Abstract
India and Sri Lanka are known to exhibit high levels of biological diversity with many endemic taxa, such as the enigmatic scorpion genus Charmus Karsch, 1879. Members of this genus are rarely encountered in the field and are also known to be morphologically very similar, which impedes their systematic assessment. Our dedicated efforts towards sampling the members of the genus resulted in the collection of important material, which allowed us to carry out a thorough systematic revision of the genus using an integrated taxonomic approach. We propose several taxonomic changes based on the results of a detailed morphological study supported by molecular data. Charmus indicus Hirst, 1915 is synonymized with Charmus laneus Karsch, 1879, owing to the lack of morphological differences and low genetic divergence between the two taxa. We designate a neotype for Charmus sinhagadensis Tikader and Bastawade, 1983, and describe a new species from Sirumalai (Tamil Nadu, India). Moreover, we provide the first molecular phylogeny of Charmus based on Cytochrome c Oxidase subunit I (COI), 16S rRNA (16S) and 28S rRNA (28S) genes. The phylogenetic position of the genus within the family Buthidae C. L. Koch, 1837 is also tested using an independent genome-wide dataset (Ultraconserved Elements). Topological congruence and discrepancies between the phylogenies generated with Sanger sequences and the Ultraconserved Elements are commented on, and the reliability of these datasets when evaluating phylogenetic relationships at different hierarchical levels is further discussed.
1. Introduction
India and Sri Lanka are regarded as major biodiversity hotspots [1], harboring a rich array of endemic species and exhibiting interesting biogeographical patterns of species distribution. While these patterns have been recently studied for various herpetofaunal groups [2,3,4], they remain largely untested in arachnids, likely owing to the elusive nature and narrow distribution range of many of these arthropods. In addition to the difficulty of collecting them, the lack of morphological diagnostic characters has hampered the recognition of species based on alpha-taxonomy alone, and in such cases, molecular phylogenetics is the only tool that can then help with species differentiation [5,6,7]. The humicolous scorpions of the genus Charmus Karsch, 1879 (Figure 1), endemic to India and Sri Lanka, are one of these problematic taxa: difficult to identify to species level and seldom encountered in the field. The material is thereby very rare in museum collections. In our continuous efforts to resolve the systematics of Indian scorpions, we have recently carried out extensive fieldwork to collect additional Charmus specimens across Peninsular India. A thorough integrated systematic study of the material collected during these surveys allows us to thoroughly reassess the taxonomy of this enigmatic genus.

Figure 1.
Charmus sinhagadensis Tikader & Bastawade, 1983 in vivo habitus at Sinhagad Fort, India. Scale bar: 5 mm. Photograph by Mihir Joshi.
1.1. Systematics of the Genus Charmus
The genus Charmus was first described based on the type species Charmus laneus Karsch, 1879 from Sri Lanka (formerly Ceylon) [8]. The species Charmus indicus Hirst, 1915 was described from Coimbatore (Tamil Nadu, India) based on a single immature specimen [9]. Reddy [10] subsequently redescribed C. indicus and documented its distribution in southern Peninsular India with the report of multiple new localities. However, none of the specimens examined in the scope of this redescription were collected from the designated type locality.
Subsequently, Charmus sinhagadensis Tikader & Bastawade, 1983 [11] was described from Sinhagad Fort (Pune, Maharashtra, India), making it the northernmost record of the genus. Lourenço [12] described Charmus brignolii Lourenço, 2000 from Puducherry (formerly Pondicherry, Tamil Nadu, India) and later Charmus minor Lourenço, 2002 [13] from Wilpattu National Park (Sri Lanka). Kovařík et al. [14] synonymized C. minor with the type species C. laneus Karsch, 1879 and described Charmus saradieli Kovarik et al., 2016 from the Central Province of the country, in a region that represents a transition area between dry and wet habitats.
Several taxonomic changes are proposed in the present study. Due to high morphological and molecular similarity, C. indicus is synonymized with C. laneus. Our investigations of the arachnological collections of the Zoological Survey of India (ZSI) also suggest that the holotype of C. sinhagadensis is lost, compelling us to designate a neotype following the recommendations of Article 75 of the International Code of Zoological Nomenclature (ICZN) [15]. Finally, a new species of Charmus is described from southern India.
1.2. Phylogenetic Analysis of the Genus Charmus
For a long time, morphological characteristics have served as the primary basis for species identification [16]. Nowadays, molecular phylogeny is used more often as a supplementary line of evidence for taxonomic amendments [17,18,19]. Interestingly, conflicts at supra-specific levels between modern molecular phylogenies and traditional morphology-based classifications have been repeatedly demonstrated not only in scorpions [20,21,22] but also in other arthropod groups [23,24,25,26,27]. Moreover, it appears that different molecular datasets are not informative at the same taxonomic scales [28]. The standard Sanger genetic markers used in phylogenetic and phylogeographic studies are usually adequate for resolving relationships at the species and population levels, but not at the generic level or above [29]. On the other hand, genome-wide datasets provide more signals for phylogenetic inference at higher taxonomic levels [30,31,32,33,34,35] but may perform poorly when assessing shallower nodes. For instance, phylogenetic information at shallow time depths, e.g., intraspecific levels, recovered by UCE loci depends entirely on the length of the flanking regions captured in the process [36]. Based on these considerations, the interspecific relationships within the genus Charmus as well as the position of the genus within the family Buthidae C. L. Koch, 1837 was assessed by phylogenetic analyses of two independent molecular datasets, one composed of Sanger-generated sequences, and one composed of UCE loci that are either newly generated or harvested from available transcriptomic data. Topological congruence and/or discrepancy between the two phylogenies obtained were then assessed. The reliability of the different datasets to reconstruct phylogenetic relationships at different hierarchical taxonomic levels was then discussed based on this comparison.
2. Materials and Methods
2.1. Specimen Collection and Morphological Data
Fieldwork was carried out in the Northern Western Ghats and Southern India, especially in the coastal region and in parts of Eastern Ghats. Scorpions were located with the help of ultraviolet light (uvBeast V3 385–395 nm UV Torch) at night and collected using forceps. Specimens were euthanized and preserved in absolute ethanol and later transferred to vials containing 70% ethanol for long-term preservation. Photographs of the preserved specimens were taken using a D500 (Nikon Corporation, Tokyo, Japan), 105 mm F2.8 microlens and Nikkor 60 mm f/2.8 microlens with a MF12 flash kit (Godox Photo Equipment Co. Ltd., Shenzhen, China). Specimens were examined and morphological measurements were recorded using a EZ4HD microscope (Leica Camera AG, Wetzlar, Germany) with the Leica application suite.
Trichobothrial terminology follows Vachon [37]; metasoma carination follows Francke [38]; pedipalp carination, chela dentition, and leg terminology follow González Santillán and Prendini [39]; morphological terminology follows Hjelle [40]; lateral ocelli terminology follows Loria and Prendini [41]. Surface morphology was examined and photographed under UV light after Volschenk [42]. Measurements following Stahnke [43] were taken (in mm) for 34 morphological characters, to the nearest 0.1 mm.
2.2. Repository
Specimens collected during this study are deposited in the collections of the Bombay Natural History Society (BNHS, Mumbai, Maharashtra, India) and of InSearch Environmental Solutions (IES, Pune, Maharashtra, India). Other specimens are housed in the British Museum of Natural History (BMNH, London, UK), Muséum National d’Histoire Naturelle (MNHN, Paris, France) and the Museum für Naturkunde der Humboldt Universität (ZMB, Berlin, Germany).
2.3. Taxon Sampling
Data for the Sanger sequence-based phylogeny (Table S4) are as follows. The ingroup includes the four species of Charmus known from the Indian subcontinent, i.e., C. brignolii, C. laneus, C. saradieli, C. sinhagadensis and the newly described Charmus dakshini sp. nov. Each of the five species is represented by more than one sample (2–4) except C. saradieli. Outgroup data were sourced from Štundlová, et al. [22] and are composed of 48 species belonging to 48 genera of the family Buthidae and two species of the genus Protoiurus Soleglad, Fet, Kovařík & Yağmur, 2012 (Iuridae Thorell, 1876).
Data for the UCE phylogeny (Table S5) are as follows. The ingroup is composed of three species of Charmus, i.e., C. brignolii, C. sinhagadensis and C. dakshini sp. nov. Each of the three species is represented by two distinct populations. The outgroups include sequences of two Buthoscorpio Werner, 1936, generated in the course of the present work, as well as additional UCE [44,45] and transcriptomic data [20] available to the scientific community through the Sequence Read Archive (SRA) accessible on the NCBI portal. The data downloaded from NCBI consist of a total of 35 species belonging to 22 genera of the family Buthidae, one species of the genus Chaerilus, two species belonging to two genera of the family Pseudochatidae Gromov, 1998 and one species of the genus Iurus (Iuridae Thorell, 1876).
2.4. Sanger Sequencing: DNA Extraction, Amplification and Sequencing
DNA extraction, amplification and sequencing protocols from Sulakhe et al. [46] were followed to generate data on the Indian Charmus species. A 550–600 base pair (bp) fragment of the cytochrome c oxidase subunit I (COI), a 450–500 bp fragment of 16S rRNA (16S) mitochondrial genes and a 700–750 bp fragment of 28S nuclear gene were amplified by polymerase chain reaction (PCR) using the primers (Table S7) as in previous studies [5,47,48,49,50]. All newly generated sequences were deposited at GenBank® [51].
2.5. Sequence Alignment of the Sanger Sequenced Data
Generated sequences were cleaned manually in MEGA v.7 [52], using chromatograms visualized in Chromas v.2.6.5 (Technelysium PTY. Ltd., South Brisbane, QLD, Australia). Cleaned sequences were aligned using MUSCLE [53], implemented in MEGA v.7 with default parameters. Separate alignments were built for COI, 16S, and 28S. The final COI alignment contained 62 sequences (each 525 bp long), the 16S alignment contained 55 sequences (each 359 bp long), and the 28S alignment contained 49 sequences (each 733 bp long). The COI, 16S and 28S datasets were concatenated, and the resultant 1617 bp long alignment was used for molecular phylogenetic analyses.
2.6. Genetic Divergence (p-Distance) of the Sanger Sequenced Data
Uncorrected p-distances were calculated separately for COI, 16S, and 28S in MEGA v.7. Missing data were partially deleted, and the site cut-off was set at 95%.
2.7. Molecular Phylogenetic Analysis of the Sanger Sequenced Data
Maximum Likelihood (ML) methods were implemented for phylogenetic analyses. The COI region was partitioned by codon position, whereas the non-coding 16S and 28S regions were not partitioned. Analyses were performed using the web implementation of IQ-TREE [54] and RAxML GUI [55]. For IQ-TREE, models of sequence evolution were determined using ModelFinder [56] which were as follows: TPM2 + FG4 for COI position 1, TIM3 + F + I + G4 for COI positions 2 and 3; TVM + F + I + G4 for 16S and TIM3 + F + I + G4 for 28S. One thousand ultrafast bootstraps (UFBoot) [57], were used to test nodal support [58]. The best substitution model for RAxML was determined using PartitionFinder v.1.1.1 [59]. Model search was performed with a greedy search algorithm [60], and models were selected using the Akaike Information Criterion (AIC). The dataset was partitioned, and the analysis was run using the GTR + I + G evolutionary model using 1000 bootstrap pseudo-replicates.
2.8. DNA Extractions and Shotgun Library Preparation
Total genomic DNA from the representatives of Charmus and Buthoscorpio was extracted from leg muscle tissues separately for UCE analyses using QIAamp DNA micro kits following the manufacturer’s protocol. The DNA length and quality were quantified using a Fragment Analyzer (FA). Samples with 50 to 600 bp DNA fragments were considered optimal for library preparation. Samples with longer DNA fragments were sheared using NEBNext® dsDNA Fragmentase® (New England Biolabs, Ipswitch, MA, USA) to reduce their size. The modified version of the protocols used by Suchan et al. [61], based upon Meyer et al. [62], were followed for the shotgun library preparation. The purified DNA was first phosphorylated using T4 polynucleotide kinase and T4 ligase buffer. The double-stranded DNA was further denatured at 95 °C for 5 min and then immediately chilled in water and ice mix. The guanine tail was added using the Terminal Transferase (TDT) reaction mix. A complementary strand was synthesized with Klenow Fragment and poly-C oligonucleotide. The blunt-end reaction was performed using T4 DNA polymerase and barcode adapters were ligated to the phosphorylated end opposite to the poly-C end using T4 DNA ligase. The adapter sequences were filled in using the Bst DNA Polymerase, and two PCR replicates were run using Phusion U Hot Start DNA Polymerase (Thermo Fisher Scientific, Waltham, MA, USA) and indexed PCR primers. The two PCR replicates were put together, and library concentrations were quantified in a Quant-iT PicoGreenR dsDNA reagent (Invitrogen, Thermo Fisher Scientific, Waltham, MA, USA) on a Hidex Sense Microplate Reader (Hidex Oy, Turku, Finland). For the capture, 12 equimolar pools of 8 libraries each were prepared and concentrated with an IR Micro-Cenvac NB-503CIR Vacuum Concentrator (N-Biotek, Bucheon, Republic of Korea).
2.9. Hybridization Capture and Sequencing of UCEs
Hybridization capture for the enrichment of shotgun libraries followed the myBaits protocol (Arbor Biosciences, Ann Arbor, MI, USA) using the UCE Arachnida 1.1Kv1 probe set [44,63]. The adaptor blockers and standard myBaits blockers were annealed to the library pools, and the resulting strands were hybridized to the arachnid probes. Two successive sets of UCE captures were performed at two different temperatures, one at 55 °C and the second at 65 °C. The second capture was performed at a higher temperature as it improved the stringency of the reaction. Library sequencing paired-end runs were performed on Illumina HiSeq X (Macrogen, Seoul, Republic of Korea) to produce 150 bp-long reads.
2.10. Data Processing and PHYLUCE Pipeline
The adapters were removed from the sequenced raw reads using a custom script. The reads were assembled using Trinity v. r2013-02-25 [64] and SPADES [65]. Contigs resulting from both assemblies were combined using a custom protocol. This allowed an increase in the number of recovered UCEs. The combined Trinity/Spades contigs were then processed using the PHYLUCE pipeline [66]. UCE loci were aligned using MAFFT [67] and trimmed with Gblocks [68,69] as implemented in the PHYLUCE pipeline. UCE loci were extracted from transcriptomic data using the PHYLUCE pipeline. The resulting individual UCE alignments were manually edited, and erroneous base pairs were treated as missing data. 87–918 UCES and 249–877 UCEs from transcriptomic data were retrieved. Each of the 1030 alignments was manually verified and potential alignment errors were corrected. The number of UCEs retrieved after sanitation of the alignments is indicated in Table S5. Four different occupancy matrices (20%, 50%, 70% and 80%) were generated with the sanitated alignments for phylogenetic analysis.
2.11. Phylogenetic Analyses of the Generated UCE Matrices
Maximum Likelihood analyses were performed with each of the assembled matrices using RAxML HPC v8.0 [70] and IQ-TREE [54] for each data matrix. For the RAxML analyses, the GTRCAT substitution model was implemented and the autoMRE option was selected, whereas, for the IQ-TREE analyses, the best-fit substitution model was automatically selected using TESTNEWMERGE as GTR + F + I + G4. One thousand ultrafast bootstrap replicates (UFBoot) [57] were used to test the nodal support [58].
3. Systematics
Family Buthidae C. L. Koch, 1837
Genus Charmus Karsch, 1879
Figure 2, Figure 3, Figure 4, Figure 5, Figure 6, Figure 7, Figure 8, Figure 9, Figure 10, Figure 11, Figure 12, Figure 13, Figure 14, Figure 15, Figure 16, Figure 17, Figure 18, Figure 19 and Figure 20, Table 1, Table 2, Table 3, Table 4, Table 5, Table 6 and Table S1–S6
Charmus Karsch, 1879: 104; Kraepelin, 1899: 39; Pocock, 1900: 31–32; Kraepelin, 1913: 131; Vachon, 1982: 79, 81; Tikader & Bastawade, 1983: 140–152; Sissom, 1990: 101; Kovařík, 1998: 120; Lourenço, 2000: 295; Kovařík, Soleglad & Fet, 2007: 201; Kovařík, 2009: 31. Heterocharmus Pocock, 1892: 46–47, type species by monotypy Heterocharmus cinctipes Pocock, 1892 (=Charmus laneus Karsch, 1879) (syn. by Kraepelin, 1899: 39; Pocock, 1900: 31) Kovařík et al., 2016: 17–29, figs. 12, 41–43, 47–119, 194, 423–426, 548, Table 1 and Table 2.
Type species: Charmus laneus Karsch, 1879
Diagnosis: Small scorpions, adult size 12 mm (male)–23.5 mm (female). Sternum type 1, subpentagonal, roughly as wide as long, exhibiting horizontal compression. Pedipalps trichobothrial pattern Aα; femur trichobothrium d2 located dorsally, patella d3 dorsal of dorsomedian carina; chela with 3 Eb trichobothria on manus. Movable finger of pedipalp longer than manus. Pectines with or without fulcra. Dentate margin of pedipalp chela movable finger with distinct granules divided into 8–9 linear rows, apical rows of 4–6 granules, and 3 terminal granules. Cheliceral fixed finger armed with two denticles on ventral surface. Tergites I–VI granular, with one clearly visible carina. Carapace granular without carinae, anterior edge with epistome present medially. Ventral surface of metasomal segments IV–V without developed carinae. Telson vesicle punctate, without subaculear tooth. Pedipalps, metasoma and telson densely hirsute. Legs III and IV with well-developed long tibial spurs, first and second tarsomeres with ventral setae.
Distribution: India and Sri Lanka.
3.1. Charmus laneus Karsch, 1879
Figure 2, Figure 3, Figure 4, Figure 10, Figure 15, Figure 16, Figure 17, Figure 18, Figure 19 and Figure 20, Table 1
Holotype (not examined): Ceylon (=Sri Lanka), leg. Hoffmeister, 1 ♀ (ZMHB No. 3051). Other material: India, Tamil Nadu, Coimbatore, 388 m, 29th June 2023, leg. Shauri Sulakhe, Shubhankar Deshpande, Mihir Joshi, 2 ♂ (IES 631, IES 633), 2 ♀ (IES 628, IES 629), one juvenile (IES 632).
Synonym:
Charmus indicus Hirst, 1915 syn. nov.
Holotype (not examined): leg. T. Bainbrigge Fletcher, ♂ (BMNH Regd. No. 1915.10.16.1). Other material (not examined): Udaygiri Fort, leg. Dr. A.P. Mathew, 1 ♂ (MNHN, Paris, R. S. 3017); Tirupati, Andhra Pradesh, 3 ♂, 1 ♀ (MNHN, Paris, R. S. 4407).
Description (♂, IES 631, measurements in Table 1):
Coloration (Figure 2A,B): body dorsal surface entirely black with dark brown stripes on the median portion of mesosoma; legs dark brown with alternate yellow and black stripes with tarsomere dark brown; metasoma almost black; pedipalp fingers dark brown. Ventral surface yellow except last mesosomal sternite black. Basal segments of chelicerae dark brown with black reticulation ending anteriorly into black transverse patch. Fingers of chelicerae blackish brown with the tip of the fingers black. Telson dark brown in color.
Carapace (Figure 3A and Figure 15A): surface with mixed and dense granulation. Carapace without carinae. Median supra-ocular area with few sparse granules. Interocular area coarsely and densely granular. A pair of median eyes situated anteriorly in the ratio 1:1.6 (ratio of median eyes to anterior margin and median eyes to posterior margin). Antero-lateral ocular tubercle granular with type 3A ocelli. Three well-developed lateral ocelli with first and third ocelli smaller than the second ocelli. Longitudinal furrow shallow anteriorly and deep posteriorly. Anterior margin finely crenulated in the median portion and straight throughout. Lateral margins weakly crenulated below lateral ocelli. Posterior margin almost entirely smooth.
Chelicerae: characteristic of the family Buthidae. Basal segments and movable fingers with tuft of short and firm setae on ventral surfaces. Dorsal surfaces smooth.
Pedipalp (Figure 2A,B and Figure 3C–I): femur with five carinae (prodorsal, retrodorsal, promedian, proventral and retroventral). All carinae crenulated. Intercarinal surfaces with mixed and sparse granulation except ventral surface smooth with a few fine granules on proximal portions. Patella almost smooth without developed carinae except promedian carina moderately developed with 7 spinoid granules. Intercarinal surfaces almost smooth. Manus smooth without carinae. Fixed fingers acarinate. Movable and fixed fingers with eight oblique rows of granules, each row (except most proximal) with one internal accessory granule. Trichobothrial pattern of type Aα, typical for the genus (chela dorsal 2, chela rerolateral 10, chela ventral 2, patella dorsal 6, patella retrolateral 7, femur dorsal 7 and femur prolateral 4).

Table 1.
Measurements and meristics of Charmus laneus Karsch, 1879 (in mm).
Table 1.
Measurements and meristics of Charmus laneus Karsch, 1879 (in mm).
| IES-631 (♂) | IES-633 (♂) | IES-628 (♀) | IES-629 (♀) | IES-632 | ||
|---|---|---|---|---|---|---|
| Carapace | L/W | 2.08/1.81 | 1.71/1.55 | 1.99/2.02 | 2.03/1.9 | 2.12/1.94 |
| Mesosoma | L | 4.48 | 3.62 | 4.6 | 5.35 | 2.47 |
| Tergite VII | L/W | 0.95/1.99 | 0.83/1.58 | 0.95/1.73 | 1.91/1.62 | 0.55/1.28 |
| Metasoma and telson | L | 8.55 | 7.83 | 10.47 | 8.84 | 6.09 |
| Segment I | L/W/D | 0.97/1.48/1.23 | 0.93/1.3/1.1 | 1.32/1.53/1.27 | 0.87/1.5/1.25 | 0.76/0.89/0.76 |
| Segment II | L/W/D | 1.21/1.4/1.2 | 0.99/1.22/1.15 | 1.61/1.57/1.42 | 1.05/1.52/1.26 | 0.86/0.87/0.80 |
| Segment III | L/W/D | 1.34/1.44/1.21 | 1.15/1.28/1.21 | 1.85/1.54/1.42 | 1.25/1.52/1.39 | 0.93/0.96/0.82 |
| Segment IV | L/W/D | 1.51/1.48/1.18 | 1.35/1.29/1.24 | 2.12/1.54/1.42 | 1.59/1.52/1.36 | 1.06/0.95/0.85 |
| Segment V | L/W/D | 1.63/1.46/1.12 | 1.7/1.31/1.16 | 2.45/1.6/1.33 | 2.25/1.5/1.26 | 1.34/1.99/0.81 |
| Telson | L/W/D | 1.14/0.8/0.9 | 0.96/0.77/0.89 | 0.63/0.50/0.99 | 0.91/0.8/0.95 | 0.62/0.46/0.54 |
| Pedipalp | L | 6.63 | 5.29 | 6.78 | 6.54 | 4.03 |
| Femur | L/W | 1.72/0.51 | 1.35/0.48 | 1.65/0.54 | 1.72/0.55 | 0.98/0.39 |
| Patella | L/W | 2.00/0.78 | 1.62/0.69 | 2.15/0.79 | 1.97/0.75 | 1.26/0.46 |
| Chela | L | 2.91 | 2.32 | 2.98 | 2.85 | 1.79 |
| Manus | W/D | 0.6/0.6 | 0.48/0.54 | 0.63/0.66 | 0.66/0.64 | 0.37/0.39 |
| Movable finger | L | 1.65 | 1.75 | 1.91 | 2.04 | 1.08 |
| Pectine | L/W | 1.72/0.43 | 1.60/0.39 | 1.74/0.47 | 1.71/0.42 | 0.99/0.26 |
| Genital Operculum | L/W | 0.43/0.62 | 0.28/0.47 | 0.38/0.72 | 0.42/0.68 | 0.22/0.36 |
| Total | L | 15.11 | 13.16 | 17.06 | 16.22 | 10.68 |
| Pectinal teeth count | PTC | 17/16 | 15/16 | 16/17 | 16/17 | 17/17 |
Legs (Figure 2A,B): femur and patellae carinated. Tibiae III and IV carinated, with long tibial spurs. All legs with a pair of pedal spurs. Tarsomere covered with long delicate setae arranged in parallel rows on ventral side. Tarsomere I (basitarsus) with a tuft of short, stout golden setae on ventral side. Tarsomere II (telotarsus) with a small bulge situated laterally on the proximal portion. Tarsomere II compressed laterally and ventrally with paired row of short, pointed, anteriorly directed, closely placed setae. Tarsomere II with a single row of granular carina on the ventral surface.
Genital operculum (Figure 3B): wider than long, elliptical, separated with a pair of short male genital papillae.
Pectines (Figure 3B and Figure 16A): Basal piece moderately notched on anterior median margin, lateral areas of posterior margin curved anteriorly. Marginal lamella of 3/3 digits and median lamella of 7/7 digits, outer margin armed with a row of stout short golden setae and few setae on surface. Fulcra 16/16, very small, roughly triangular, each armed with few short golden setae, placed in between adjacent pectinal teeth. Teeth 16/16, strong and stout.

Figure 2.
Charmus laneus Karsch, 1879; adult ♂ (IES-631); (A) Dorsal view (B) Ventral view. Scale bar: 5 mm. Remark: Metasomal segments II-IV are broken and this individual was photographed due to unavailability of additional intact adult male specimens.
Mesosoma (Figure 2A,B): tergites I–VI with mixed granulation with median carina moderately developed throughout except vestigial on the first segment. Posterior margin and lateral margins almost smooth. Tergite VII strongly and densely granular, narrowed posteriorly, with two pairs of lateral granular carinae. Sternites III–VI smooth, each with a pair of spiracles. Sternite VII with two pairs of weakly developed granular carinae present on two-thirds posterior portion. Intercarinal surfaces strongly and sparsely granular.
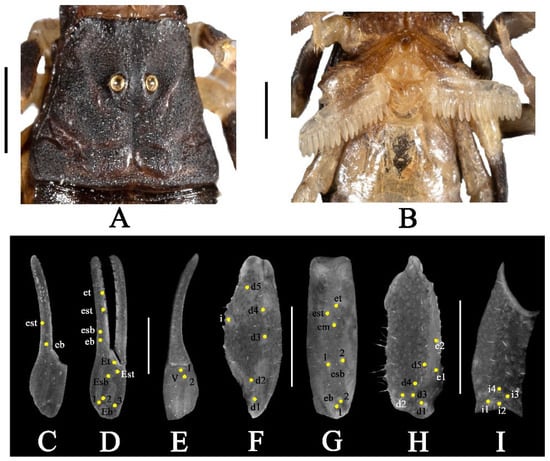
Figure 3.
Charmus laneus Karsch, 1879; adult ♂ (IES-631); (A) carapace, white light (B) sternopectinal area (C–I) trichobothrial pattern on (C–E) Chela, (C) dorsal view. (D) External view, (E) ventral view; (F,G) patella, (F) dorsal view, (G) external view; (H,I) femur, (H) dorsal view, (I) internal view. Scale bars: 1 mm.
Metasoma (Figure 2A,B, Figure 17A, Figure 18A, Figure 19A and Figure 20A): all segments longer than wide. All segments punctate and strongly hairy with few long and few short setae. Segment I with four pairs of granular carinae (dorsal lateral, lateral supramedian, ventral lateral and ventral submedian). Segment II with five pairs of carinae (dorsal lateral, lateral supramedian, lateral inframedian, ventral lateral and ventral submedian). Segment III with three pairs of carinae (dorsal lateral, ventral lateral and ventral submedian). Dorsal lateral carinae of segment I–III ending posteriorly into a short tubercle. Ventral submedian carina on segments I–III strongly developed. Segment IV and V heavily punctate and hairy. Segment IV with a pair of moderately developed dorsal lateral carina present on the anterior and posterior portions. Segment V with a pair of moderately developed dorsal lateral carina present on the anterior portion. Anterior ventral portion of Segment IV and posterior ventral portion of segment V with few scattered granules. Intercarinal surfaces of segments I–III strongly and densely granular except surface between dorsal lateral carinae and ventral lateral carinae of segment III almost smooth. Anal rim with two margins, lower margin moderately crenulated and upper margin weakly crenulated.
Telson (Figure 2A,B, Figure 17A, Figure 18A, Figure 19A and Figure 20A): telson punctate and hairy. Stout and globular without carinae. A depression present on dorsal lateral surface of vesicle. Proximal portion of vesicle with a pair of weakly developed nodule. Aculeus elongated and strongly curved.
Distribution, habitat and ecology (Figure 4): the specimens of C. laneus were collected from low-lying open fields surrounding the city of Coimbatore, Tamil Nadu, India. The individuals were found active at night, hiding under short grasses and were seen to be sympatric with other buthids such as Buthoscorpio sp. and Hottentota sp.

Figure 4.
Scrub forests surrounding Coimbatore, India, from where the specimens of Charmus laneus Karsch, 1879 were collected. Photo by Gopan Madathil.
Remarks:
The species Charmus indicus Hirst, 1915 syn. nov. was collected and described from Coimbatore (Tamil Nadu) as the first occurrence of the genus from India. This description was incomplete and based on an immature specimen of unknown sex. Subsequently, Reddy [9] redescribed C. indicus based on adult specimens of both sexes from new localities (Tirupati, Andhra Pradesh; Udayagiri Fort and Puducherry, Tamil Nadu). However, no specimens from the type locality were present in his large sampling. Hirst [8] distinguished C.indicus from C. laneus based on the following diagnostic characteristics: (1) shortness and stoutness of tail, (2) presence of punctures (instead of granules) on the sides of the metasomal segment III (3) numerous punctures on metasomal segments IV and V and (4) absence of granules from the ventral surfaces of metasomal segments IV and V (Figure 17A, Figure 18A and Figure 19A). However, the morphology of the specimens collected during our own field work from the original type locality of C. indicus as well as the morphology of C. laneus specimens from Sri Lanka photographed by Kovarik et al. [13] (Figures 71–73, p. 19) are congruent with all the abovementioned diagnostic characters. This strongly suggest that Charmus sampled from Coimbatore are not C. indicus but rather C. laneus. These specimens show the following combination of characters that are typical of C. laneus: (1) surface of carapace with mixed and dense granulation; (2) tergites I–VI with mixed granulation; (3) strongly developed ventral submedian carina on metasomal segments I-III. Therefore, according to Article 23 of the ICZN, we hereby propose to transfer C. indicus under the junior synonymy of C. laneus.
3.2. Charmus sinhagadensis Tikader and Bastawade, 1983
Figure 5, Figure 6, Figure 7, Figure 10, Figure 15, Figure 16, Figure 17, Figure 18, Figure 19 and Figure 20, Table 2
Holotype (considered lost): India, Maharashtra, Pune City, Sinhagad Fort, leg. D. B. Bastawade, ♀ (ZSI No. 5079/18). Paratype (considered lost): same data as holotype, ♀ (ZSI No. 5080/18). Neotype (designated here): India, Maharashtra, Pune City, Sinhagad Fort, 18.3663° N, 73.7559° E, 1315 m a.s.l., 5 May 2023, leg. Mihir Joshi, Aarya Gramopadhye, Aditya Soman, Akash Joshi, ♂ (BNHS SC 403). Other material examined: Same data as neotype, 2 ♂ (IES 587, IES 602). India, Maharashtra, Kolhapur District, Amba Ghat, 17.00° N, 73.777° E, 455 m a.s.l., 21 October 2019, leg. Makarand Ketkar, Akshay Marathe, ♂ (IES-405); India, Maharashtra State, Pune District, Bhimashankar Wildlife Sanctuary, 19.0728° N, 73.5565° E, 997 m a.s.l., 10 December 2022, leg. Makarand Ketkar, Akshay Marathe, ♂ (IES 561).

Table 2.
Measurements and meristics of Charmus sinhagadensis Tikader and Bastawade, 1983 (in mm).
Table 2.
Measurements and meristics of Charmus sinhagadensis Tikader and Bastawade, 1983 (in mm).
| BNHS SC 403 (♂) (Neotype) | IES-602(♂) | IES-587 (♂) | ||
|---|---|---|---|---|
| Carapace | L/W | 2.27/2.28 | 1.8/1.56 | 1.82/1.58 |
| Mesosoma | L | 4.78 | 3.32 | 3.23 |
| Tergite VII | L/W | 1.21/2.52 | 0.94/1.63 | 0.86/1.62 |
| Metasoma and telson | L | 10.62 | 8.53 | 8.48 |
| Segment I | L/W/D | 0.88/1.50/1.26 | 0.96/1.11/1.05 | 0.94/1.1/1.13 |
| Segment II | L/W/D | 1.29/1.31/1.27 | 1.12/1.11/1.05 | 1.15/1.1/1.14 |
| Segment III | L/W/D | 1.50/1.35/1.38 | 1.39/1.11/1.09 | 1.26/1.11/1.13 |
| Segment IV | L/W/D | 2.18/1.36/1.31 | 1.45/1.14/1.1 | 1.52/1.1/1.12 |
| Segment V | L/W/D | 2.56/1.36/1.26 | 1.87/1.15/1.08 | 1.78/1.1/1.07 |
| Telson | L/W/D | 1.24/0.84/1.05 | 0.93/0.81/0.73 | 1.17/0.76/0.93 |
| Pedipalp | L | 9.08 | 5.95 | 6.14 |
| Femur | L/W | 2.19/0.67 | 1.5/0.48 | 1.57/0.46 |
| Patella | L/W | 2.81/0.98 | 1.83/0.63 | 1.83/0.66 |
| Chela | L | 4.08 | 2.62 | 2.74 |
| Manus | W/D | 0.86/0.84 | 0.54/0.53 | 0.54/0.55 |
| Movable finger | L | 2.81 | 1.78 | 2.02 |
| Pectine | L/W | 2.04/0.52 | 1.54/0.37 | 1.6/0.36 |
| Genital Operculum | L/W | 0.43/0.96 | 0.35/0.56 | 0.37/0.55 |
| Total | L | 17.67 | 13.65 | 13.53 |
| Pectinal teeth count | PTC | 14/14 | 15/15 | 15/15 |
Description (♂ neotype, BNHS SC 403, measurements in Table 2):
Coloration (Figure 5A,B): body dorsal surface entirely dark brownish to blackish, except tergites with a brick red median longitudinal patch; legs dark brown with tarsomere yellowish brown; metasoma dark brownish to blackish; pedipalp fingers dark yellow. Ventral surface light yellow except last mesosomal sternite light brown. Basal segments of chelicerae yellow with black reticulation ending anteriorly into black transverse patch. Fingers of chelicerae blackish brown with tip of the fingers brown. Telson dark reddish brown in color.
Carapace (Figure 6A and Figure 15B): surface finely and densely granular. Carapace without carinae. Median supra-ocular area smooth with few sparse granules. Interocular area finely and sparsely granular. A pair of median eyes situated anteriorly in the ratio 1:2.1 (ratio of median eyes to anterior margin and median eyes to posterior margin). Antero-lateral ocular tubercle granular with type 3A ocelli. Three well developed lateral ocelli, all three ocelli of same size. Longitudinal furrow shallow anteriorly and deep posteriorly. Anterior margin smooth throughout and slightly curved. Lateral margins smooth below lateral ocelli. Posterior margin almost entirely smooth.

Figure 5.
Charmus sinhagadensis Tikader & Bastawade, 1983; neotype, adult ♂ (BNHS SC 403); (A) Dorsal view (B) Ventral view. Scale bar: 5 mm.
Chelicerae: characteristic of the family Buthidae. Basal segments and movable fingers with tuft of short and firm setae on ventral surfaces. Dorsal surfaces smooth.
Pedipalp (Figure 5A,B, Figure 6C–I and Figure 10B): femur with five carinae (prodorsal, retrodorsal, promedian, proventral and retroventral). All carinae crenulated. Intercarinal surfaces with mixed and sparse granulation except ventral surface smooth with a few fine granules on proximal portions. Patella almost smooth without developed carinae except promedian carina weakly developed with 7 spinoid granules. Intercarinal surfaces almost smooth. Manus smooth without carinae. Fixed fingers acarinate. Movable and fixed fingers with eight oblique rows of granules, each row (except most proximal) with one internal accessory granule. Trichobothrial pattern of type Aα, typical for the genus (chela dorsal 2, chela retrolateral 10, chela ventral 2, patella dorsal 6, patella retrolateral 7, femur dorsal 7 and femur prolateral 4).
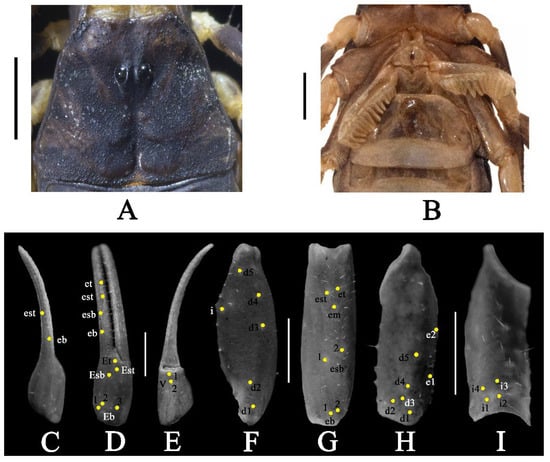
Figure 6.
Charmus sinhagadensis Tikader & Bastawade, 1983; neotype, adult ♂ (BNHS SC 403); (A) aarapace, white light, (B) sternopectinal area, (C–I) trichobothrial pattern on (C–E) chela, (C) dorsal view. (D) External view, (E) ventral view; (F,G) patella, (F) dorsal view, (G) external view; (H,I) femur, (H) dorsal view, (I) internal view. Scale bars: 1 mm.
Legs (Figure 5A,B): femur and patellae carinated. Tibiae III and IV carinated, with long tibial spurs. All legs with a pair of pedal spurs. Tarsomere covered with long delicate setae arranged in parallel rows on ventral side. Tarsomere I (basitarsus) with a tuft of short, stout golden setae on ventral side. Tarsomere II (telotarsus) with a small bulge situated laterally on the proximal portion. Tarsomere II compressed laterally and ventrally with paired row of short, pointed, anteriorly directed, closely placed setae. Tarsomere II with a single row of granular carina on the ventral surface.
Genital operculum (Figure 6B): Wider than long, elliptical, separated by a pair of short male genital papillae.
Pectines (Figure 6B and Figure 16B): basal piece deeply notched on anterior median margin, lateral areas of posterior margin curved anteriorly. Marginal lamella of 3/3 digits and median lamella of 7/7 digits, outer margin armed with a row of stout short golden setae and few setae on surface. Fulcra 17/17, very small, roughly triangular, each armed with few short golden setae, placed in between adjacent pectinal teeth. Teeth 14/16, strong and stout.
Mesosoma (Figure 5A,B): tergites I–VI finely granular with median carina moderately developed throughout. Posterior margin finely crenulated and lateral margins almost smooth. Tergite VII strongly and densely granular, narrowed posteriorly, with two pairs of lateral granular carinae. Sternites III–VI almost smooth, each with a pair of spiracles. Lateral margin of Sternite VI with a pair of weakly granular carinae. Sternite VII with two pairs of weakly developed granular carinae present on two-thirds posterior portion. Intercarinal space strongly and sparsely granular.
Metasoma (Figure 5A,B, Figure 17B, Figure 18B, Figure 19B and Figure 20B): all segments longer than wide. All segments punctate and strongly hairy with few long and few short setae. Segment I with four pairs of granular carinae (dorsal lateral, lateral supramedian, ventral lateral and ventral submedian). Segment II with five pairs of carinae (dorsal lateral, lateral supramedian, lateral inframedian, ventral lateral and ventral submedian). Segment III with three pairs of carinae (dorsal lateral, ventral lateral and ventral submedian). Dorsal lateral carinae of segment I–III ending posteriorly into a short tubercle. Ventral submedian carina of segment I–III moderately developed. Segment IV acarinate, heavily punctate and hairy. Segment V with a pair of weakly developed dorsal lateral carina present on the anterior portion. Anterior ventral portion of Segment IV and posterior ventral portion of segment V with few scattered granules. Intercarinal surfaces of segments I–II strongly and densely granular, segment III weakly granular. Anal rim with two margins, lower margin moderately crenulated and upper margin weakly crenulated.

Figure 7.
Dry deciduous forests around Sinhagad Fort, India; the type locality of Charmus sinhagadensis Tikader and Bastawade, 1983. Photo by Mihir Joshi.
Telson (Figure 5A,B, Figure 17B, Figure 18B, Figure 19B and Figure 20B): telson punctate and hairy. Stout and globular without carinae. A depression present on dorsal lateral surface of vesicle. Proximal portion of vesicle with a pair of strongly developed nodule. Aculeus elongated and strongly curved.
Distribution, habitat and ecology (Figure 7): the types of C. sinhagadensis were collected from the trail to Sinhagad Fort, Pune, Maharashtra. The individuals were mainly observed scurrying across basaltic boulders while others were found under the dense undergrowth of Strobilanthes sp. Blume, 1826 shrubs, which occupy much of the hill slopes in the Northern Western Ghats of Maharashtra. Individuals were actively seen and collected only during the dry seasons between April and June, and their ecology remains unknown.
Remarks:
The authors believe that the holotype of C. sinhagadensis (female) (ZSI 5079/18) is lost, as it could not be found in any ZSI centers despite an extensive search. To stabilize the taxonomy of the genus, we found it necessary to designate a neotype using the specimen under the voucher number BNHS SC 403. The neotype meets all the requirements of Article 75 of ICZN as it was collected from the exact type locality and the collected specimens exactly match the descriptions as per Tikader and Bastawade [6].
3.3. Charmus brignolii (Lourenço, 2000)
Figure 8, Figure 9, Figure 10, Figure 11, Figure 15, Figure 16, Figure 17, Figure 18, Figure 19 and Figure 20, Table 3 and Table 4
Holotype (not examined): India, Tamil Nadu, Puducherry, 1899, ♂ (MNHN RS-1247). Other material: India, Tamil Nadu, Puducherry, Auroville, 12.0052° N, 79.8069° E, 84 m a.s.l., 26 June 2023, leg. Shauri Sulakhe, Shubhankar Deshpande, Mihir Joshi; 3 ♂ (IES 614, IES 615, IES 616), 7 ♀ (IES 609, IES 609, IES 610, IES 611, IES 612, IES 613); India, Andhra Pradesh, Nellore District, Sangam town, 14.5900N, 79.7472E, 17m a.s.l., 28 June 2022, leg. Shauri Sulakhe, Shubhankar Deshpande, Rohit Karmalkar;, 3♂ (IES 475, IES 479, IES 483), 6 ♀ (IES 474, IES 476, IES 477, IES 480, IES 481, IES 482); India, Tamil Nadu, Salem District, Salem, 11°34′36.9″ N 78°02′31.4″ E, 278 m a.s.l., 30 September 2021, leg. Shauri Sulakhe, Shubhankar Deshpande, Rohit Karmalkar, 1 individual (unknown sex) (IES 586).
Coloration (Figure 8A,B): body dorsal surface entirely black; legs blackish yellow with tarsomere yellow; metasoma black to dark brown; pedipalp fingers light brown. Ventral surface light yellow except last sternite dark brown. Basal segments of chelicerae yellow with black reticulation ending anteriorly into black transverse patch. Fingers of chelicerae blackish brown with tip of the fingers brown. Telson dark reddish brown in color.
Carapace (Figure 9A and Figure 15C). Surface with mixed and dense granulation. Carapace without carinae. Median supra-ocular area densely granular. Interocular area with mixed and dense granulation. A pair of median eyes situated anteriorly in the ratio 1:1.4 (Ratio of median eyes to anterior margin and median eyes to posterior margin). Antero-lateral ocular tubercle granular with type 3A ocelli. Three well-developed lateral ocelli, all three ocelli of same size. Longitudinal furrow shallow anteriorly and deep posteriorly. Anterior margin finely crenulated in the median portion and slightly curved. Lateral margins smooth below lateral ocelli. Posterior margin almost entirely smooth.

Table 3.
Measurements and meristics of Charmus brignolii Lourenco, 2000 (in mm).
Table 3.
Measurements and meristics of Charmus brignolii Lourenco, 2000 (in mm).
| IES-614 (♂) | IES-615 (♂) | IES-616 (♂) | IES-611 (♀) | IES-612 (♀) | ||
|---|---|---|---|---|---|---|
| Carapace | L/W | 1.96/1.73 | 2.09/1.83 | 2.08/1.87 | 1.98/1.89 | 2.08/1.80 |
| Mesosoma | L | 4.06 | 3.97 | 3.84 | 2.36 | 4.67 |
| Tergite VII | L/W | 0.92/2.04 | 0.86/2.04 | 0.83/2.26 | 0.55/1.24 | 1.16/2.33 |
| Metasoma and telson | L | 8.47 | 8.40 | 9.05 | 6.05 | 8.39 |
| Segment I | L/W/D | 0.79/1.32/0.96 | 0.82/1.41/1.1 | 0.85/1.53/1.04 | 1.09/1.4/1.19 | 0.81/1.39/1.06 |
| Segment II | L/W/D | 1.05/1.36/0.90 | 1.13/1.46/1.26 | 1.2/1.36/1.13 | 1.19/1.41/1.19 | 1.07/1.43/1.04 |
| Segment III | L/W/D | 1.19/1.42/1.05 | 1.33/1.54/1.32 | 1.43/1.48/1.2 | 1.83/1.41/1.18 | 1.26/1.45/1.27 |
| Segment IV | L/W/D | 1.52/1.42/1.20 | 1.5/1.51/1.24 | 1.57/1.62/1.24 | 1.94/1.41/1.17 | 1.44/1.57/1.33 |
| Segment V | L/W/D | 2.12/1.53/1.12 | 1.8/1.56/1.21 | 2.2/1.55/1.15 | - | 1.8/1.59/1.35 |
| Telson | L/W/D | 0.91/0.71/0.83 | 1.13/0.82/0.76 | 1.04/0.78/0.90 | - | 1.14/0.81/0.84 |
| Pedipalp | L | 6.47 | 6.37 | 6.68 | 7.42 | 8.36 |
| Femur | L/W | 1.66/0.48 | 1.68/0.46 | 1.66/0.47 | 1.51/0.79 | 1.71/0.52 |
| Patella | L/W | 1.94/0.71 | 1.95/0.76 | 2.07/0.74 | 3.08/1.11 | 2/0.76 |
| Chela | L | 2.87 | 2.74 | 2.95 | 2.83 | 4.65 |
| Manus | W/D | 0.57/0.54 | 0.56/0.53 | 0.65/0.59 | 0.93/0.62 | 0.58/0.55 |
| Movable finger | L | 2 | 1.64 | 2.02 | 1.91 | 2.01 |
| Pectine | L/W | 1.78/0.45 | 1.77/0.4 | 1.82/0.41 | 1.63/0.4 | 1.97/0.39 |
| Genital Operculum | L/W | 0.43/0.54 | 0.45/0.65 | 0.38/0.59 | 0.36/0.64 | 0.48/0.66 |
| Total | L | 14.49 | 14.46 | 14.97 | 10.39 | 15.14 |
| Pectinal teeth count | PTC | 16/16 | 16/16 | 17/17 | 16/17 | 16/16 |
| IES-608 (unidentified sex) | IES-609 (♀) | IES-610 (♀) | IES-613 (♀) | IES-617 (♀) | ||
| Carapace | L/W | 2.04/1.83 | 2.06/1.75 | 2.08/1.81 | 1.87/1.70 | 1.99/1.83 |
| Mesosoma | L | 3.94 | 4.15 | 3.81 | 3.92 | 3.64 |
| Tergite VII | L/W | 0.85/1.65 | 0.95/2.19 | 0.83/1.81 | 0.89/1.89 | 0.76/1.96 |
| Metasoma and telson | L | 8.86 | 8.63 | 7.93 | 8.76 | 8.54 |
| Segment I | L/W/D | 0.83/1.37/1.12 | 0.8/1.36/1.19 | 0.81/1.39/1.16 | 0.9/1.36/1.09 | 0.82/1.36/1.15 |
| Segment II | L/W/D | 1.15/1.4/1.18 | 1.00/1.4/1.27 | 1.17/1.37/1.22 | 1.07/1.44/1.1 | 1.03/1.44/1.19 |
| Segment III | L/W/D | 1.27/1.46/1.24 | 1.18/1.46/1.39 | 1.26/1.48/1.32 | 1.19/1.51/1.22 | 1.20/1.42/1.22 |
| Segment IV | L/W/D | 1.52/1.49/1.23 | 1.54/1.44/1.39 | 1.34/1.5/1.3 | 1.48/1.56/1.14 | 1.52/1.48/1.24 |
| Segment V | L/W/D | 2.22/1.49/1.17 | 2.25/1.54/1.35 | 1.73/1.55/1.28 | 2.22/1.57/1.18 | 2.18/1.43/1.25 |
| Telson | L/W/D | 1.1/0.75/0.84 | 1.09/0.75/0.89 | 0.99/0.76/0.79 | 0.99/0.79/0.96 | 1/0.71/0.94 |
| Pedipalp | L | 6.09 | 6.42 | 6.17 | 6.08 | 6.16 |
| Femur | L/W | 1.61/0.51 | 1.65/0.51 | 1.65/0.49 | 1.6/0.48 | 1.54/0.53 |
| Patella | L/W | 1.77/0.71 | 1.94/0.75 | 1.85/0.68 | 1.89/0.70 | 1.93/0.75 |
| Chela | L | 2.71 | 2.83 | 2.67 | 2.59 | 2.69 |
| Manus | W/D | 0.52/0.61 | 0.56/0.5 | 0.57/0.61 | 0.56/0.6 | 0.61/0.58 |
| Movable finger | L | 2.09 | 1.84 | 1.88 | 1.72 | 1.96 |
| Pectine | L/W | 1.67/0.39 | 1.57/0.37 | 1.61/0.4 | 1.55/0.3 | 1.55/0.34 |
| Genital Operculum | L/W | 0.4/0.59 | 0.44/0.62 | 0.44/0.6 | 0.33/0.68 | 0.36/0.53 |
| Total | L | 14.84 | 14.84 | 13.82 | 14.55 | 14.17 |
| Pectinal teeth count | PTC | 17/17 | 16/17 | 16/16 | 16/17 | 16/16 |

Table 4.
Measurements and meristics of Charmus brignolii Lourenço, 2000 (in mm).
Table 4.
Measurements and meristics of Charmus brignolii Lourenço, 2000 (in mm).
| IES-475 (♂) | IES-479(♂) | IES-483(♂) | IES-474(♀) | |||
|---|---|---|---|---|---|---|
| Carapace | L/W | 2.1/1.85 | 2.06/1.85 | 1.64/1.42 | 1.69/1.49 | |
| Mesosoma | L/W | 4.55 | 4.10 | 2.98 | 2.48 | |
| Tergite VII | L/W | 1.13/2.43 | 0.93/1.95 | 0.80/1.47 | 0.7/1.78 | |
| Metasoma and telson | L | 8.88 | 8.41 | 7.59 | 7.32 | |
| Segment I | L/W/D | 0.84/1.47/1.26 | 0.94/1.41/1.11 | 0.85/1.21/0.98 | 0.73/1.17/1.03 | |
| Segment II | L/W/D | 1.15/1.53/1.37 | 1.12/1.46/1.19 | 1.00/1.12/1.09 | 1.01/1.26/1.12 | |
| Segment III | L/W/D | 1.24/1.58/1.50 | 1.2/1.5/1.29 | 1.07/1.2/1.12 | 1.16/1.2/1.21 | |
| Segment IV | L/W/D | 1.65/1.6/1.50 | 1.141/1.52/1.36 | 1.18/1.26/1.14 | 1.25/1.22/1.15 | |
| Segment V | L/W/D | 2.06/1.58/1.51 | 1.95/1.57/1.42 | 1.86/1.27/1.16 | 1.74/1.22/1.13 | |
| Telson | L/W/D | 1.16/0.89/0.94 | 1.07/0.82/0.95 | 0.97/0.68/0.90 | 0.78/0.66/0.79 | |
| Pedipalp | L | 6.72 | 6.48 | 5.29 | 4.19 | |
| Femur | L/W | 1.61/0.59 | 1.60/0.48 | 1.34/0.47 | 0.91/0.43 | |
| Patella | L/W | 2.09/0.78 | 1.98/0.77 | 1.64/0.58 | 1.02/0.59 | |
| Chela | L | 3.02 | 2.90 | 2.31 | 2.26 | |
| Manus | W/D | 0.61/0.64 | 0.67/0.75 | 0.47/0.52 | 0.43/0.47 | |
| Movable finger | L | 2.08 | 1.91 | 1.53 | 1.56 | |
| Pectine | L/W | 1.57/0.36 | 1.46/0.35 | 1.11/0.26 | 1.31/0.31 | |
| Genital Operculum | L/W | 0.28/0.47 | 0.44/0.58 | 0.29/0.38 | 0.32/0.42 | |
| Total | L | 15.53 | 14.57 | 12.21 | 11.49 | |
| Pectinal teeth count | PTC | 16/16 | 15/16 | 15/17 | 15/15 | |
| IES-476(♀) | IES-477(♀) | IES-481 (♀) | IES-482 (♀) | IES-586 (unidentidied sex) | ||
| Carapace | L/W | 1.67/1.41 | 2.06/1.8 | 2.14/1.9 | 1.65/1.4 | 1.88/1.70 |
| Mesosoma | L/W | 3.03 | 6.21 | 4.26 | 2.62 | 3.23 |
| Tergite VII | L/W | 0.8/1.45 | 1.18/2.37 | 0.93/2.24 | 0.72/1.57 | 0.91/1.8 |
| Metasoma and telson | L | 7.40 | 8.70 | 9.39 | 7.11 | 8.42 |
| Segment I | L/W/D | 0.67/1.05/0.98 | 0.69/1.47/1.27 | 0.91/1.43/1.26 | 0.73/1.19/0.93 | 0.83/1.15/1.06 |
| Segment II | L/W/D | 0.9/1.12/1.01 | 1.35/1.5/1.32 | 1.12/1.32/1.22 | 0.96/1.14/0.93 | 1.12/1.11/1.08 |
| Segment III | L/W/D | 0.96/1.16/1.04 | 1.44/1.58/1.39 | 1.31/1.51/1.32 | 1.00/1.81/1.14 | 1.19/1.16/1.07 |
| Segment IV | L/W/D | 1.36/1.22/1.05 | 1.49/1.53/1.42 | 1.61/1.47/1.45 | 1.06/1.18/1.15 | 1.48/1.13/1.05 |
| Segment V | L/W/D | 2.01/1.23/1.13 | 1.96/1.62/1.27 | 2.26/1.56/1.44 | 1.71/1.16/0.99 | 1.91/1.17/1.04 |
| Telson | L/W/D | 0.89/0.64/0.74 | 1.17/0.82/0.96 | 1.09/0.85/0.96 | 0.96/0.63/0.78 | 1.1/0.79/0.86 |
| Pedipalp | L | 4.86 | 6.05 | 5.98 | 4.76 | 5.98 |
| Femur | L/W | 1.31/0.43 | 1.75/0.55 | 1.62/0.51 | 1.16/0.51 | 1.6/0.44 |
| Patella | L/W | 1.48/0.53 | 1.87/0.78 | 1.69/0.7 | 1.53/0.57 | 1.86/0.63 |
| Chela | L | 2.07 | 2.43 | 2.67 | 2.07 | 2.52 |
| Manus | W/D | 0.43/0.45 | 0.60/0.63 | 0.58/0.63 | 0.39/0.45 | 0.49/0.54 |
| Movable finger | L | 1.50 | 2.00 | 1.8 | 1.44 | 1.93 |
| Pectine | L/W | 1.59/0.40 | 1.76/0.35 | 1.62/0.37 | 1.31/0.32 | 1.66/0.32 |
| Genital Operculum | L/W | 0.32/0.47 | 0.47/0.62 | 0.45/0.57 | 0.29/0.57 | 0.32/0.48 |
| Total | L | 12.1 | 16.97 | 15.79 | 11.38 | 13.53 |
| Pectinal teeth count | PTC | 16/17 | 18/16 | 16/16 | 16/15 | - |
Chelicerae: characteristic of the family Buthidae. Basal segments and movable fingers with tuft of short and firm setae on ventral surfaces. Dorsal surfaces smooth.
Pedipalp (Figure 8A,B, Figure 9C–I and Figure 10C): femur with five carinae (prodorsal, retrodorsal, promedian, proventral and retroventral). All carinae crenulated. Intercarinal surfaces with mixed and dense granulation except ventral surface smooth with a few fine granules on proximal portions. Patella almost smooth without developed carinae except promedian carina moderately developed with 7 spinoid granules. Intercarinal surface almost smooth. Manus smooth without carinae. Fixed fingers acarinate. Movable and fixed fingers with eight oblique rows of granules, each row (except most proximal) with one internal accessory granule. Trichobothrial pattern of type Aα, typical for the genus (chela dorsal 2, chela retrolateral 10, chela ventral 2, patella dorsal 6, patella retrolateral 7, femur dorsal 7 and femur prolateral 4).
Legs (Figure 8A,B): femur and patellae carinated. Tibiae III and IV carinated, with long tibial spurs. All legs with a pair of pedal spurs. Tarsomere covered with long delicate setae arranged in parallel rows on ventral side. Tarsomere I (basitarsus) with a tuft of short, stout golden setae on ventral side. Tarsomere II (telotarsus) with a small bulge situated laterally on the proximal portion. Tarsomere II compressed laterally and ventrally with a paired row of short, pointed, anteriorly directed, closely placed setae. Tarsomere II with a single row of granular carina on the ventral surface.

Figure 8.
Charmus brignolii Lourenço, 2000; adult ♂ (IES-616); (A) dorsal view (B) ventral view. Scale bar: 5 mm.
Pectines (Figure 9B and Figure 16C): basal piece moderately notched on anterior median margin, lateral areas of posterior margin almost straight. Marginal lamella of 3/3 digits and median lamella of 6/6 digits, outer margin armed with a row of stout short golden setae and few setae on surface. Fulcra 16/16, very small, roughly triangular, each armed with few short golden setae, placed in between adjacent pectinal teeth. Teeth 17/17, strong and stout.
Mesosoma (Figure 8A,B): tergites I–VI coarsely granular with median carina moderately developed throughout, except vestigial on first segment. Posterior margin strongly crenulated and lateral margins almost smooth. Tergite VII strongly and densely granular, narrowed posteriorly, with two pairs of lateral granular carinae. Sternites III–VI smooth, each with a pair of spiracles. Sternite VII acarinate with scattered strong and sparse granules.
Genital operculum (Figure 9B): wider than long, elliptical, separated by a pair of short male genital papillae.
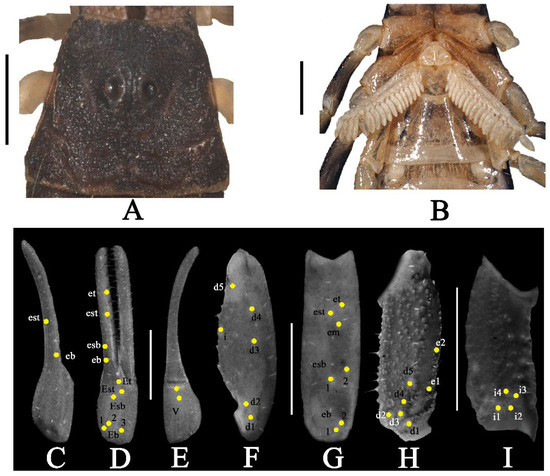
Figure 9.
Charmus brignolii Lourenço, 2000; adult ♂ (IES-616); (A) carapace, white light (B) sternopectinal area, (C–I) trichobothrial pattern on (C–E) chela, (C) dorsal view. (D) External view, (E) ventral view; (F,G) patella, (F) dorsal view, (G) external view; (H,I) femur, (H) dorsal view, (I) internal view. Scale bars: 1 mm.
Metasoma (Figure 8A,B, Figure 17C, Figure 18C, Figure 19C and Figure 20C): all segments longer than wide except Segment I wider than long. All segments punctate and strongly hairy with few long and few short setae. Segment I with four pairs of granular carinae (dorsal lateral, lateral supramedian, ventral lateral and ventral submedian). Segment II with five pairs of carinae (dorsal lateral, lateral supramedian, lateral inframedian, ventral lateral and ventral submedian). Segment III with three pairs of carinae (dorsal lateral, ventral lateral and ventral submedian). Dorsal lateral carinae of segment I–III ending posteriorly into a short tubercle. Ventral submedian carina of segment I–III strongly developed. Segment IV and V heavily punctate and hairy. Segment IV with a pair of strongly developed dorsal lateral carina present on the anterior portion. Segment V with a pair of strongly developed dorsal lateral carina present on the anterior portion. Anterior ventral portion of Segment IV and posterior ventral portion of segment V with few scattered granules. Intercarinal surfaces of segments I–III strongly and densely granular. Anal rim with two margins, lower margin moderately crenulated and upper margin weakly crenulated.
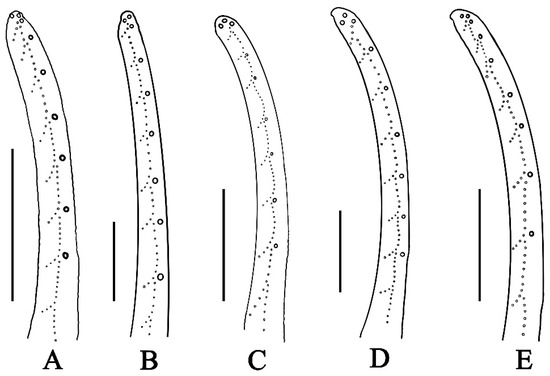
Figure 10.
Illustration showing number of rows of granules on movable finger of pedipalp chela; (A–E); (A) Charmus laneus Karsch, 1879, adult ♂ (IES-631); (B) Charmus sinhagadensis Tikader & Bastawade, 1983; neotype, adult ♂ (BNHS SC 403); (C) Charmus brignolii Lourenço, 2000; adult ♂ (IES-616); (D) Charmus dakshini sp. nov.; holotype, adult ♂ (BNHS SC 404), (E) Charmus saradieli Kovarik et al. 2016 adult ♂ (Locality 18 CO) from Sri Lanka. Scale bars: 1 mm. Illustrations by Rohit Karmalkar.
Telson (Figure 8A,B, Figure 17C, Figure 18C, Figure 19C and Figure 20C): telson punctate and hairy. Stout and globular without carinae. A depression present on dorsal lateral surface of vesicle. Proximal portion of vesicle with a pair of moderately developed nodules. Aculeus elongated and strongly curved.
Distribution, habitat, and ecology (Figure 11): specimens of C. brignolii were collected from the low-lying coastal plains of Auroville, Puducherry, India. The individuals were observed active at night, hiding under leaf litter or grass.
Remarks:
Reddy [9], in his redescription of C. indicus, cites a male specimen collected from Puducherry around the year 1900, which was deposited at the MNHN. The same individual was identified as C. annulipes by E. Simon and finally described by Lourenço [6] as Charmus brignolii. Several of the differences that Lourenço used to separate C. indicus from C. brignolii such as overall granulation on the body, hairiness of metasomal segments, the number of rows of granules on the movable finger of the chela, the nature of the median carina on the tergites, the size of the lateral eyes, and the presence of spinoid granules on the promedian carina of the patella seem rather arguable given the high amounts of morphological plasticity within the genus.
We examined multiple specimens of C. brignolii from various localities and compared them with all the existing Charmus species from India. All populations showed significant intraspecific variation in characters like the size of lateral eyes, hairiness of metasoma, nature of median carina on tergites, and the number of spinoid granules on promedian carina of the patella. The only character of importance that showed significant stability throughout the examined material is the number of rows of granules on the pedipalp chela, which happens to be constant across all known species of Charmus (Figure 10, Table S6).
These findings demonstrate that the character diagnosis used by Lourenço to separate C. brignolii from C. laneus is inaccurate because the intraspecific variation was not taken into account. Nevertheless, C. brignolii remains a valid species, exhibiting few but reliable morphological differences, such as median supra ocular region densely granular and mesosomal tergites I–VI coarsely granular (characters that were not recognized as diagnostically important by Lourenço), as well as significant molecular divergence from C. laneus.
In his description, Lourenço explicitly mentions that the exact geographic locality of C. brignolii is unknown, as Puducherry functioned as a French trading post, where goods were typically packaged and shipped to France. Therefore, the type specimen of C. brignolii could have been collected anywhere in India before being shipped to France from Puducherry. In order to remove this ambiguity, we consider that among the localities where the species has been collected (Sangam, Andhra Pradesh and Salem), Auroville (12.0052° N, 79.8069° E) is the closest to Puducherry and therefore the most appropriate proxy for the type locality of Charmus brignolii Lourenco, 2000.

Figure 11.
Dry scrubland around Aurovile, Puducherry from where Charmus brignolii Lourenço, 2000 were collected. Photo by Neil Kakkar.
3.4. Charmus dakshini sp. nov.
urn:lsid:zoobank.org:pub:4DD30BD9-355F-4837-9053-C51489896E88
Figure 12, Figure 13, Figure 14, Figure 15, Figure 16, Figure 17, Figure 18, Figure 19 and Figure 20, Table 5 and Table 6
Holotype: India, Tamil Nadu, Sirumalai, Sirumalai Road, 10.1942° N, 77.9967° E, 1600 m a.s.l., 27 June 2023, leg. Shauri Sulakhe, Shubhankar Deshpande, Mihir Joshi; Holotype ♂ (BNHS SC 404). Paratypes: Same data as holotype, 3♂ (IES 624, IES 626, IES 627), 3♀ (IES 621, IES 622, IES 623); 28 September 2021, leg. Shauri Sulakhe, Shubhankar Deshpande, Mukta Nachare, 1♂ (IES 385), 1♀ (IES 386). Other material examined: India, Tamil Nadu, Palani Hills, Palani-Kodaikanal Road, 10.36° N 77.54° E, 813 m a.s.l., 26th October 2023, leg. Shauri Sulakhe, Shubhankar Deshpande, Mihir Joshi, 2♂ (IES 618, IES 619), 2♀ (IES 620, IES 621), 30 September 2021, Shauri Sulakhe, Shubhankar Deshpande, Mukta Nachare, 1♀ (IES 401).
Etymology: the specific epithet is derived from the Sanskrit word “Dakshin” (=south) indicating the distribution of the species in the southernmost regions of India. It is an invariable noun in apposition.
Coloration (Figure 12A–D): body dorsal surface entirely dark brownish to blackish; legs dark brown with tarsomere yellowish brown; metasoma dark brownish to blackish; pedipalp fingers dark brown. Ventral surface brown except last sternite dark brown. Basal segments of chelicerae dark brown with black reticulation ending anteriorly into black transverse patch. Fingers of chelicerae blackish brown with tip of the fingers brown. Telson dark reddish brown in color.

Table 5.
Measurements and meristics of Charmus dakshini sp. nov. from Sirumalai (in mm).
Table 5.
Measurements and meristics of Charmus dakshini sp. nov. from Sirumalai (in mm).
| BNHS SC-404 (♂) | IES-624 (♂) | IES-626 (♂) | IES-627 (♂) | |||
|---|---|---|---|---|---|---|
| Holotype | Paratype | Paratype | Paratype | |||
| Carapace | L/W | 2.15/1.84 | 2.09/1.75 | 2.01/1.77 | 2.09/1.74 | |
| Mesosoma | L | 3.68 | 3.61 | 3.18 | 3.63 | |
| Tergite VII | L/W | 1.06/1.80 | 1.01/1.9 | 0.92/1.85 | 1.04/1.8 | |
| Metasoma and telson | L | 9.46 | 8.93 | 9.50 | 5.53 | |
| Segment I | L/W/D | 1.01/1.36/0.93 | 1.08/1.37/1.18 | 1.1/1.35/0.94 | 1.07/1.31/0.9 | |
| Segment II | L/W/D | 1.05/1.33/0.94 | 1.11/1.36/1.25 | 1.17/1.3/1.08 | 1.04/1.31/0.98 | |
| Segment III | L/W/D | 1.31/1.38/1.06 | 1.29/1.37/1.33 | 1.37/1.34/1.13 | 1.24/1.3/1.08 | |
| Segment IV | L/W/D | 1.62/1.36/1.04 | 1.59/1.35/1.35 | 1.62/1.35/1.15 | - | |
| Segment V | L/W/D | 2.41/1.35/1.07 | 1.87/1.33/1.25 | 2.14/1.35/1.16 | - | |
| Telson | L/W/D | 1.15/0.79/0.95 | 1.27/0.97/0.94 | 1.32/0.84/0.92 | 1.35/0.94/0.98 | |
| Pedipalp | L | 6.97 | 7.88 | 6.48 | 6.76 | |
| Femur | L/W | 1.81/0.58 | 2.03/0.64 | 1.8/0.57 | 1.68/0.64 | |
| Patella | L/W | 2.19/0.79 | 2.37/0.92 | 2.16/0.76 | 2.12/0.89 | |
| Chela | L | 2.97 | 3.48 | 2.52 | 2.96 | |
| Manus | W/D | 0.62/0.70 | 0.70/0.62 | 0.61/0.64 | 0.64/0.74 | |
| Movable finger | L | 1.90 | 2.02 | 1.88 | 2.36 | |
| Pectine | L/W | 1.98/0.4 | 2.07/0.41 | 1.91/0.38 | 1.22/0.44 | |
| Genital Operculum | L/W | 0.38/0.7 | 0.32/0.53 | 0.37/0.55 | 0.32/0.34 | |
| Total | L | 15.29 | 14.63 | 14.69 | 11.25 | |
| Pectinal teeth count | PTC | 18/18 | 18/18 | 18/19 | 18/19 | |
| IES-385 (♂) | IES-621 (♀) | IES-622 (♀) | IES-623 (♀) | IES-386 (♀) | ||
| Paratype | Paratype | Paratype | Paratype | Paratype | ||
| Carapace | L/W | 2.01/1.88 | 2.04/1.76 | 2.01/1.78 | 1.95/1.7 | 2.23/1.66 |
| Mesosoma | L | 4.36 | 4.40 | 3.15 | 3.34 | 4.07 |
| Tergite VII | L/W | 1.19/1.80 | 1.1/1.73 | 0.86/1.99 | 0.94/1.78 | 1.13/2.02 |
| Metasoma and telson | L | 9.01 | 9.50 | 8.40 | 8.56 | 10.05 |
| Segment I | L/W/D | 0.84/1.31/1.19 | 1.02/1.28/1.02 | 0.91/1.18/1.28 | 0.82/1.26/1.23 | 1.03/1.44.1.33 |
| Segment II | L/W/D | 1.06/1.22/1.13 | 1.15/1.25/1.04 | 1.01/1.29/1.29 | 0.98/1.26/1.16 | 1.22/1.43/1.3 |
| Segment III | L/W/D | 1.4/1.23/1.17 | 1.34/1.28/1.04 | 1.16/1.26/1.31 | 1.15/1.23/1.31 | 1.38/1.4/1.35 |
| Segment IV | L/W/D | 1.53/1.23/1.16 | 1.66/1.38/1.04 | 1.37/1.33/1.35 | 1.49/1.26/1.3 | 1.66/1.42/1.33 |
| Segment V | L/W/D | 2.25/1.28/1.12 | 2.1/1.36/1.04 | 2.15/1.35/1.21 | 2.17/1.28/1.25 | 2.70/1.57/1.26 |
| Telson | L/W/D | 1.09/0.77/0.9 | 1.41/0.85/1.00 | 1.2/0.85/0.99 | 1.21/0.77/0.94 | 1.18/0.98/1.21 |
| Pedipalp | L | 6.77 | 7.09 | 6.64 | 6.42 | 7.37 |
| Femur | L/W | 1.65/0.51 | 1.85/0.59 | 1.76/0.54 | 1.67/0.54 | 1.99/0.64 |
| Patella | L/W | 2.14/0.68 | 2.21/0.79 | 2.10/0.72 | 1.95/0.75 | 2.28/0.92 |
| Chela | L | 2.98 | 3.03 | 2.78 | 2.80 | 3.10 |
| Manus | W/D | 0.54/0.58 | 0.62/0.59 | 0.61/0.58 | 0.6/0.61 | 0.73/0.71 |
| Movable finger | L | 1.76 | 2.15 | 2.16 | 2.10 | 2.47 |
| Pectine | L/W | 1.86/0.37 | 2.01/0.41 | 1.74/0.31 | 1.82/0.32 | 2.05/0.47 |
| Genital Operculum | L/W | 0.37/0.53 | 0.44/0.64 | 0.41/0.58 | 0.37/0.61 | 0.37/0.68 |
| Total | L | 15.38 | 15.94 | 13.56 | 13.85 | 16.35 |
| Pectinal teeth count | PTC | - | 18/19 | 18/18 | 17/18 | 18/18 |

Table 6.
Measurements and meristics of Charmus dakshini sp. nov. from Palani Hills (in mm).
Table 6.
Measurements and meristics of Charmus dakshini sp. nov. from Palani Hills (in mm).
| IES-618 (♂) | IES-619 (♂) | IES-620 (♀) | IES-401 (♀) | ||
|---|---|---|---|---|---|
| Paratype | Paratype | Paratype | Paratype | ||
| Carapace | L/W | 2.22/1.85 | 1.96/1.64 | 2.03/1.73 | 2.1/2.06 |
| Mesosoma | L | 3.96 | 3.20 | 3.69 | 3.27 |
| Tergite VII | L/W | 1.05/2.06 | 0.9/1.71 | 1/1.79 | 0.95/1.93 |
| Metasoma and telson | L | 9.93 | 9.09 | 9.53 | 7.88 |
| Segment I | L/W/D | 1.25/1.47/1.22 | 1.13/1.18/1.05 | 1.01/1.24/1.23 | 1.13/1.32/1.22 |
| Segment II | L/W/D | 1.28/1.41/1.23 | 1.17/1.16/1.06 | 1.2/1.14/1.2 | 1.41/1.29/1.22 |
| Segment III | L/W/D | 1.46/1.39/1.27 | 1.24/1.17/1.05 | 1.36/1.2/1.24 | 1.47/1.32/1.21 |
| Segment IV | L/W/D | 1.52/1.42/1.22 | 1.38/1.2/1.05 | 1.58/1.24/1.2 | 1.61/1.3/1.22 |
| Segment V | L/W/D | 2.38/1.46/1.23 | 2.18/1.19/1.08 | 2.34/1.27/1.14 | 2.26/1.32/1.21 |
| Telson | L/W/D | 1.29/0.84/1.03 | 1.21/0.77/0.91 | 1.26/0.77/0.93 | - |
| Pedipalp | L | 7.23 | 5.69 | 7.14 | 6.86 |
| Femur | L/W | 1.80/0.58 | 1.53/0.51 | 1.69/0.55 | 1.63/0.56 |
| Patella | L/W | 2.25/0.85 | 1.78/0.68 | 2.35/0.75 | 2.10/0.76 |
| Chela | L | 3.18 | 2.38 | 3.10 | 3.13 |
| Manus | W/D | 0.64/0.65 | 0.53/0.56 | 0.59/0.58 | 0.63/0.63 |
| Movable finger | L | 2.00 | 2.15 | 2.14 | 2.10 |
| Pectine | L/W | 2.07/0.41 | 1.79/0.40 | 1.7/0.33 | 1.97/0.42 |
| Genital Operculum | L/W | 0.37/0.57 | 0.38/0.58 | 0.35/0.54 | 0.35/0.52 |
| Total | L | 16.11 | 14.25 | 15.25 | 13.25 |
| Pectinal teeth count | PTC | 18/17 | 19/19 | 16/17 | 19/19 |
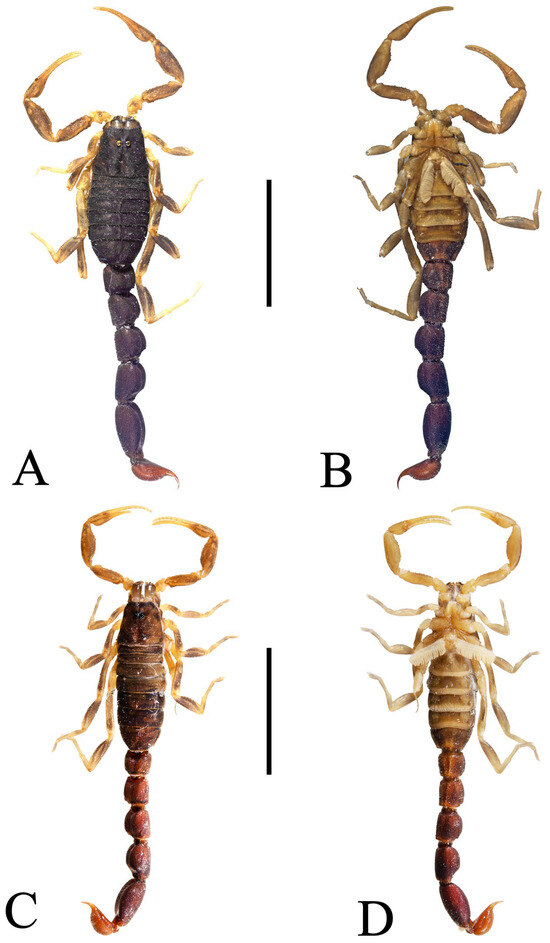
Figure 12.
Charmus dakshini sp. nov. holotype, adult ♂ (BNHS SC 404); (A) dorsal view (B) ventral view; paratype, adult ♀ (IES-621); (C) dorsal view, (D) ventral view. Scale bars: 5 mm.
Carapace (Figure 13A,B and Figure 15D): surface finely and densely granular. Carapace without carinae. Median supra-ocular area with few sparse granules. Interocular area finely and densely granular. A pair of median eyes situated anteriorly in the ratio 1:2.3 (ratio of median eyes to anterior margin and median eyes to posterior margin). Antero-lateral ocular tubercle granular with type 3A ocelli. Three well developed lateral ocelli with first and third ocelli smaller than the second ocelli. Longitudinal furrow shallow anteriorly and deep posteriorly. Anterior margin finely crenulated in the median portion and slightly curved. Lateral margins weakly crenulated below lateral ocelli. Posterior margin almost entirely smooth.
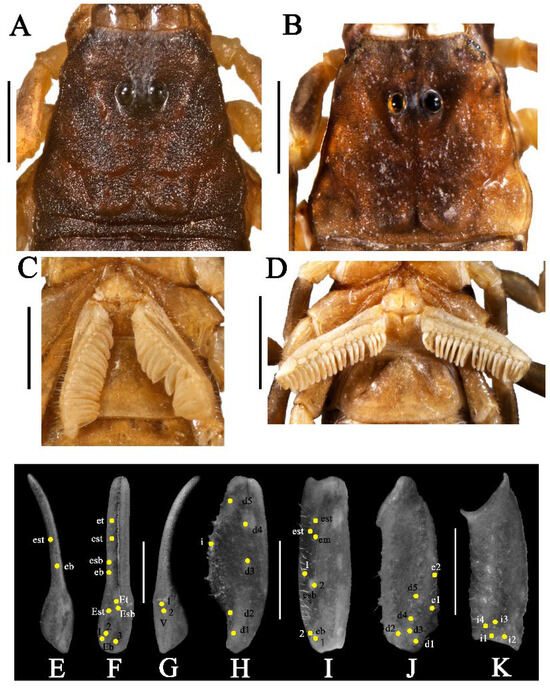
Figure 13.
Charmus dakshini sp. nov. holotype, adult ♂ (BNHS SC 404); (A–C); (A) carapace, white light, (C) sternopectinal area. (B–D) Paratype, adult ♀ (IES-621); (B) carapace, white light, (D) sternopectinal area; (E–K) trichobothrial pattern on (E–G) chela, (E) dorsal view. (F) External view, (G) ventral view; (H,I) patella, (H) dorsal view (I).
Chelicerae: characteristic of the family Buthidae. Basal segments and movable fingers with tuft of short and firm setae on ventral surfaces. Dorsal surfaces smooth.
Pedipalp (Figure 12A–D and Figure 13E–K): femur with five carinae (prodorsal, retrodorsal, promedian, proventral and retroventral). All carinae crenulated. Intercarinal surfaces with mixed and sparse granulation except ventral surface smooth with a few fine granules on proximal portions. Patella almost smooth without developed carinae except promedian carina moderately developed with 7 spinoid granules. Intercarinal surfaces almost smooth. Manus smooth without carinae. Fixed fingers acarinate. Movable and fixed fingers with eight oblique rows of granules, each row (except most proximal) with one internal accessory granule. Trichobothrial pattern of type Aα, typical for the genus (chela dorsal 2, chela retrolateral 10, chela ventral 2, patella dorsal 6, patella retrolateral 7, femur dorsal 7 and femur prolateral 4).
Legs (Figure 12A–D): femur and patellae carinated. Tibiae III and IV carinated, with long tibial spurs. All legs with a pair of pedal spurs. Tarsomere covered with long delicate setae arranged in parallel rows on ventral side. Tarsomere I (basitarsus) with a tuft of short, stout golden setae on ventral side. Tarsomere II (telotarsus) with a small bulge situated laterally on the proximal portion. Tarsomere II compressed laterally and ventrally with paired row of short, pointed, anteriorly directed, closely placed setae. Tarsomere II with a single row of granular carina on the ventral surface.
Genital operculum (Figure 13C,D): wider than long, elliptical, separated with a pair of short male genital papillae.
Pectines (Figure 13C,D and Figure 16D): basal piece deeply notched on anterior median margin, lateral areas of posterior margin curved anteriorly. Marginal lamella of 3/3 digits and median lamella of 7/7 digits, outer margin armed with a row of stout short golden setae and few setae on surface. Fulcra 17/17, very small, roughly triangular, each armed with few short golden setae, placed in between adjacent pectinal teeth. Teeth 18/18, strong and stout.
Mesosoma (Figure 12A–D): tergites I–VI finely granular with median carina moderately developed throughout except vestigial on the first segment. Posterior margin finely crenulated and lateral margins almost smooth. Tergite VII strongly and densely granular, narrowed posteriorly, with two pairs of lateral granular carinae. Sternites III–VI almost smooth, each with a pair of spiracles. Sternite VII with two pairs of weakly developed granular carinae present on two-thirds posterior portion. Intercarinal space strongly and sparsely granular.
Metasoma (Figure 12A–D, Figure 17D, Figure 18D, Figure 19D and Figure 20D): all segments longer than wide. All segments punctate and strongly hairy with few long and few short setae. Segment I with five pairs of granular carinae (dorsal lateral, lateral supramedian, lateral inframedian, ventral lateral and ventral submedian). Segment II with four pairs of carinae (dorsal lateral, lateral supramedian, ventral lateral and ventral submedian). Segment III with two pairs of carinae (dorsal lateral and ventral submedian). Dorsal lateral carinae of Segment I–III ending posteriorly into a short tubercle. Ventral submedian carina of Segment I–III weakly developed. Segment IV–V acarinated, heavily punctate and hairy. Intercarinal surfaces of Segments I–II strongly and densely granular, segment III weakly granular. Anal rim with two margins, both lower margin and upper margin weakly crenulated.
Telson (Figure 12A–D, Figure 17D, Figure 18D, Figure 19D and Figure 20D): telson punctate and hairy. Stout and globular without carinae. A depression present on dorsal lateral surface of vesicle. Proximal portion of vesicle with a pair of weakly developed nodule. Aculeus elongated and strongly curved.
Distribution, habitat and ecology (Figure 14): specimens of Charmus dakshini sp. nov. were observed to be active at night, usually scurrying across open soil or under dense undergrowth. The species was also observed on Palani-Kodaikanal Road, Palani Hills, Tamil Nadu, about 50 km west from the type locality.

Figure 14.
Dense evergreen forests of Sirumalai, India; the type locality of Charmus dakshini sp. nov. Photo by Mihir Joshi.
3.5. Affinities (Figure 2, Figure 3, Figure 5, Figure 6, Figure 8, Figure 9, Figure 10, Figure 12, Figure 13, Figure 15, Figure 16, Figure 17, Figure 18, Figure 19 and Figure 20)
Species of the genus Charmus can be differentiated by the following combination of characters:
- Surface of carapace finely and densely granular in C. sinhagadensis (Figure 6A and Figure 15B) and C. dakshini sp. nov. (Figure 13A and Figure 15D) as opposed to surface with mixed and dense granulation in C. laneus (Figure 3A and Figure 15A,E), C. brignolii (Figure 9A and Figure 15C) and C. saradieli (Figure 15F).
- Anterior margin of carapace finely crenulated in the median portion in C. laneus (Figure 3A and Figure 15A,E), C. brignolii (Figure 9A and Figure 15C), C. saradieli (Figure 15F), and C. dakshini sp. nov. (Figure 13A and Figure 15D) as opposed to smooth throughout in C. sinhagadensis (Figure 6A and Figure 15B).
- Ventral submedian carina of metasomal segment (Figure 19) I–III weakly developed in C. dakshini sp. nov. as opposed to moderately developed in C. sinhagadensis and strongly developed in C. laneus C. brignolii and C. saradieli.
- Metasomal segment V acarinate (Figure 17, Figure 18, Figure 19 and Figure 20) in C. dakshini sp. nov. as opposed to segment with a pair of weakly developed dorsal lateral carina present on the proximal portion in C. saradieli, moderately developed and present on the proximal portion in C. sinhagadensis and strongly developed and present on the proximal portion in C. laneus and C. brignolii.
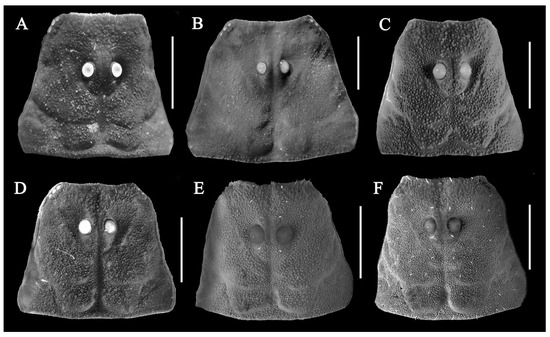
Figure 15.
Carapace dorsal views under UV light. (A) Charmus laneus Karsch, 1879, adult ♂ (IES-631); (B) Charmus sinhagadensis Tikader & Bastawade, 1983; neotype, adult ♂ (BNHS SC 403); (C) Charmus brignolii Lourenço, 2000; adult ♂ (IES-616); (D) Charmus dakshini sp. nov.; holotype, adult ♂ (BNHS SC 404), (E) Charmus laneus Karsch, 1879; adult ♂ (Locality 15CN) from Sri Lanka; (F) Charmus saradieli Kovarik et al. 2016 adult ♂ (Locality 18 CO) from Sri Lanka. Scale bars: 1 mm.
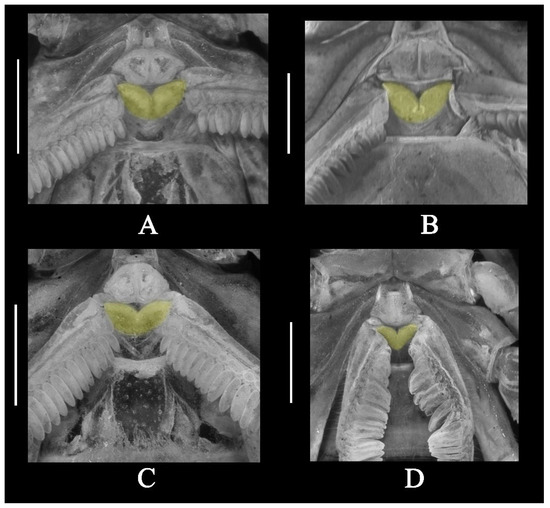
Figure 16.
Sternopectinal area under UV light; basal piece highlighted in yellow. (A) Charmus laneus Karsch, 1879, adult ♂ (IES-631); (B) Charmus sinhagadensis Tikader & Bastawade, 1983; neotype, adult ♂ (BNHS SC 403); (C) Charmus brignolii Lourenço, 2000; adult ♂ (IES-616); (D) Charmus dakshini sp. nov.; holotype, adult ♂ (BNHS SC 404).
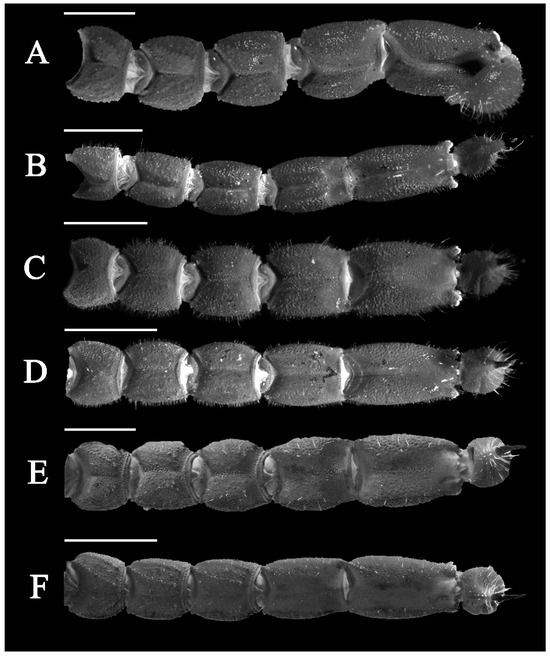
Figure 17.
Metasomal segments dorsal view under UV light. (A) Charmus laneus Karsch, 1879, adult ♀ (IES-628); (B) Charmus sinhagadensis Tikader & Bastawade, 1983; neotype, adult ♂ (BNHS SC 403); (C) Charmus brignolii Lourenço, 2000; adult ♂ (IES-616); (D) Charmus dakshini sp. nov.; holotype, adult ♂ (BNHS SC 404), (E) Charmus laneus Karsch, 1879; adult ♂ (Locality 15CN) from Sri Lanka; (F) Charmus saradieli Kovarik et al. 2016 adult ♂ (Locality 18 CO) from Sri Lanka. Scale bars: 2 mm. Remark: an adult female specimen of Charmus laneus was photographed due to unavailability of an intact adult male specimen.
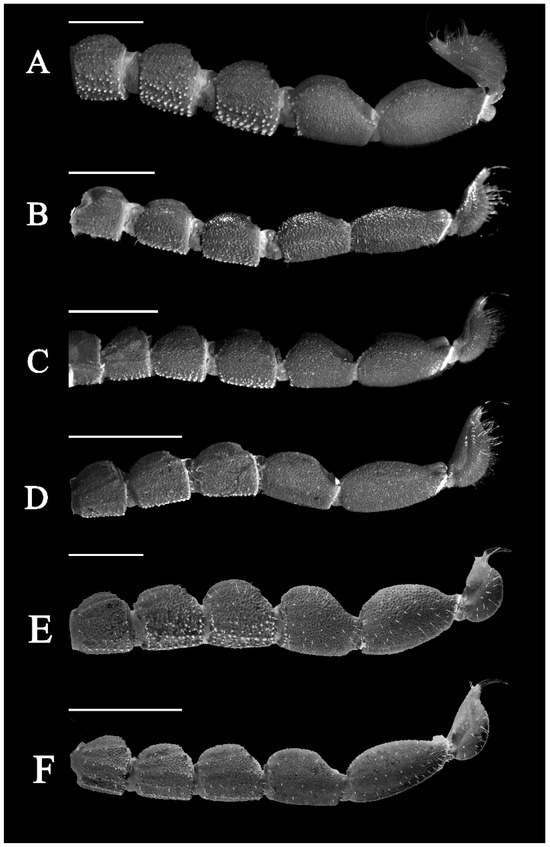
Figure 18.
Metasomal segments lateral view under UV light. (A) Charmus laneus Karsch, 1879, adult ♀ (IES-628); (B) Charmus sinhagadensis Tikader & Bastawade, 1983; neotype, adult ♂ (BNHS SC 403); (C) Charmus brignolii Lourenço, 2000; adult ♂ (IES-616); (D) Charmus dakshini sp. nov.; holotype, adult ♂ (BNHS SC 404), (E) Charmus laneus Karsch, 1879; adult ♂ (Locality 15CN) from Sri Lanka; (F) Charmus saradieli Kovarik et al. 2016 adult ♂ (Locality 18 CO) from Sri Lanka. Scale bars: 2 mm. Remark: an adult female specimen of Charmus laneus was photographed due to the unavailability of an intact adult male specimen.
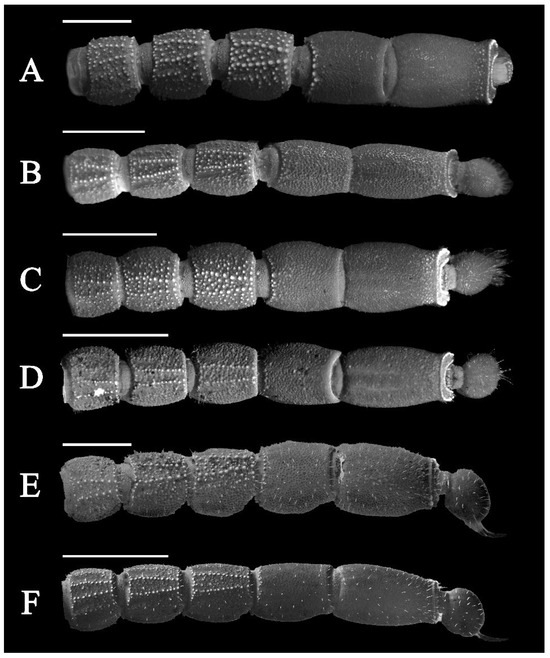
Figure 19.
Metasomal segments ventral view under UV light. (A) Charmus laneus Karsch, 1879, adult ♀ (IES-628); (B) Charmus sinhagadensis Tikader & Bastawade, 1983; neotype, adult ♂ (BNHS SC 403); (C) Charmus brignolii Lourenço, 2000; adult ♂ (IES-616); (D) Charmus dakshini sp. nov.; holotype, adult ♂ (BNHS SC 404), (E) Charmus laneus Karsch, 1879; adult ♂ (Locality 15CN) from Sri Lanka; (F) Charmus saradieli Kovarik et al. 2016 adult ♂ (Locality 18 CO) from Sri Lanka. Scale bars: 2 mm. Remark: an adult female specimen of Charmus laneus was photographed due to the unavailability of an intact adult male specimen.
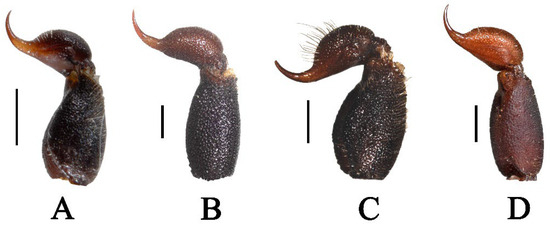
Figure 20.
Metasomal segment V and telson lateral view. (A) Charmus laneus, adult ♂ (IES-631); (B) Charmus sinhagadensis; neotype, adult ♂ (BNHS SC 403); (C) Charmus brignolii; adult ♂ (IES-616); (D) Charmus dakshini sp. nov.; holotype, adult ♂ (BNHS SC 404) Scale bars: 1 mm.
4. Results
4.1. Molecular Phylogeny of Charmus Based on Sanger-Sequenced Data (Figure 21 and Figure S1)
The genus Charmus is recovered as monophyletic and sister to the clade ((Parabuthus + Karasbergia) ((Thaicharmus + Buthoscorpio) (Grosphus + Teruelius)) in the IQ-TREE analysis, whereas it clusters with Uroplectes gracilior in a clade sister to ((Parabuthus + Karasbergia) ((Thaicharmus + Buthoscorpio) (Grosphus + Teruelius)) in the RAxML analysis. Charmus brignolii is recovered as the more basal element in Charmus and sister to the clade comprising all other species in both analyses. Charmus sinhagadensis is recovered as monophyletic and sister to the clade (C. saradieli (C. laneus + C. dakshini sp. nov.)) in both analyses. Charmus saradieli is sister to the clade (C. laneus + C. dakshini sp. nov.) in both analyses. Finally, Charmus dakshini sp. nov. is always recovered as the sister species to C. laneus.
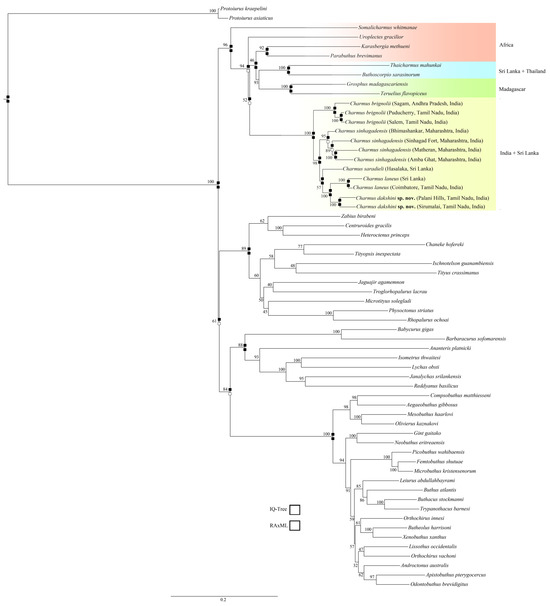
Figure 21.
Tree topology based on Maximum Likelihood (ML) analysis of Sanger-sequencing data (IQ-Tree). Values at the nodes are bootstraps for 1000 iterations. Navajo rugs indicate recovery of nodes for IQ Tree and RAxML analysis.
4.2. Comparisons of p-Distance with Closest Related Congeners (Tables S1–S3)
The new species C. dakshini sp. nov. is more closely related to C. laneus and differs by a raw genetic divergence of 9.4–11.4% and 6.4–7.1% on the COI and 16S genes, respectively. The new species is separated from C. brignolii by a raw genetic divergence of 10.1–12.4% and 9.4–10.7% on the COI and 16S genes, respectively. Charmus brignolii differs from C. laneus by a high raw genetic divergence of 10.3–13.6% and 9.4–10.7% on the COI and 16S genes, respectively. Specimens of C. laneus collected from Coimbatore, Tamil Nadu differ by a raw genetic divergence of 6.7% and 1.5% from C. laneus samples collected at the type locality on the COI and 16S genes, respectively. Based on the COI gene, the population of C. dakshini sp. nov. from Palani Hills, Tamil Nadu differs by a raw genetic divergence of 6.7% from the ones collected at the type locality. The populations of C. brignolii from Salem (Tamil Nadu), and Sangam (Andhra Pradesh) differ from C. brignolii collected at the type locality by a raw genetic divergence of 1.6% and 6.2% on the COI gene. The populations of C. sinhagadensis from Matheran, Amba Ghat, and Bhimashankar in Maharashtra show raw genetic divergences from C. sinhagadensis collected at the type locality of 8.6%, 8.4%, and 8.3%, respectively, on the COI gene. Additionally, the Bhimashankar population exhibits a divergence of 4.8% on the 16S gene from the population at the type locality. Based on the 28S gene data, C. laneus from Sri Lanka shows no genetic divergence from C. laneus collected from Coimbatore. Charmus saradieli differs from C. laneus by a raw genetic divergence of 0.6%. The populations of C. brignolii from Puducherry and Sangam (Andhra Pradesh) show an intraspecific divergence of 0.2% and differ from C. laneus by a raw genetic divergence of 0.6–0.8%. The populations of C. sinhagadensis from Sinhagad Fort, Bhimashankar and Amba Ghat show no intraspecific genetic divergence based on the 28S gene data. Charmus sinhagadensis shows a raw interspecific genetic divergence of 0.8–1.1% with the populations of C. brignolii and 0.2% with C. laneus.
4.3. Molecular Phylogenetics Based on the UCE Data (Figure 22)
Analyses of the various matrices recovered the same topology. The genus Charmus is recovered to be monophyletic and the sister group of Buthoscorpio Werner, 1936. The clade (Charmus + Buthoscorpio) is observed to be sister to Grosphus Simon, 1880. The clade (Grosphus (Charmus + Buthoscorpio) is found to be a sister to Parabuthus Pocock, 1890 and (Parabuthus (Grosphus (Charmus + Buthoscorpio) is itself sister to Uroplectes Peters, 1861. Charmus sinhagadensis from Amba Ghat and Bhimashankar (Maharashtra) is recovered to be a sister to Charmus dakshini sp. nov. Charmus brignolii is recovered to be a sister to the clade (C. sinhagadensis + C. dakshini sp. nov.).
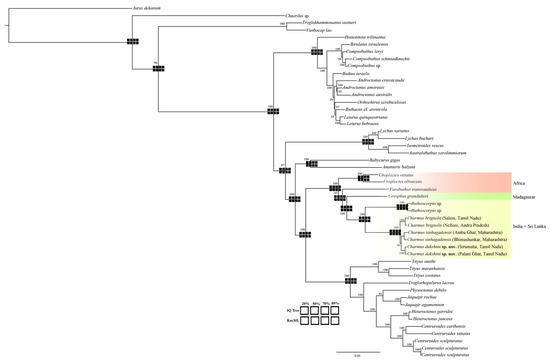
Figure 22.
Tree topology based on Maximum likelihood (ML) analysis of the UCE data. Navajo rugs indicate the recovery of nodes given the different occupancy matrices (20%, 50%, 70% and 80%).
5. Discussion
5.1. Congruence Between Morphology, Genetic Divergence, Phylogeny and Geography
Based on the genetic divergence computed for COI and 16S genes, the intraspecific variation is determined to be 8.6% or below for the COI gene and 4.8% or below for the 16S gene. Interestingly, the intraspecific variations reported for C. sinhagadensis samples (8.3–8.6% on COI and 4.8% on 16S) are much higher than those reported for C. brignolii (1.6–6.2% on COI), C. dakshini sp. nov. (6.7% on COI) and C. laneus (6.7% on COI and 1.5% on 16S). On the other hand, interspecific genetic divergence is at least 9.4% and 6.4% for COI and 16S, respectively. We hereby consider that a sample exhibiting a genetic divergence of 9.4% or more on the COI gene and/or a genetic divergence of 6.4% or more on the 16S gene, is to be considered as a distinct species, provided its distinctness holds against other lines of evidence.
The Sanger phylogeny generated here is congruent with the different morpho-species defined and the different genetic divergences calculated (inter- and intraspecific). Moreover, the geographical structure of the data is also in agreement with the phylogeny. Charmus is divided into three clades with distinct non-overlapping distribution ranges, i.e., the C. brignolii clade occurring in the plains of south-eastern India, the C. sinhagandensis clade occurring in the northern Western Ghats and the C. laneus clade (C. dakshini sp. nov., C. laneus, C. saradieli) occurring in the southern Western Ghats and Sri Lanka (Figure 23).
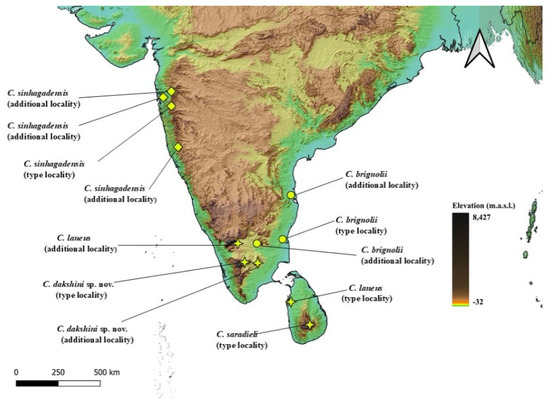
Figure 23.
Distribution of Charmus Karsch, 1879 in India and Sri Lanka with elevation data. The diamonds represent the C. sinhagadensis clade, circles represent the C. brignolii clade and the stars represent the C. laneus clade.
The alpha taxonomic study of the genus is strongly reinforced here by the analysis of genetic data, both comparatively and phylogenetically, as well as by the geographical pattern observed. The congruence between results from these various analyses allows us to propose a solid systematics assessment of Charmus.
5.2. Morphological Difference Between Lowland and Highland Species
Charmus brignolii and C. laneus are confined to the lowland grassland patches of Tamil Nadu and Andhra Pradesh (84–388 m). C. dakshini sp. nov. is found at comparatively higher elevations (813–1600 m), inhabiting the semi-evergreen forests of Palani Hills and Sirumalai. Similarly, C. sinhagadensis is also found at high elevations in the moist deciduous forests of the northern Western Ghats (455–1315 m). A feature worth mentioning is the reduced carination and overall granulation on metasomal segments observed in these two highland species (>700 m a.s.l.) when compared to the lowland species. Moreover, while highland species are bound to moist habitats, lowland species occur in much drier ecosystems.
We here hypothesize that the different degrees of cuticular carination and granulation observed in Charmus species are directly linked to the air temperatures and humidity levels. In arthropods, cuticular structures and patterning are known to influence the surface interaction with water [71]. For instance, on textured surfaces, water droplets will be suspended on top of the observed structures, creating a three-phase liquid–air–solid interface known as the Cassie–Baxter model [72,73]. In this model, an air pocket trapped between the droplets and the structured surface prevents the surface from getting wet. This interface will then have a cooling effect on the underlying surface. Moreover, dropwise condensation increases with the roughness of the surface, resulting in more heat transfer and cooling. Conversely, smooth surfaces are not able to accumulate the water as much as textured surfaces, and thus will not generate the same cooling effect. The textured cuticle of the lowland species may help them cope with low levels of air humidity and high temperatures—environmental constraints that highland species do not have.
5.3. Topological Differences Between Phylogenies
The genus Charmus is consistently grouped with South African and Malagasy taxa in both UCE and Sanger analyses, which is consistent with the Gondwana break-up history [74,75]. The UCE tree consistently reveals stable phylogenetic relationships with strong nodal supports across various analyses of several occupancy matrices. The following topological sequence is retrieved in all analyses: (Uroplectes (Parabuthus (Grosphus + (Buthoscorpio + Charmus)))). However, analyses of Sanger data with IQ-TREE and RAxML yield slightly different topologies with some weakly supported nodes: (Uroplectes ((Parabuthus + Karasbergia) (Charmus ((Thaicharmus + Buthoscorpio) (Grosphus + Teruelius))))) in the RAxML analysis, ((Uroplectes + Charmus) ((Parabuthus + Karasbergia) ((Thaicharmus + Buthoscorpio) (Grosphus + Teruelius)))). The assumption that the UCEs are more suitable for resolving higher-level relationships is confirmed here. The Sanger phylogenies do not provide a consensus topology, and the branch supports remain insufficient to consider it reliable. On the other hand, the topology computed by analyses of the UCE dataset remains constant and well-supported.
Whereas Charmus is sister to Buthoscorpio in UCE analysis, it is retrieved as sister to a clade comprising Buthoscorpio, Thaicharmus, Grosphus, Teruelius, Karasbergia and Parabuthus in the IQ-TREE analysis of Sanger data and as sister to Uroplectes in the RAxML analysis of Sanger data, albeit with low branch support. The morphology and geographical distribution of Buthoscorpio and Thaicharmus suggest that they are more closely related to Charmus than to Grosphus/Teruelius. The UCE topology is congruent with this assumption and is considered as more reliable than those generated with Sanger data. However, this cannot be presently unambiguously proven because a sample of Thaicharmus is currently missing in the UCE dataset. Therefore, additional UCE phylogenetic analyses with an updated dataset need to be conducted before the status of Charmus within the family Buthidae is further clarified.
5.4. Biogeographic Considerations
The Southern Western Ghats (SWG) and the Central Highlands of Sri Lanka acted as refugia for many groups of the Indian wet evergreen fauna and flora during the KT Mass Extinction [76,77,78,79]. It is expected from this pattern that the lineages occurring in the SWG are ancient and more diverse, whereas those occurring in the northern and central Western Ghats (NWG and CWG) are less speciose, relatively younger and phylogenetically nested [80]. However, the phylogeny of Charmus partially contradicts this biogeographic pattern. The basal species C. brignolii is not present in the Western Ghats but rather occurs on the southeastern plains. Furthermore, C. sinhagadensis seems to be restricted to the NWG whereas its sister group (C. saradieli (C. laneus + C. dakshini sp. nov.)) is probably endemic to the SWG and Sri Lanka. Interestingly, no Charmus species are currently known from the CWG suggesting that either Charmus is present in the area but has not been collected yet or that, contrary to the NWG and SWG, the CWG were not sufficiently stable environmentally to ensure the survival of Charmus during range contractions. This interesting biogeographic pattern needs to be tested separately using dated phylogenies and dispersal-vicariance analyses.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/d17050354/s1.
Author Contributions
Conceptualization, S.S., L.M., S.D., D.B. and M.J.; methodology, S.S., L.M., S.D., M.J., S.U., J.B. and H.M.; software, L.M., S.D., M.J. and S.U.; validation, S.S., L.M., S.D., D.B., G.G., F.K. and F.Š.; formal analysis, S.S., L.M., S.D., G.G. and M.J.; investigation, S.S., S.D., M.J. and S.U.; resources, L.M. and S.S.; data curation, S.D., M.J. and S.U.; writing—original draft preparation, M.J. and S.D.; writing—review and editing, S.S., L.M., S.D., G.G., M.J., S.U., F.K. and F.Š.; visualization, S.S., L.M., S.D. and M.J.; supervision, S.S., L.M., S.D., D.B. and G.G.; project administration, S.S. and L.M.; funding acquisition, L.M. and S.S. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
Not applicable.
Data Availability Statement
Data have been submitted to the NCBI portal and will be publicly available at a later data (Submission numbers are available on the Supplementary Tables).
Acknowledgments
The authors are grateful to the National Biodiversity Authority (NBA); Ministry of Environment, Forest and Climate Change (MoEFCC), Government of India for their cooperation and for providing the necessary permits for transporting tissue samples from India to Switzerland. We are grateful to Scorpion Systematics Laboratory (SSL), and InSearch Environmental Solutions (IES) for the partial funding and institutional support. We thank Hemant Ghate for providing access to his photographic facilities. We also thank Aarya Gramopadhye, Aditya Soman, Akash Joshi, Akshay Marathe, Makarand Ketkar, Mukta Nachare, Avrajjal Ghosh, Rohit Karmalkar and Vivek Surase for their help during fieldwork. Additionally, we are grateful to Rohit Karmalkar for making the illustration in Figure 10. We are thankful to Rahul Khot for his help during the registration of specimens at the Bombay Natural History Society (BNHS), Mumbai. We thank Neil Kakkar for providing photographs of Auroville, Puduchery, India. We also appreciate the valuable comments and suggestions from two anonymous reviewers that helped improve the manuscript.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Perera, S.J.; Suranjanfernando, R.H.S. Physiography, Climate, and Historical Biogeography of Sri Lanka in Making a Biodiversity Hotspot. In Biodiversity Hotspots of the Western Ghats and Sri Lanka; Apple Academic Press: Palm Bay, FL, USA, 2022; pp. 405–445. [Google Scholar]
- Dissanayake, C.B.; Chandrajith, R. Sri Lanka–Madagascar Gondwana Linkage: Evidence for a Pan-African Mineral Belt. J. Geol. 1999, 107, 223–235. [Google Scholar] [CrossRef]
- Deepak, V.; Karanth, P. Aridification Driven Diversification of Fan-Throated Lizards from the Indian Subcontinent. Mol. Phylogenet. Evol. 2018, 120, 53–62. [Google Scholar] [CrossRef] [PubMed]
- Gorin, V.A.; Solovyeva, E.N.; Hasan, M.; Okamiya, H.; Karunarathna, D.M.S.S.; Pawangkhanant, P.; de Silva, A.; Juthong, W.; Milto, K.D.; Nguyen, L.T.; et al. A little frog leaps a long way: Compounded colonizations of the Indian Subcontinent discovered in the tiny Oriental frog genus Microhyla (Amphibia: Microhylidae). PeerJ 2020, 8, e9411. [Google Scholar] [CrossRef] [PubMed]
- Prendini, L.; Weygoldt, P.; Wheeler, W.C. Systematics of the Damon variegatus group of African whip spiders (Chelicerata: Amblypygi): Evidence from behavior, morphology, and DNA. Org. Divers. Evol. 2005, 5, 203–236. [Google Scholar] [CrossRef]
- Seiter, M.; Lerma, A.; Král, J.; Sember, A.; Divišová, K.; Palacios-Vargas, J.; Colmenares, P.A.; Loria, S.F.; Prendini, L. Cryptic diversity in the whip spider genus Paraphrynus (Amblypygi: Phrynidae): Integrating morphology, karyotype, and DNA. Arthropod Syst. Phylogeny 2020, 78, 265–285. [Google Scholar]
- Schramm, F.D.; Valdez-Mondragón, A.; Prendini, L. Volcanism and palaeoclimate change drive diversification of the world’s largest whip spider (Amblypygi). Mol. Ecol. 2021, 12, 2872–2890. [Google Scholar] [CrossRef]
- Karsch, F. Skorpionologische Beiträge I. und II. Mitt. Münch. Entomol. Ver. 1879, 3, 6–22, 97–136. [Google Scholar]
- Hirst, S. Description of a new Indian Scorpion (Charmus indicus sp. n.). Ann. Mag. Nat. Hist. 1915, 15, 224–225. [Google Scholar] [CrossRef]
- Sreenivasa-Reddy, R.P. Contribution à la connaissance des Scorpions de l’Inde. I. Charmus indicus Hirst, 1915 (Fam. Buthidae). Bull. Mus. Natl. Hist. 1966, 38, 247–256. [Google Scholar]
- Tikader, B.K.; Bastawade, D.B. The Fauna of India; Zoological Survey of India: Calcutta, India, 1983; Volume 3, pp. 146–152. [Google Scholar]
- Lourenço, W.R. Taxonomic considerations about the genus Charmus Karsch, 1879 with description of a new species to India (Scorpiones: Buthidae). Mem. Soc. Entomol. 2000, 78, 295–303. [Google Scholar]
- Lourenço, W.R. Further taxonomic considerations about the genus Charmus Karsch, 1879 (Scorpiones, Buthidae), with the description of a new species from Sri Lanka. Entomol. Mitt. Zool. Mus. Hamb. 2002, 14, 17–25. [Google Scholar]
- Kovarik, F.; Lowe, G.; Ranawana, K.B.; Hoferek, D.; Jayarathne, V.A.S.; Pliskova, J. Scorpions of Sri Lanka (Arachnida, Scorpiones: Buthidae, Chaerilidae, Scorpionidae) with Description of Four New Species of the Genera Charmus Karsch, 1879 and Reddyanus Vachon, 1972, stat. n. Euscorpius 2016, 220, 1–133. [Google Scholar] [CrossRef]
- International Commission on Zoological Nomenclature (ICZN). International Code of Zoological Nomenclature; Online Edition; International Commission on Zoological Nomenclature (ICZN): Singapore, 2000; Available online: https://www.iczn.org/the-code/the-international-code-of-zoological-nomenclature/the-code-online/ (accessed on 7 December 2024).
- Wong, E.S.; Dahlem, G.A.; Stamper, T.I.; DeBry, R.W. Discordance between Morphological Species Identification and mtDNA Phylogeny in the Flesh Fly Genus Ravinia (Diptera: Sarcophagidae). Invertebr. Syst. 2015, 29, 1–11. [Google Scholar] [CrossRef]
- Andriaholinirina, N.; Fausser, J.L.; Roos, C.; Zinner, D.; Thalmann, U.; Rabarivola, C.; Ravoarimanana, I.; Ganzhorn, J.U.; Meier, B.; Hilgartner, R.; et al. Molecular Phylogeny and Taxonomic Revision of the Sportive Lemurs (Lepilemur, Primates). BMC Evol. Biol. 2006, 6, 1–13. [Google Scholar] [CrossRef]
- Cook, L.G.; Edwards, R.D.; Crisp, M.D.; Hardy, N.B. Need Morphology Always Be Required for New Species Descriptions? Invertebr. Syst. 2010, 24, 322–326. [Google Scholar] [CrossRef]
- Gullan, P.J.; Downie, D.A.; Steffan, S.A. A New Pest Species of the Mealybug Genus Ferrisia Fullaway (Hemiptera: Pseudococcidae) from the United States. Ann. Entomol. Soc. Am. 2003, 96, 723–737. [Google Scholar] [CrossRef]
- Sharma, P.K.; Fernández, R.M.N.; Esposito, L.A.; González-Santillán, E.; Monod, L. Phylogenomic Resolution of Scorpions Reveals Multilevel Discordance with Morphological Phylogenetic Signal. Proc. R. Soc. B Biol. Sci. 2015, 282, 20142953. [Google Scholar] [CrossRef]
- Santibáñez-López, C.E.; Ojanguren-Affilastro, A.A.; Sharma, P.P. Another One Bites the Dust: Taxonomic Sampling of a Key Genus in Phylogenomic Datasets Reveals More Non-Monophyletic Groups in Traditional Scorpion Classification. Invertebr. Syst. 2020, 34, 133–143. [Google Scholar] [CrossRef]
- Štundlová, J.; Šťáhlavský, F.; Opatova, V.; Stundl, J.; Kovařík, F.; Dolejš, P.; Šmíd, J. Molecular Data Do Not Support the Traditional Morphology-Based Groupings in the Scorpion Family Buthidae (Arachnida: Scorpiones). Mol. Phylogenet. Evol. 2022, 173, 107511. [Google Scholar] [CrossRef]
- Svenson, G.J.; Whiting, M.F. Reconstructing the origins of praying mantises (Dictyoptera: Mantodea): The roles of Gondwanan vicariance and morphological convergence. Cladistics 2009, 25, 468–514. [Google Scholar] [CrossRef]
- Hendrixson, E.; Bond, J.E. Evaluating the efficacy of continuous quantitative characters for reconstructing the phylogeny of a morphologically homogeneous spider taxon (Araneae, Mygalomorphae, Antrodiaetidae, Antrodiaetus). Mol. Phylogenet. Evol. 2009, 53, 300–313. [Google Scholar] [CrossRef] [PubMed]
- Ortiz, D.; Francke, O.F.; Bond, J.E. A tangle of forms and phylogeny: Extensive morphological homoplasy and molecular clock heterogeneity in Bonnetina and related tarantulas. Mol. Phylogenet. Evol. 2018, 127, 55–73. [Google Scholar] [CrossRef] [PubMed]
- Opatova, V.; Hamilton, C.A.; Hedin, M.; Montes De Oca, L.; Král, J.; Bond, J.E. Phylogenetic systematics and evolution of the spider infraorder Mygalomorphae using genomic scale data. Syst. Biol. 2020, 69, 671707. [Google Scholar] [CrossRef] [PubMed]
- Masonick, P.K.; Knyshov, A.; Gordon, E.R.L.; Forero, D.; Hwang, W.S.; Hoey-Chamberlain, R.; Bush, T.; Castillo, S.; Hernandez, M.; Ramirez, J.; et al. A revised classification of the assassin bugs (Hemiptera: Heteroptera: Reduviidae) based on combined analysis of phylogenomic and morphological data. Syst. Entomol. 2025, 50, 102–138. [Google Scholar] [CrossRef]
- Sharma, P.P.; Kaluziak, S.T.; Pérez-Porro, A.R.; González, V.L.; Hormiga, G.; Wheeler, W.C.; Giribet, G. Phylogenomic interrogation of Arachnida reveals systemic conflicts in phylogenetic signal. Mol. Biol. Evol. 2014, 31, 2963–2984. [Google Scholar] [CrossRef]
- Agnarsson, I.; Coddington, J.A.; Kuntner, M. Systematics: Progress in the study of spider diversity and evolution. In Spider Research in the 21st Century: Trends and Perspectives; Penney, D., Ed.; Siri Scientific Press: Greater Manchester, UK, 2013; pp. 58–111. [Google Scholar]
- Bond, J.E.; Garrison, N.L.; Hamilton, C.A.; Godwin, R.L.; Hedin, M.; Agnarsson, I. Phylogenomics resolves a spider backbone phylogeny and rejects a prevailing paradigm for orb web evolution. Curr. Biol. 2014, 24, 1765–1771. [Google Scholar] [CrossRef] [PubMed]
- Fernández, R.; Hormiga, G.; Giribet, G. Phylogenomic analysis of spiders reveals nonmonophyly of orb weavers. Curr. Biol. 2014, 24, 1772–1777. [Google Scholar] [CrossRef]
- Godwin, R.L.; Opatova, V.; Garrison, N.L.; Hamilton, C.A.; Bond, J.E. Phylogeny of a cosmopolitan family of morphologically conserved trapdoor spiders (Mygalomorphae, Ctenizidae) using Anchored Hybrid Enrichment, with a description of the family Halonoproctidae Pocock 1901. Mol. Phylogenet. Evol. 2018, 126, 303–313. [Google Scholar] [CrossRef]
- Hedin, M.; Derkarabetian, S.; Ramírez, M.J.; Vink, C.; Bond, J.E. Phylogenomic reclassification of the world’s most venomous spiders (Mygalomorphae, Atracinae), with implications for venom evolution. Sci. Rep. 2018, 8, 1636. [Google Scholar] [CrossRef]
- Wood, H.M.; González, V.L.; Lloyd, M.; Coddington, J.; Scharff, N. Next-generation Museum genomics: Phylogenetic relationships among palpimanoid spiders using sequence capture techniques (Araneae: Palpimanoidea). Mol. Phylogenet. Evol. 2018, 127, 907–918. [Google Scholar] [CrossRef]
- Kulkarni, S.; Kallal, B.; Wood, H.; Dimitrov, D.; Giribet, G.; Hormiga, G. Interrogating genomic-scale data to resolve recalcitrant nodes in the spider tree of life. Mol. Biol. Evol. 2020, 38, 891–903. [Google Scholar] [CrossRef] [PubMed]
- Faircloth, B.C.; McCormack, J.E.; Crawford, N.G.; Harvey, M.G.; Brumfield, R.T.; Glenn, T.C. Ultraconserved elements anchor thousands of genetic markers spanning multiple evolutionary timescales. Syst. Biol. 2012, 61, 717–726. [Google Scholar] [CrossRef] [PubMed]
- Vachon, M. Études des Caractères Utilisés pour Classer les Familles et les Genres de Scorpions (Arachnides). 1. La Trichobothriotaxie en Arachnologie. Sigles Trichobothriaux et Types de Trichobothriotaxie chez les Scorpions. Bull. Mus. Natl. Hist. Nat. 1974, 104, 857–958. [Google Scholar]
- Francke, O.F. Scorpions of the Genus Diplocentrus from Oaxaca, Mexico (Scorpionida, Diplocentridae). J. Arachnol. 1977, 4, 145–200. [Google Scholar]
- González-Santillán, E.; Prendini, L. Redefinition and Generic Revision of the North American Vaejovid Scorpion Subfamily Syntropinae Kraepelin, 1905, with Descriptions of Six New Genera. Bull. Am. Mus. Nat. Hist. 2013, 382, 1–71. [Google Scholar] [CrossRef]
- Hjelle, J.T. Anatomy and morphology. In The Biology of Scorpions; Polis, G.A., Cloudsley-Thompson, J., Eds.; Stanford University Press: Stanford, CA, USA, 1990; pp. 9–63. [Google Scholar]
- Loria, S.F.; Prendini, L. Homology of the Lateral Eyes of Scorpiones: A Six-Ocellus Model. PLoS ONE 2014, 9, 1–30. [Google Scholar] [CrossRef]
- Volschenk, E.S. A New Technique for Examining Surface Morphosculpture of Scorpions. J. Arachnol. 2005, 33, 820–825. [Google Scholar] [CrossRef]
- Stahnke, H.L. Scorpion Nomenclature and Mensuration. Entomol. News 1970, 81, 297–316. [Google Scholar]
- Starrett, J.; Derkarabetian, S.; Hedin, M.; Bryson, R.W., Jr.; McCormack, J.E.; Faircloth, B.C. High phylogenetic utility of an ultraconserved element probe set designed for Arachnida. Mol. Ecol. Resour. 2017, 17, 812–823. [Google Scholar] [CrossRef]
- Santibáñez-López, C.E.; Aharon, S.; Ballesteros, J.A.; Gainett, G.; Baker, C.M.; González-Santillán, E.; Harvey, M.S.; Hassan, M.K.; Abu-Almaaty, A.H.; Aldeyarbi, S.M.; et al. Phylogenomics of scorpions reveal a co-diversification of scorpion mammalian predators and mammal-specific sodium channel toxins. Syst. Biol. 2022, 71, 1281–1289. [Google Scholar] [CrossRef]
- Sulakhe, S.; Deshpande, S.; Gowande, G.; Dandekar, N.; Ketkar, M. Arboreal Gems: Resurrection of Isometrus sankeriensis Tikader & Bastawade, 1983 and Descriptions of Two New Species of Isometrus Ehrenberg, 1828 (Scorpiones: Buthidae) from the Western Ghats, India. Eur. J. Taxon. 2022, 811, 1–50. [Google Scholar] [CrossRef]
- Folmer, O.; Black, M.; Hoeh, W.; Lutz, R.; Vrijenhoek, R. DNA primers for amplification of mitochondrial cytochrome c oxidase subunit I from diverse metazoan invertebrates. Mol. Mar. Biol. Biotechnol. 1994, 3, 294–299. [Google Scholar] [CrossRef] [PubMed]
- Simon, C.; Frati, F.; Beckenbach, A.; Crespi, B.; Liu, H.; Flook, P. Evolution, weighting, and phylogenetic utility of mitochondrial gene sequences and a compilation of conserved polymerase chain reaction primers. Ann. Entomol. Soc. Am. 1994, 87, 651–701. [Google Scholar] [CrossRef]
- Giribet, G.; Carranza, S.; Baguñà, J.; Riutort, M.; Ribera, C. First molecular evidence for the existence of a Tardigrada + Arthropoda clade. Mol. Biol. Evol. 1996, 13, 76–84. [Google Scholar] [CrossRef]
- Barrett, R.D.H.; Hebert, P.D.N. Identifying spiders through DNA barcodes. Can. J. Zool. 2005, 83, 481–491. [Google Scholar] [CrossRef]
- Benson, D.A.; Cavanaugh, M.; Clark, K.; Karsch-Mizrachi, I.; Lipman, D.J.; Ostell, J.; Sayers, E.W. GenBank. Nucleic Acids Res. 2017, 45, D37–D42. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef]
- Edgar, R.C. MUSCLE: Multiple Sequence Alignment with High Accuracy and High Throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef]
- Nguyen, L.T.; Schmidt, H.A.; Von Haeseler, A.; Minh, B.Q. IQ-TREE: A Fast and Effective Stochastic Algorithm for Estimating Maximum-Likelihood Phylogenies. Mol. Biol. Evol. 2015, 32, 268–274. [Google Scholar] [CrossRef]
- Edler, D.; Klein, J.; Antonelli, A.; Silvestro, D. raxmlGUI 2.0: A graphical interface and toolkit for phylogenetic analyses using RAxML. Methods Ecol. Evol. 2021, 12, 373–377. [Google Scholar] [CrossRef]
- Kalyaanamoorthy, S.; Minh, B.Q.; Wong, T.K.; Von Haeseler, A.; Jermiin, L.S. ModelFinder: Fast Model Selection for Accurate Phylogenetic Estimates. Nat. Methods 2017, 14, 587–589. [Google Scholar] [CrossRef] [PubMed]
- Hoang, D.T.; Chernomor, O.; Von Haeseler, A.; Minh, B.Q.; Vinh, L.S. UFBoot2: Improving the Ultrafast Bootstrap Approximation. Mol. Biol. Evol. 2018, 35, 518–522. [Google Scholar] [CrossRef] [PubMed]
- Minh, B.Q.; Schmidt, H.A.; Chernomor, O.; Schrempf, D.; Woodhams, M.D.; Von Haeseler, A.; Lanfear, R. IQ-TREE 2: New Models and Efficient Methods for Phylogenetic Inference in the Genomic Era. Mol. Biol. Evol. 2020, 37, 1530–1534. [Google Scholar] [CrossRef] [PubMed]
- Lanfear, R.; Calcott, B.; Ho, S.Y.W.; Guindon, S. PartitionFinder: Combined Selection of Partitioning Schemes and Substitution Models for Phylogenetic Analyses. Mol. Biol. Evol. 2012, 29, 1695–1701. [Google Scholar] [CrossRef]
- Schwarz, G. Estimating the Dimension of a Model. Ann. Stat. 1978, 6, 461–464. [Google Scholar] [CrossRef]
- Suchan, T.; Pitteloud, C.; Gerasimova, N.S.; Kostikova, A.; Schmid, S.; Arrigo, N.; Pajkovic, M.; Ronikier, M.; Alvarez, N. Hybridization Capture Using RAD Probes (hyRAD), a New Tool for Performing Genomic Analyses on Collection Specimens. PLoS ONE 2016, 11, e0151651. [Google Scholar] [CrossRef]
- Meyer, M.; Kircher, M. Illumina Sequencing Library Preparation for Highly Multiplexed Target Capture and Sequencing. Cold Spring Harb. Protoc. 2010, 2010, pdb-prot5448. [Google Scholar] [CrossRef]
- Faircloth, B.C. Identifying conserved genomic elements and designing universal bait sets to enrich them. Methods Ecol. Evol. 2017, 8, 1103–1112. [Google Scholar] [CrossRef]
- Grabherr, M.G.; Haas, B.J.; Yassour, M.; Levin, J.Z.; Thompson, D.A.; Amit, I.; Adiconis, X. Trinity: Reconstructing a Full-Length Transcriptome Without a Genome from RNA-Seq Data. Nat. Biotechnol. 2011, 29, 644. [Google Scholar] [CrossRef]
- Bankevich, A.; Nurk, S.; Antipov, D.; Gurevich, A.A.; Dvorkin, M.; Kulikov, A.S.; Lesin, V.M.; Nikolenko, S.I.; Pham, S.; Prjibelski, A.D.; et al. SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 2012, 19, 455–477. [Google Scholar] [CrossRef]
- Faircloth, B.C. PHYLUCE is a software package for the analysis of conserved genomic loci. Bioinformatics 2016, 32, 786–788. [Google Scholar] [CrossRef] [PubMed]
- Katoh, K.; Standley, D.M. MAFFT Multiple Sequence Alignment Software Version 7: Improvements in Performance and Usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef]
- Castresana, J. Selection of Conserved Blocks from Multiple Alignments for Their Use in Phylogenetic Analysis. Mol. Biol. Evol. 2000, 17, 540–552. [Google Scholar] [CrossRef] [PubMed]
- Talavera, G.; Casteresana, J. Improvement of phylogenies after removing divergent and ambiguously aligned blocks from protein sequence alignments. Syst. Biol. 2007, 56, 564–577. [Google Scholar] [CrossRef] [PubMed]
- Stamatakis, A.; Hoover, P.; Rougemont, J. A Rapid Bootstrap Algorithm for the RAxML Web Servers. Syst. Biol. 2008, 57, 758–771. [Google Scholar] [CrossRef]
- Bello, E.; Chen, Y.; Alleyne, M. Staying Dry and Clean: An Insect’s Guide to Hydrophobicity. Insects 2023, 14, 42. [Google Scholar] [CrossRef]
- Wenzel, R.N. Resistance of Solid Surfaces to Wetting by Water. Ind. Eng. Chem. 1936, 28, 988–994. [Google Scholar] [CrossRef]
- Cassie, A.B.D.; Baxter, S. Wettability of Porous Surfaces. Trans. Faraday Soc. 1944, 40, 546–551. [Google Scholar] [CrossRef]
- Mani, M.S. (Ed.) Ecology and Biogeography in India, 3rd ed.; Dr. W. Junk, B.V., Publishers: The Hague, The Netherlands, 1974; pp. 154–196. [Google Scholar]
- Aitchison, J.C.; Ali, J.R.; Davis, A.M. When and where did India and Asia collide? J. Geophys. Res. Solid Earth 2007, 112, B05423. [Google Scholar] [CrossRef]
- Conti, E.; Eriksson, T.; Schönenberger, J.; Sytsma, K.J.; Baum, D.A. Early Tertiary Out-of-India dispersal of Crypteroniaceae: Evidence from phylogeny and molecular dating. Evolution 2002, 56, 1931–1942. [Google Scholar]
- Bossuyt, F.; Brown, R.M.; Hillis, D.M.; Cannatella, D.C.; Milinkovitch, M.C. Phylogeny and biogeography of a cosmopolitan frog radiation: Late Cretaceous diversification resulted in continent-scale endemism in the family Ranidae. Syst. Biol. 2006, 55, 579–594. [Google Scholar] [CrossRef] [PubMed]
- Joshi, J.; Karanth, P. Did southern Western Ghats of peninsular India serve as refugia for its endemic biota during the Cretaceous volcanism? Ecol. Evol. 2013, 3, 3275–3282. [Google Scholar] [CrossRef] [PubMed]
- Loria, S.F.; Prendini, L. Out of India, thrice: Diversification of Asian forest scorpions reveals three colonizations of Southeast Asia. Sci. Rep. 2020, 10, 22301. [Google Scholar] [CrossRef] [PubMed]
- Bharti, D.K.; Edgecombe, G.D.; Karanth, K.P.; Joshi, J. Spatial patterns of phylogenetic diversity and endemism in the Western Ghats, India: A case study using ancient predatory arthropods. Ecol. Evol. 2021, 11, 16499–16513. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).