. J. Protistol. 2016, 53, 11–19. [Google Scholar] [CrossRef]
- Charvet, S.; Vincent, W.F.; Lovejoy, C. Chrysophytes and other protists in High Arctic lakes: Molecular gene surveys, pigment signatures and microscopy. Polar Biol. 2012, 35, 733–748. [Google Scholar] [CrossRef]
- Gentekaki, E.; Lynn, D.H. Evidence for cryptic speciation in Carchesium polypinum Linnaeus, 1758 (Ciliophora: Peritrichia) inferred from mitochondrial, nuclear, and morphological markers. J. Eukaryot. Microbiol. 2010, 57, 508–519. [Google Scholar] [CrossRef]
- Monchy, S.; Sanciu, G.; Jobard, M.; Rasconi, S.; Gerphagnon, M.; Chabé, M.; Cian, A.; Meloni, D.; Niquil, N.; Christaki, U.; et al. Exploring and quantifying fungal diversity in freshwater lake ecosystems using rDNA cloning/sequencing and SSU tag pyrosequencing. Environ. Microbiol. 2011, 13, 1433–1453. [Google Scholar] [CrossRef]
- Dunthorn, M.; Stoeck, T.; Wolf, K.; Breiner, H.-W.; Foissner, W. Diversity and endemism of ciliates inhabiting Neotropical phytotelmata. Syst. Biodivers. 2012, 10, 195–205. [Google Scholar] [CrossRef]
- Rad-Menéndez, C.; Stanley, M.; Green, D.H.; Cox, E.J.; Day, J.G. Exploring cryptic diversity in publicly available strains of the model diatom Thalassiosira pseudonana (Bacillariophyceae). J. Mar. Biol. Assoc. United Kingd. 2015, 95, 1081–1090. [Google Scholar] [CrossRef]
- Andersen, R.A. Molecular systematics of the Chrysophyceae and Synurophyceae. In Unravelling the Algae—The Past, Present, and Future of Algal Systematics, Systematics Association Special Volume; Brodie, J., Lewis, J., Eds.; CRC Press: Boca Raton, FL, USA, 2007; pp. 285–313. [Google Scholar]
- Luo, W.; Bock, C.; Li, H.R.; Padisak, J.; Krienitz, L. Molecular and microscopic diversity of planktonic eukaryotes in the oligotrophic Lake Stechlin (Germany). Hydrobiologia 2011, 661, 133–143. [Google Scholar] [CrossRef]
- Oikonomou, A.; Katsiapi, M.; Karayanni, H.; Moustaka-Gouni, M.; Kormas, K.A. Plankton microorganisms coinciding with two consecutive mass fish kills in a newly reconstructed lake. Sci. World J. 2012, 2012, 504135. [Google Scholar] [CrossRef]
- Choi, B.; Son, M.; Kim, J.I.; Shin, W. Taxonomy and phylogeny of the genus Cryptomonas (Cryptophyceae, Cryptophyta) from Korea. Algae 2013, 28, 307–330. [Google Scholar] [CrossRef]
- Andersen, R.A.; Van de Peer, Y.; Potter, D.; Sexton, J.P.; Kawachi, M.; LaJeunesse, T. Phylogenetic analysis of the SSU rRNA from members of the Chrysophyceae. Protist 1999, 150, 71–84. [Google Scholar] [CrossRef]
- Richards, T.A.; Vepritskiy, A.A.; Gouliamova, D.E.; Nierzwicki-Bauer, S.A. The molecular diversity of freshwater picoeukaryotes from an oligotrophic lake reveals diverse, distinctive and globally dispersed lineages. Environ Microbiol. 2005, 7, 1413–1425. [Google Scholar] [CrossRef]
- Kahn, P.; Herfort, L.; Peterson, T.D.; Zuber, P. Discovery of a Katablepharis sp. in the Columbia River estuary that is abundant during the spring and bears a unique large ribosomal subunit sequence element. MicrobiologyOpen 2014, 3, 764–776. [Google Scholar] [CrossRef] [PubMed]
- Weisse, T.; Strüder-Kypke, M.C.; Berger, H.; Foissner, W. Genetic, morphological, and ecological diversity of spatially separated clones of Meseres corlissi Petz & Foissner, 1992 (Ciliophora, Spirotrichea). J. Eukaryot Microbiol. 2008, 55, 257–270. [Google Scholar] [CrossRef]
- Tarbe, A.; Stenuite, S.; Balagué, V.; Sinyinza, D.; Descy, J.; Massana, R. Molecular characterisation of the small-eukaryote community in a tropical Great Lake (Lake Tanganyika, East Africa). Aquat. Microb. Ecol. 2011, 62, 177–190. [Google Scholar] [CrossRef][Green Version]
- Gao, F.; Li, J.; Song, W.; Xu, D.; Warren, A.; Yi, Z.; Gao, S. Multi-gene-based phylogenetic analysis of oligotrich ciliates with emphasis on two dominant groups: Cyrtostrombidiids and strombidiids (Protozoa, Ciliophora). Mol. Phylogenet. Evol. 2016, 105, 241–250. [Google Scholar] [CrossRef]
- Karpov, S.A.; Tcvetkova, V.S.; Mamkaeva, M.A.; Torruella, G.; Timpano, H.; Moreira, D.; Mamanazarova, K.S.; López-García, P. Morphological and Genetic Diversity of Opisthosporidia: New Aphelid Paraphelidium tribonemae gen. et sp. Nov. J. Eukaryot. Microbiol. 2017, 64, 204–212. [Google Scholar] [CrossRef] [PubMed]
- Lefranc, M.; Thénot, A.; Lepère, C.; Debroas, D. Genetic diversity of small eukaryotes in lakes differing by their trophic status. Appl. Environ. Microbiol. 2005, 71, 5935–5942. [Google Scholar] [CrossRef]
- Tuji, A.; Mohri, Y.; Ki, J.-S.; Jung, S.W.; Julius, M.L. Phylogeny of Praestephanos gen. nov. (Thalassiosirales, Bacillariophyceae) based on Stephanodiscus suzukii, and related freshwater thalassiosiroid diatoms. Plankton Benthos Res. 2014, 9, 132–140. [Google Scholar] [CrossRef][Green Version]
- Kaczmarska, I.; Beaton, M.; Benoit, A.C.; Medlin, L.K. Molecular phylogeny of selected members of the order Thalassiosirales (Bacillariophyta) and evolution of the fultoportula. J. Phycol. 2006, 42, 121–138. [Google Scholar] [CrossRef]
- Yamamoto, M.; Nozaki, H.; Kawano, S. Evolutionary relationships among multiple modes of cell division in the genus Nannochloris (Chlorophyta) revealed by genome size, actin gene multiplicity, and phylogeny. J. Phycol. 2001, 37, 106–120. [Google Scholar] [CrossRef]
- Pröschold, T.; Darienko, T.; Silva, P.C.; Reisser, W.; Krienitz, L. The systematics of Zoochlorella revisited employing an integrative approach. Environ. Microbiol. 2011, 13, 350–364. [Google Scholar] [CrossRef]
- Bhattacharya, D.; Medlin, L.; Wainright, P.O.; Ariztia, E.V.; Bibeau, C.; Stickel, S.K.; Sogin, M.L. Algae containing chlorophylls a + c are paraphyletic: Molecular evolutionary analysis of the CHROMOPHYTA. Evolution 1993, 47, 1801–1817. [Google Scholar] [CrossRef]
- Jeong, M.; Kim, J.I.; Nam, S.W.; Shin, W. Molecular Phylogeny and Taxonomy of the Genus Spumella (Chrysophyceae) Based on Morphological and Molecular Evidence. Front. Plant Sci. 2021, 26, 758067. [Google Scholar] [CrossRef]
- Song, W.; Xu, D.; Zhang, Q.; Liu, W.; Warren, A.; Song, W. Taxonomy and phylogeny of two poorly studied genera of marine oligotrich ciliates including descriptions of two new species: Cyrtostrombidium paraboreale sp. N. and Apostrombidium orientale sp. N. (Ciliophora: Spirotrichea). Eur. J. Protistol. 2019, 70, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Bachy, C.; Gómez, F.; López-García, P.; Dolan, J.R.; Moreira, D. Molecular phylogeny of tintinnid ciliates (Tintinnida, Ciliophora). Protist 2012, 163, 873–887. [Google Scholar] [CrossRef]
- Nakai, R.; Abe, T.; Baba, T.; Imura, S.; Kagoshima, H.; Kanda, H.; Kohara, Y.; Koi, A.; Niki, H.; Yanagihara, K.; et al. Eukaryotic phylotypes in aquatic moss pillars inhabiting a freshwater lake in East Antarctica, based on 18S rRNA gene analysis. Polar Biol. 2012, 35, 1495–1504. [Google Scholar] [CrossRef]
- Newbold, L.K.; Oliver, A.E.; Booth, T.; Tiwari, B.; Desantis, T.; Maguire, M.; Andersen, G.; van der Gast, C.J.; Whiteley, A.S. The response of marine picoplankton to ocean acidification. Environ. Microbiol. 2012, 14, 2293–2307. [Google Scholar] [CrossRef]
- Medlin, L.; Jung, I.; Bahulikar, R.; Mendgen, K.; Kroth, P.; Kooistra, W.H.C.F. Evolution of the diatoms. VI. Assessment of the new genera in the araphids using molecular data. Nova Hedwig. 2008, 133, 81–100. [Google Scholar]
- Vd’ačný, P.; Bourland, W.A.; Orsi, W.; Epstein, S.S.; Foissner, W. Phylogeny and classification of the Litostomatea (Protista, Ciliophora), with emphasis on free-living taxa and the 18S rRNA gene. Mol. Phylogenet Evol. 2011, 59, 510–522. [Google Scholar] [CrossRef]
- Chen, M.; Chen, F.; Yu, Y.; Ji, J.; Kong, F. Genetic diversity of eukaryotic microorganisms in Lake Taihu, a large shallow subtropical lake in china. Microb. Ecol. 2008, 56, 572–583. [Google Scholar] [CrossRef]
- Boenigk, J.; Pfandl, K.; Stadler, P.; Chatzinotas, A. High diversity of the ‘Spumella-like’ flagellates: An investigation based on the SSU rRNA gene sequences of isolates from habitats located in six different geographic regions. Environ. Microbiol. 2005, 7, 685–697. [Google Scholar] [CrossRef]
- Atkins, M.S.; Teske, A.P.; Anderson, O.R. A survey of flagellate diversity at four deep-sea hydrothermal vents in the Eastern Pacific Ocean using structural and molecular approaches. J. Eukaryot. Microbiol. 2000, 47, 400–411. [Google Scholar] [CrossRef] [PubMed]

| IndepVar1 | IndepVar2 | DepVar | ANOVA | Kruskal–Wallis | ||
|---|---|---|---|---|---|---|
| padj | padj Sign | padj | padj Sign | |||
| Environmental variables | ||||||
| SB | Month | Si | 4.27 × 10−2 | * | 4.22 × 10−2 | * |
| IR | Month | Si | 1.76 × 10−6 | *** | 1.69 × 10−3 | ** |
| SB | Month | PO43− | 2.57 × 10−5 | *** | 2.42 × 10−3 | ** |
| SB | Month | NO2− | 3.60 × 10−2 | * | 2.48 × 10−2 | * |
| IR | Month | NO2− | 1.42 × 10−2 | * | 2.48 × 10−2 | * |
| SB | Month | NO3− | 3.65 × 10−6 | *** | 1.69 × 10−3 | ** |
| SB | Month | NH4+ | 3.61 × 10−2 | * | 2.48 × 10−2 | * |
| IR | Month | NH4+ | 1.52 × 10−3 | ** | 6.30 × 10−3 | ** |
| SB | Month | S | 3.25 × 10−3 | ** | 4.57 × 10−2 | * |
| SB | Month | pH | 6.76 × 10−2 | . | 2.48 × 10−2 | * |
| IR | Month | pH | 4.75 × 10−4 | *** | 5.49 × 10−3 | ** |
| SB | Month | Temp | 1.94 × 10−5 | *** | 1.04 × 10−3 | ** |
| IR | Month | Temp | 1.76 × 10−6 | *** | 2.79 × 10−3 | ** |
| SB | Month | abundance | 2.56 × 10−1 | 7.37 × 10−2 | . | |
| IR | Month | abundance | 4.15 × 10−5 | *** | 3.62 × 10−3 | ** |
| SB | Month | biomass | 2.21 × 10−2 | * | 2.79 × 10−3 | ** |
| IR | Month | biomass | 1.65 × 10−4 | *** | 2.77 × 10−3 | ** |
| Alpha-diversity of bacterial communities | ||||||
| SB | Month | ACE | 8.89 × 10−3 | ** | 1.87 × 10−2 | * |
| Alpha-diversity of microeukaryotic communities | ||||||
| SB | Month | Observed | 3.14 × 10−2 | * | 2.58 × 10−2 | * |
| SB | Month | ACE | 1.11 × 10−2 | * | 1.47 × 10−2 | * |
| SB | Month | Shannon | 4.75 × 10−3 | ** | 1.20 × 10−2 | * |
| IR | Month | Shannon | 6.06 × 10−1 | 2.80 × 10−2 | * | |
| SB | Month | Simpson | 1.89 × 10−2 | * | 2.15 × 10−2 | * |
| IR | Month | Simpson | 8.88 × 10−1 | 2.57 × 10−2 | * | |
| SB | Month | InvSimpson | 8.65 × 10−3 | ** | 2.15 × 10−2 | * |
| IR | Month | InvSimpson | 4.75 × 10−3 | ** | 2.57 × 10−2 | * |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Galachyants, Y.; Petrova, D.; Marchenkov, A.; Nalimova, M.; Likhoshway, Y. Dynamics of Phyto- and Bacterioplankton in Southern Baikal and Irkutsk Reservoir During the Open Water Period of 2023 According to Metabarcoding Data. Diversity 2025, 17, 369. https://doi.org/10.3390/d17060369
Galachyants Y, Petrova D, Marchenkov A, Nalimova M, Likhoshway Y. Dynamics of Phyto- and Bacterioplankton in Southern Baikal and Irkutsk Reservoir During the Open Water Period of 2023 According to Metabarcoding Data. Diversity. 2025; 17(6):369. https://doi.org/10.3390/d17060369
Chicago/Turabian StyleGalachyants, Yuri, Darya Petrova, Artem Marchenkov, Maria Nalimova, and Yelena Likhoshway. 2025. "Dynamics of Phyto- and Bacterioplankton in Southern Baikal and Irkutsk Reservoir During the Open Water Period of 2023 According to Metabarcoding Data" Diversity 17, no. 6: 369. https://doi.org/10.3390/d17060369
APA StyleGalachyants, Y., Petrova, D., Marchenkov, A., Nalimova, M., & Likhoshway, Y. (2025). Dynamics of Phyto- and Bacterioplankton in Southern Baikal and Irkutsk Reservoir During the Open Water Period of 2023 According to Metabarcoding Data. Diversity, 17(6), 369. https://doi.org/10.3390/d17060369







