Environmental DNA for Assessing Population and Spatial Distribution of Spinibarbus caldwelli in the Liuxi River
Abstract
1. Introduction
- Tank experiment: exploring the relationship between environmental DNA and abundance/biomass.
- Pool cage fish experiment: analyzing the diffusion distance of eDNA in a lentic ecosystem.
- Field cage fish experiment: studying the diffusion distance of eDNA in a lotic ecosystem.
- Experiment on monitoring the resources of S. caldwelli in the Liuxi River: investigating the distribution and population size of S. caldwelli by using environmental DNA technology and comparing the data with those obtained by traditional methods.
2. Materials and Methods
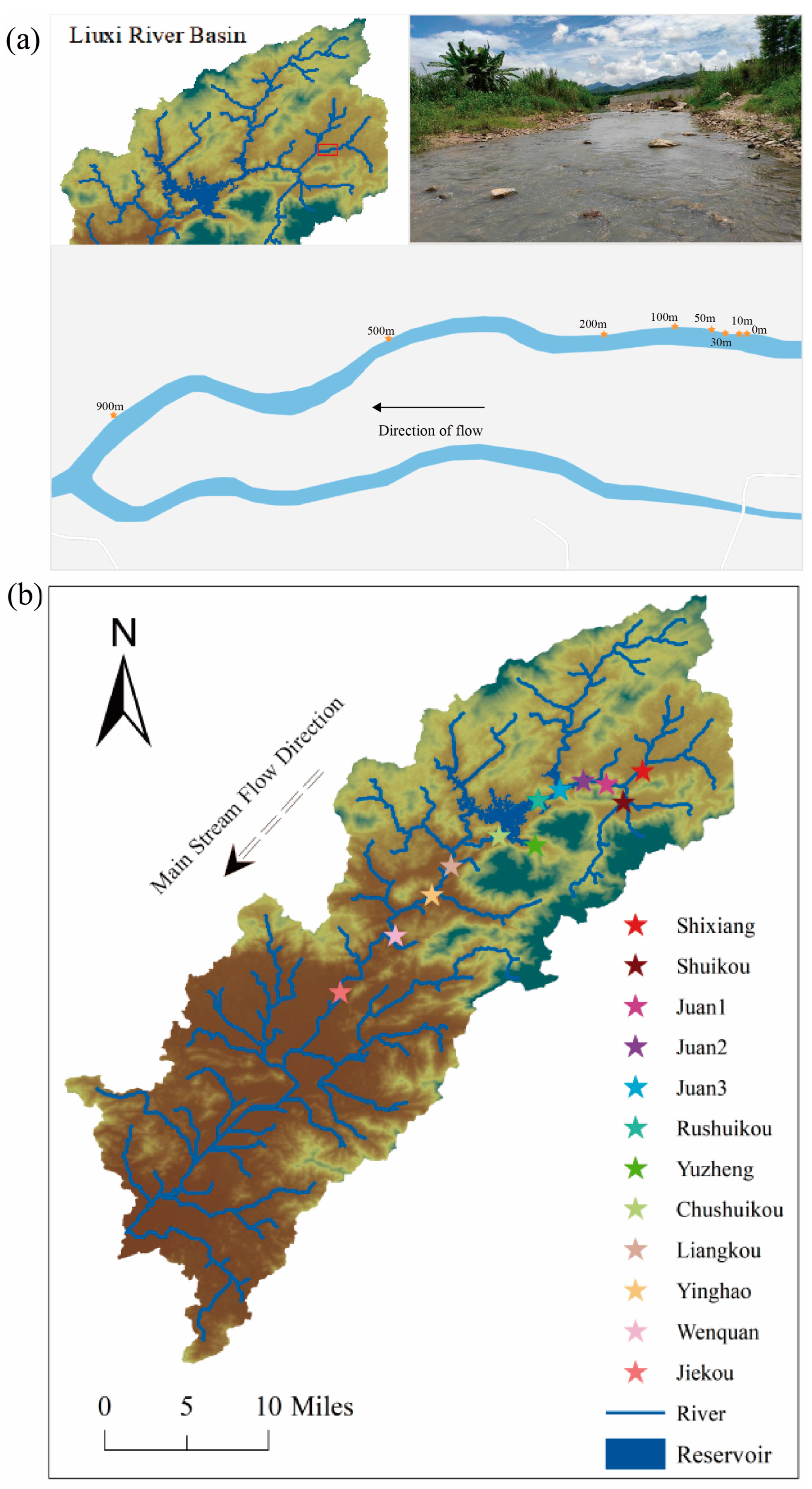
2.1. Experimental Strategy
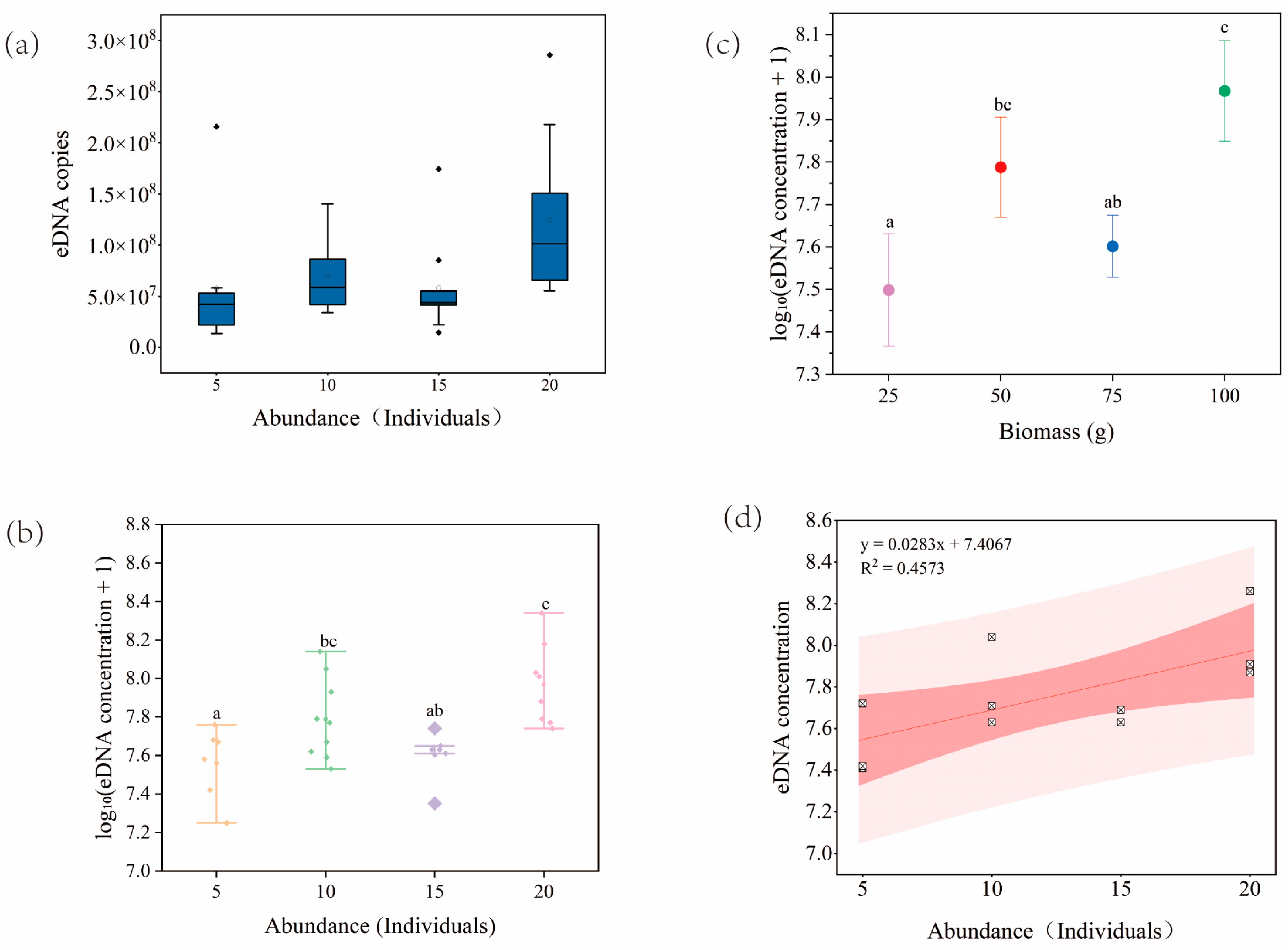
2.1.1. Tank Experiment: Relationship Between eDNA Concentration and Abundance/Biomass
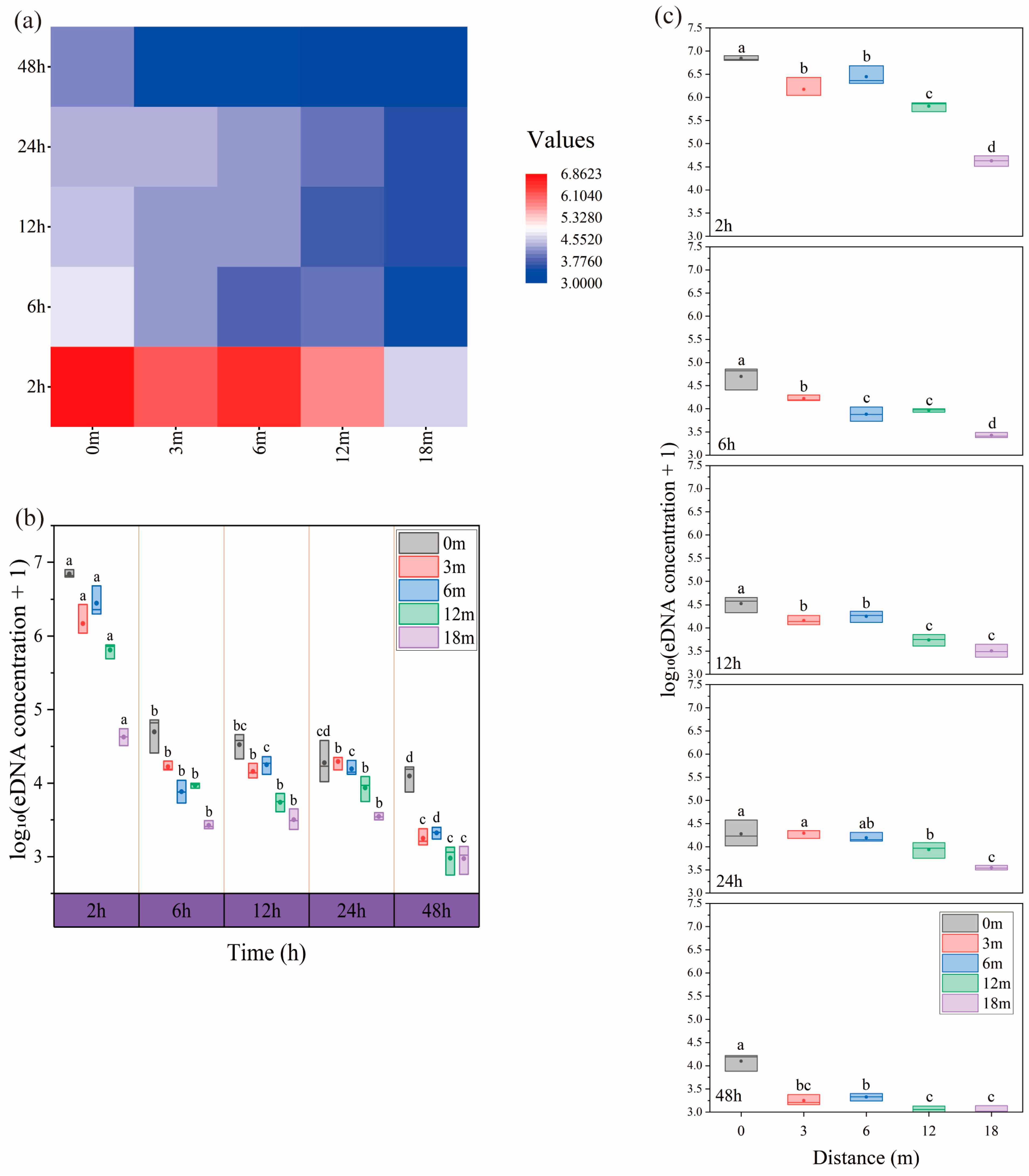
2.1.2. Lentic Ecosystem: Diffusion of eDNA
2.1.3. Lotic Ecosystem: Diffusion of eDNA
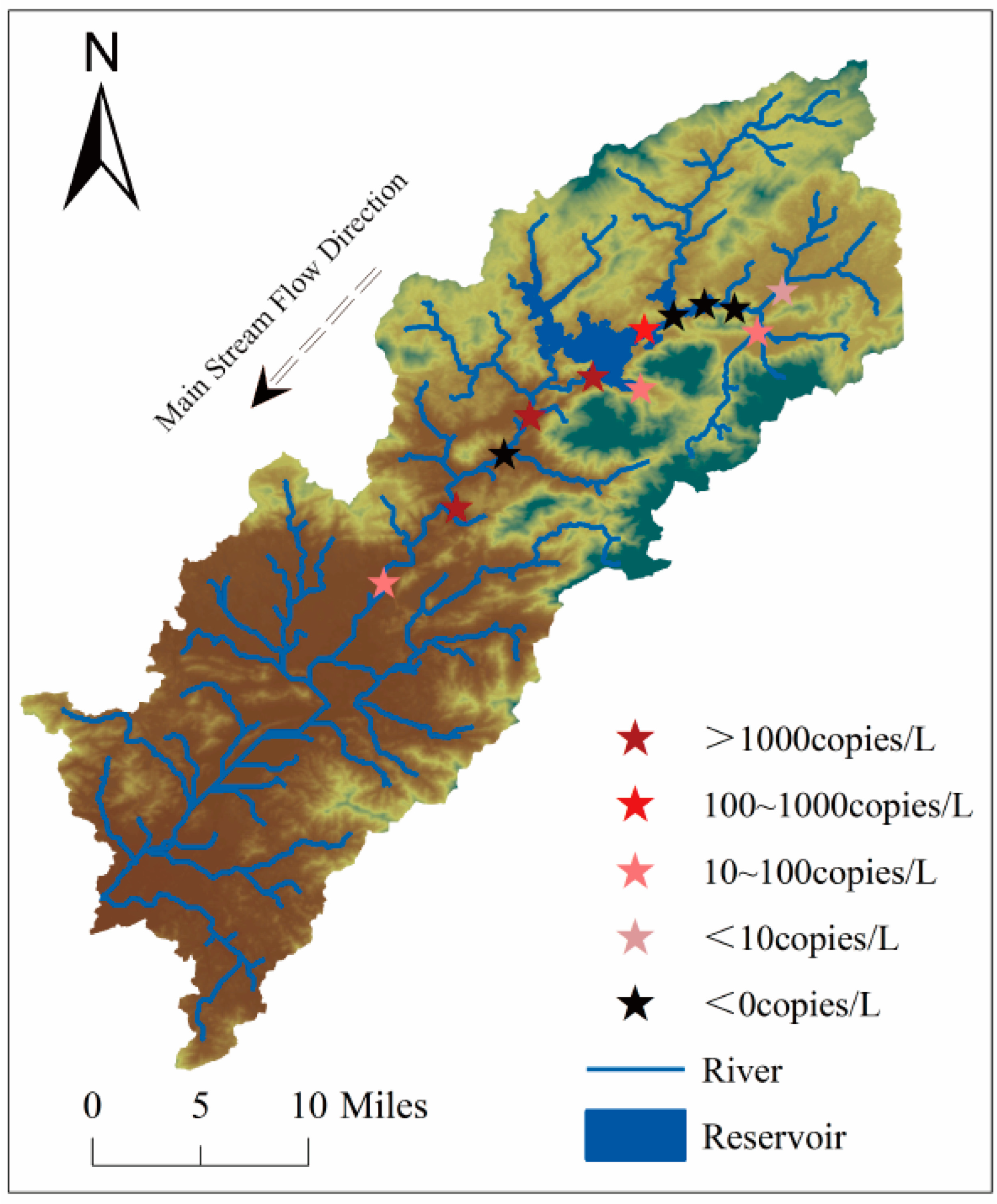
2.1.4. Liuxi River Survey: Monitoring Effectiveness of eDNA Methodology
2.2. Primer/Probe Design
2.3. Plasmid Standard Preparation
2.4. eDNA Sample Collection/Filtration
2.4.1. Tank Experiment
2.4.2. Lentic Ecosystem
2.4.3. Lotic Ecosystem
2.4.4. Liuxi River Survey
2.5. eDNA Extraction
2.6. Sensitivity Testing
2.7. Statistical Processing and Analysis of Data
2.7.1. Tank Experiment
2.7.2. Lentic Ecosystem
2.7.3. Lotic Ecosystem
2.7.4. Liuxi River Survey
3. Results
3.1. Tank Experiment: Relationship Between eDNA Concentration and Abundance/Biomass
3.2. Lentic Ecosystem: Diffusion of eDNA
3.2.1. Lentic Ecosystem: Relationship Between eDNA Concentration and Time Factor
3.2.2. Lentic Ecosystem: Relationship Between eDNA Concentration and Distance Factors
3.3. Lotic Ecosystem: Relationship Between eDNA Concentration and Sampling Distance
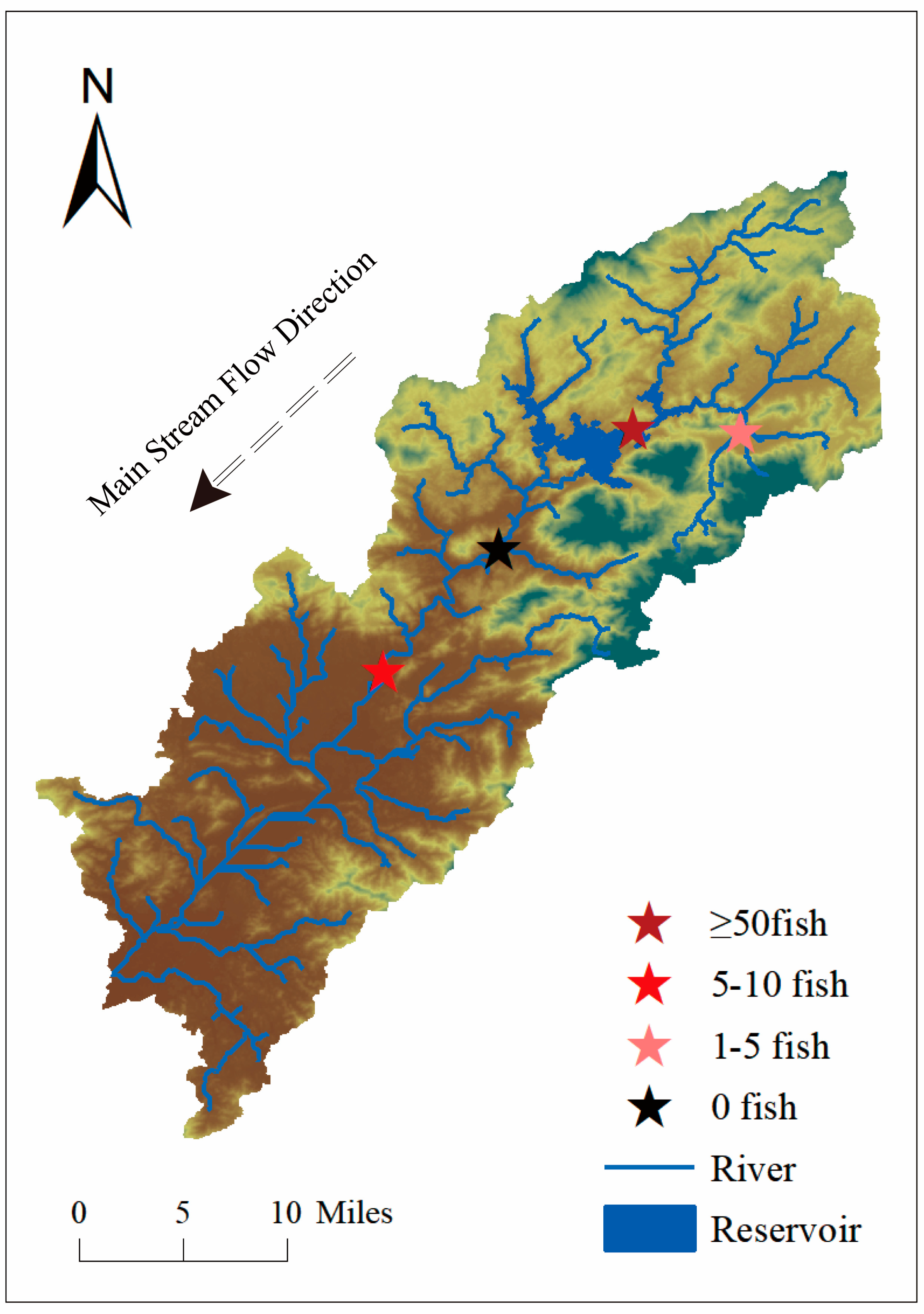
3.4. Liuxi River Survey
3.4.1. eDNA Methods for Detecting the Distribution of S. caldwelli in the Liuxi River
3.4.2. Detecting the Distribution of S. caldwelli in the Liuxi River Using Traditional Methods
3.4.3. Comparison of eDNA Methods with Traditional Methods
4. Discussion
4.1. Relationship of eDNA to Abundance/Biomass
4.2. Lentic Ecosystem
4.2.1. Relationship Between eDNA Concentration and Time
4.2.2. Relationship Between eDNA Concentration and Distance
4.3. Lotic Ecosystem: Relationship Between eDNA Concentration and Sampling Distance
4.4. Liuxi River Survey
4.4.1. Relationship Between eDNA and Traditional Methods of Monitoring Results
4.4.2. Comparison of eDNA and Traditional Methods
5. Conclusions
- (1)
- In the cultured tank experiment, despite the low level of model fit, some correlation was observed between eDNA concentration and S. caldwelli abundance/biomass, suggesting a potential resource assessment capability of the eDNA technique.
- (2)
- An experiment with caged fish in still water showed that eDNA could spread up to 18 m in 2 h, with concentrations decreasing gradually with distance and time. This result provides a reference for the assessment of fish distribution in lentic ecosystems such as lakes and reservoirs.
- (3)
- An experiment with caged fish in natural rivers has shown that eDNA released by 10 S. caldwelli can be detected up to 900 m. There were differences in detection results at different sampling locations for the same sampling distance, and a cross-sectional sampling strategy could improve the representativeness of the monitoring results. This provides methodological support for the assessment of fish stock distribution in river systems.
- (4)
- Using the eDNA method to detect the distribution of S. caldwelli in the Liuxi River, the results show that environmental DNA is extremely sensitive and reliable in detecting the presence of species. Environmental DNA and traditional methods have their own advantages and disadvantages, and using them to complement each other can improve monitoring efficiency.
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
References
- Butchart, S.H.M.; Walpole, M.; Collen, B.; van Strien, A.; Scharlemann, J.P.W.; Almond, R.E.A.; Baillie, J.E.M.; Bomhard, B.; Brown, C.; Bruno, J.; et al. Global Biodiversity: Indicators of Recent Declines. Science 2010, 328, 1164–1168. [Google Scholar] [CrossRef] [PubMed]
- WWF. Living Planet Report—2018: Aiming Higher; Grooten, M., Almond, R.E.A., Eds.; WWF: Gland, Switzerland, 2018. [Google Scholar]
- Williams, M.-A.; O’Grady, J.; Ball, B.; Carlsson, J.; de Eyto, E.; McGinnity, P.; Jennings, E.; Regan, F.; Parle-McDermott, A. The Application of CRISPR-Cas for Single Species Identification from Environmental DNA. Mol. Ecol. Resour. 2019, 19, 1106–1114. [Google Scholar] [CrossRef]
- Vié, J.-C.; Hilton-Taylor, C.; Stuart, S.N. Wildlife in a Changing World: An Analysis of the 2008 IUCN Red List of Threatened Species; IUCN: Gland, Switzerland, 2009; ISBN 2-8317-1063-4. [Google Scholar]
- Ge, Y.; Cheng, Q. Environmental DNA and Its Applications in Aquatic Biodiversity Surveys. Fish. Inf. Strategy 2020, 35, 55–62. [Google Scholar] [CrossRef]
- Zhang, H.; Xian, W. Applications of Environmental DNA Technology in Ecological Conservation and Monitoring. Mar. Sci. 2020, 44, 96–102. [Google Scholar] [CrossRef]
- Hao, Y.; Zhang, A.; Liu, J.; Gu, Z. The Applications of Environmental DNA Technology in Fish Resource Research. Biotechnol. Bull. 2018, 34, 56. [Google Scholar] [CrossRef]
- Bohmann, K.; Evans, A.; Gilbert, M.T.P.; Carvalho, G.R.; Creer, S.; Knapp, M.; Douglas, W.Y.; De Bruyn, M. Environmental DNA for Wildlife Biology and Biodiversity Monitoring. Trends Ecol. Evol. 2014, 29, 358–367. [Google Scholar] [CrossRef] [PubMed]
- Taberlet, P.; Coissac, E.; Hajibabaei, M.; Rieseberg, L.H. Environmental DNA. Mol. Ecol. 2012, 21, 1789–1793. [Google Scholar] [CrossRef] [PubMed]
- Rees, H.C.; Maddison, B.C.; Middleditch, D.J.; Patmore, J.R.; Gough, K.C. The Detection of Aquatic Animal Species Using Environmental DNA—A Review of eDNA as a Survey Tool in Ecology. J. Appl. Ecol. 2014, 51, 1450–1459. [Google Scholar] [CrossRef]
- Tillotson, M.D.; Kelly, R.P.; Duda, J.J.; Hoy, M.; Kralj, J.; Quinn, T.P. Concentrations of Environmental DNA (eDNA) Reflect Spawning Salmon Abundance at Fine Spatial and Temporal Scales. Biol. Conserv. 2018, 220, 1–11. [Google Scholar] [CrossRef]
- Ficetola, G.F.; Miaud, C.; Pompanon, F.; Taberlet, P. Species Detection Using Environmental DNA from Water Samples. Biol. Lett. 2008, 4, 423–425. [Google Scholar] [CrossRef]
- Dejean, T.; Valentini, A.; Miquel, C.; Taberlet, P.; Bellemain, E.; Miaud, C. Improved Detection of an Alien Invasive Species through Environmental DNA Barcoding: The Example of the American Bullfrog Lithobates Catesbeianus. J. Appl. Ecol. 2012, 49, 953–959. [Google Scholar] [CrossRef]
- Martellini, A.; Payment, P.; Villemur, R. Use of Eukaryotic Mitochondrial DNA to Differentiate Human, Bovine, Porcine and Ovine Sources in Fecally Contaminated Surface Water. Water Res. 2005, 39, 541–548. [Google Scholar] [CrossRef]
- Doi, H.; Inui, R.; Akamatsu, Y.; Kanno, K.; Yamanaka, H.; Takahara, T.; Minamoto, T. Environmental DNA Analysis for Estimating the Abundance and Biomass of Stream Fish. Freshw. Biol. 2017, 62, 30–39. [Google Scholar] [CrossRef]
- Dejean, T.; Valentini, A.; Duparc, A.; Pellier-Cuit, S.; Pompanon, F.; Taberlet, P.; Miaud, C. Persistence of Environmental DNA in Freshwater Ecosystems. PLoS ONE 2011, 6, e23398. [Google Scholar] [CrossRef] [PubMed]
- Thomsen, P.F.; Kielgast, J.O.S.; Iversen, L.L.; Wiuf, C.; Rasmussen, M.; Gilbert, M.T.P.; Orlando, L.; Willerslev, E. Monitoring Endangered Freshwater Biodiversity Using Environmental DNA. Mol. Ecol. 2012, 21, 2565–2573. [Google Scholar] [CrossRef] [PubMed]
- Takahara, T.; Minamoto, T.; Yamanaka, H.; Doi, H.; Kawabata, Z. Estimation of Fish Biomass Using Environmental DNA. PLoS ONE 2012, 7, e35868. [Google Scholar] [CrossRef]
- Baldigo, B.P.; Sporn, L.A.; George, S.D.; Ball, J.A. Efficacy of Environmental DNA to Detect and Quantify Brook Trout Populations in Headwater Streams of the Adirondack Mountains, New York. Trans. Am. Fish. Soc. 2017, 146, 99–111. [Google Scholar] [CrossRef]
- Doi, H.; Uchii, K.; Takahara, T.; Matsuhashi, S.; Yamanaka, H.; Minamoto, T. Use of Droplet Digital PCR for Estimation of Fish Abundance and Biomass in Environmental DNA Surveys. PLoS ONE 2015, 10, e0122763. [Google Scholar] [CrossRef]
- Wilcox, T.M.; McKelvey, K.S.; Young, M.K.; Sepulveda, A.J.; Shepard, B.B.; Jane, S.F.; Whiteley, A.R.; Lowe, W.H.; Schwartz, M.K. Understanding Environmental DNA Detection Probabilities: A Case Study Using a Stream-Dwelling Char Salvelinus Fontinalis. Biol. Conserv. 2016, 194, 209–216. [Google Scholar] [CrossRef]
- Jerde, C.L.; Olds, B.P.; Shogren, A.J.; Andruszkiewicz, E.A.; Mahon, A.R.; Bolster, D.; Tank, J.L. Influence of Stream Bottom Substrate on Retention and Transport of Vertebrate Environmental DNA. Environ. Sci. Technol. 2016, 50, 8770–8779. [Google Scholar] [CrossRef]
- Jane, S.F.; Wilcox, T.M.; McKelvey, K.S.; Young, M.K.; Schwartz, M.K.; Lowe, W.H.; Letcher, B.H.; Whiteley, A.R. Distance, Flow and PCR Inhibition: E DNA Dynamics in Two Headwater Streams. Mol. Ecol. Resour. 2015, 15, 216–227. [Google Scholar] [CrossRef]
- Shogren, A.J.; Tank, J.L.; Andruszkiewicz, E.; Olds, B.; Mahon, A.R.; Jerde, C.L.; Bolster, D. Controls on eDNA Movement in Streams: Transport, Retention, and Resuspension. Sci. Rep. 2017, 7, 5065. [Google Scholar] [CrossRef] [PubMed]
- Shogren, A.J.; Tank, J.L.; Egan, S.P.; August, O.; Rosi, E.J.; Hanrahan, B.R.; Renshaw, M.A.; Gantz, C.A.; Bolster, D. Water Flow and Biofilm Cover Influence Environmental DNA Detection in Recirculating Streams. Environ. Sci. Technol. 2018, 52, 8530–8537. [Google Scholar] [CrossRef] [PubMed]
- Pilliod, D.S.; Goldberg, C.S.; Arkle, R.S.; Waits, L.P. Factors Influencing Detection of eDNA from a Stream-dwelling Amphibian. Mol. Ecol. Resour. 2014, 14, 109–116. [Google Scholar] [CrossRef] [PubMed]
- Robinson, A.T.; Paroz, Y.M.; Clement, M.J.; Franklin, T.W.; Dysthe, J.C.; Young, M.K.; McKelvey, K.S.; Carim, K.J. Environmental DNA Sampling of Small-bodied Minnows: Performance Relative to Location, Species, and Traditional Sampling. N. Am. J. Fish. Manag. 2019, 39, 1073–1085. [Google Scholar] [CrossRef]
- Wood, Z.T.; Lacoursière-Roussel, A.; LeBlanc, F.; Trudel, M.; Kinnison, M.T.; Garry McBrine, C.; Pavey, S.A.; Gagné, N. Spatial Heterogeneity of eDNA Transport Improves Stream Assessment of Threatened Salmon Presence, Abundance, and Location. Front. Ecol. Evol. 2021, 9, 650717. [Google Scholar] [CrossRef]
- Pan, J.; Zhong, L.; Zheng, C.; Wu, H.; Liu, J. Records of Freshwater Fishes in Guangdong; Guangdong Science Publishing House: Guangzhou, China, 1991. [Google Scholar]
- Huckstorf, V. Spinibarbus caldwelli. The IUCN Red List of Threatened Species 2013:e.T187914A1834637. 2013. Available online: https://www.iucnredlist.org/species/187914/1834637 (accessed on 21 April 2025).
- Yamamoto, S.; Minami, K.; Fukaya, K.; Takahashi, K.; Sawada, H.; Murakami, H.; Tsuji, S.; Hashizume, H.; Kubonaga, S.; Horiuchi, T. Environmental DNA as a ‘Snapshot’of Fish Distribution: A Case Study of Japanese Jack Mackerel in Maizuru Bay, Sea of Japan. PLoS ONE 2016, 11, e0149786. [Google Scholar] [CrossRef]
- Li, H. Studies on the Conservation Biology of Spinibarbus caldwelli D; Huazhong Agricultural University: Wuhan, China, 2016. [Google Scholar]
- Lacoursière-Roussel, A.; Rosabal, M.; Bernatchez, L. Estimating Fish Abundance and Biomass from eDNA Concentrations: Variability among Capture Methods and Environmental Conditions. Mol. Ecol. Resour. 2016, 16, 1401–1414. [Google Scholar] [CrossRef]
- Maruyama, A.; Sugatani, K.; Watanabe, K.; Yamanaka, H.; Imamura, A. Environmental DNA Analysis as a Non-invasive Quantitative Tool for Reproductive Migration of a Threatened Endemic Fish in Rivers. Ecol. Evol. 2018, 8, 11964–11974. [Google Scholar] [CrossRef]
- Tsuji, S.; Takahara, T.; Doi, H.; Shibata, N.; Yamanaka, H. The Detection of Aquatic Macroorganisms Using Environmental DNA Analysis—A Review of Methods for Collection, Extraction, and Detection. Environ. DNA 2019, 1, 99–108. [Google Scholar] [CrossRef]
- Sakata, M.K.; Yamamoto, S.; Gotoh, R.O.; Miya, M.; Yamanaka, H.; Minamoto, T. Sedimentary eDNA Provides Different Information on Timescale and Fish Species Composition Compared with Aqueous eDNA. Environ. DNA 2020, 2, 505–518. [Google Scholar] [CrossRef]
- Wang, S.; Yan, Z.; Hänfling, B.; Zheng, X.; Wang, P.; Fan, J.; Li, J. Methodology of Fish eDNA and Its Applications in Ecology and Environment. Sci. Total Environ. 2021, 755, 142622. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Zou, Y.; Xiao, S.; Hou, J. Environmental DNA Metabarcoding Serves as a Promising Method for Aquatic Species Monitoring and Management: A Review Focused on Its Workflow, Applications, Challenges and Prospects. Mar. Pollut. Bull. 2023, 194, 115430. [Google Scholar] [CrossRef]
- Yamanaka, H.; Minamoto, T.; Matsuura, J.; Sakurai, S.; Tsuji, S.; Motozawa, H.; Hongo, M.; Sogo, Y.; Kakimi, N.; Teramura, I. A Simple Method for Preserving Environmental DNA in Water Samples at Ambient Temperature by Addition of Cationic Surfactant. Limnology 2017, 18, 233–241. [Google Scholar] [CrossRef]
- Thomsen, P.F.; Willerslev, E. Environmental DNA–An Emerging Tool in Conservation for Monitoring Past and Present Biodiversity. Biol. Conserv. 2015, 183, 4–18. [Google Scholar] [CrossRef]
- Farrington, H.L.; Edwards, C.E.; Guan, X.; Carr, M.R.; Baerwaldt, K.; Lance, R.F. Mitochondrial Genome Sequencing and Development of Genetic Markers for the Detection of DNA of Invasive Bighead and Silver Carp (Hypophthalmichthys nobilis and H. molitrix) in Environmental Water Samples from the United States. PLoS ONE 2015, 10, e0117803. [Google Scholar] [CrossRef]
- Shu, L.; Ludwig, A.; Peng, Z. Standards for Methods Utilizing Environmental DNA for Detection of Fish Species. Genes 2020, 11, 296. [Google Scholar] [CrossRef]
- Bustin, S.A.; Benes, V.; Garson, J.A.; Hellemans, J.; Huggett, J.; Kubista, M.; Mueller, R.; Nolan, T.; Pfaffl, M.W.; Shipley, G.L.; et al. The MIQE Guidelines: Minimum Information for Publication of Quantitative Real-Time PCR Experiments. Clin. Chem. 2009, 55, 611–622. [Google Scholar] [CrossRef]
- Benoit, N.P.; Robinson, K.M.; Kellogg, C.T.E.; Lemay, M.A.; Hunt, B.P.V. Using qPCR of Environmental DNA (eDNA) to Estimate the Biomass of Juvenile Pacific Salmon (Oncorhynchus spp.). Environ. DNA 2023, 5, 683–696. [Google Scholar] [CrossRef]
- Buxton, A.S.; Groombridge, J.J.; Zakaria, N.B.; Griffiths, R.A. Seasonal Variation in Environmental DNA in Relation to Population Size and Environmental Factors. Sci. Rep. 2017, 7, 46294. [Google Scholar] [CrossRef]
- Erickson, R.A.; Rees, C.B.; Coulter, A.A.; Merkes, C.M.; McCalla, S.G.; Touzinsky, K.F.; Walleser, L.; Goforth, R.R.; Amberg, J.J. Detecting the Movement and Spawning Activity of Bigheaded Carps with Environmental DNA. Mol. Ecol. Resour. 2016, 16, 957–965. [Google Scholar] [CrossRef] [PubMed]
- Klobucar, S.L.; Rodgers, T.W.; Budy, P. At the Forefront: Evidence of the Applicability of Using Environmental DNA to Quantify the Abundance of Fish Populations in Natural Lentic Waters with Additional Sampling Considerations. Can. J. Fish. Aquat. Sci. 2017, 74, 2030–2034. [Google Scholar] [CrossRef]
- Hinlo, R.; Lintermans, M.; Gleeson, D.; Broadhurst, B.; Furlan, E. Performance of eDNA Assays to Detect and Quantify an Elusive Benthic Fish in Upland Streams. Biol. Invasions 2018, 20, 3079–3093. [Google Scholar] [CrossRef]
- Grant, I.-I.S. Evaluating Environmental DNA Detection alongside Standard Fish Sampling in Great Lakes Coastal Wetland Monitoring (Seed Project); Illinois-Indiana Sea Grant: Urbana, IL, USA, 2012. [Google Scholar]
- Goldberg, C.S.; Sepulveda, A.; Ray, A.; Baumgardt, J.; Waits, L.P. Environmental DNA as a New Method for Early Detection of New Zealand Mudsnails (Potamopyrgus antipodarum). Freshw. Sci. 2013, 32, 792–800. [Google Scholar] [CrossRef]
- Deiner, K.; Altermatt, F. Transport Distance of Invertebrate Environmental DNA in a Natural River. PLoS ONE 2014, 9, e88786. [Google Scholar] [CrossRef]
- Maruyama, A.; Nakamura, K.; Yamanaka, H.; Kondoh, M.; Minamoto, T. The Release Rate of Environmental DNA from Juvenile and Adult Fish. PLoS ONE 2014, 9, e114639. [Google Scholar] [CrossRef]
- Jo, T.S. Pooling of Intra-Site Measurements Inflates Variability of the Correlation between Environmental DNA Concentration and Organism Abundance. Environ. Monit. Assess. 2023, 195, 936. [Google Scholar] [CrossRef]
- Hong, X.; Wang, K.; Ji, L.; Liu, X.; Yu, L.; Wei, J.; Wang, Y.; Wei, C.; Li, W.; Zhu, X. Exploring the Relationship between Environmental DNA Concentration and Biomass in Asian Giant Softshell Turtle (Pelochelys cantorii). PeerJ 2023, 11, e16218. [Google Scholar] [CrossRef]
- Yan, H.; Dong, Z.; Ma, T.; Zhang, L.; Wang, X.; Ye, H.; Yao, W.; He, W. Detection and Biomass Assessment of Procypris Rabaudi Based on Environmental DNA. J. Fish. China 2022, 46, 1018–1026. [Google Scholar] [CrossRef]
- Wood, Z.T.; Erdman, B.F.; York, G.; Trial, J.G.; Kinnison, M.T. Experimental Assessment of Optimal Lotic eDNA Sampling and Assay Multiplexing for a Critically Endangered Fish. Environ. DNA 2020, 2, 407–417. [Google Scholar] [CrossRef]
- Pont, D.; Rocle, M.; Valentini, A.; Civade, R.; Jean, P.; Maire, A.; Roset, N.; Schabuss, M.; Zornig, H.; Dejean, T. Environmental DNA Reveals Quantitative Patterns of Fish Biodiversity in Large Rivers despite Its Downstream Transportation. Sci. Rep. 2018, 8, 10361. [Google Scholar] [CrossRef]
- Barnes, M.A.; Turner, C.R.; Jerde, C.L.; Renshaw, M.A.; Chadderton, W.L.; Lodge, D.M. Environmental Conditions Influence eDNA Persistence in Aquatic Systems. Environ. Sci. Technol. 2014, 48, 1819–1827. [Google Scholar] [CrossRef]
- Valentini, A.; Taberlet, P.; Miaud, C.; Civade, R.; Herder, J.; Thomsen, P.F.; Bellemain, E.; Besnard, A.; Coissac, E.; Boyer, F.; et al. Next-generation Monitoring of Aquatic Biodiversity Using Environmental DNA Metabarcoding. Mol. Ecol. 2016, 25, 929–942. [Google Scholar] [CrossRef]
- Knudsen, S.W.; Ebert, R.B.; Hesselsøe, M.; Kuntke, F.; Hassingboe, J.; Mortensen, P.B.; Thomsen, P.F.; Sigsgaard, E.E.; Hansen, B.K.; Nielsen, E.E.; et al. Species-Specific Detection and Quantification of Environmental DNA from Marine Fishes in the Baltic Sea. J. Exp. Mar. Biol. Ecol. 2019, 510, 31–45. [Google Scholar] [CrossRef]
- Qin, C.; Zuo, T.; Yu, G.; Zhou, W.; Li, C. Advances in environmental DNA for biomass assessment in aquatic ecosystems. South China Fish. Sci. 2020, 16, 123–128. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, J.; Lin, H.; Xiao, J.; Tan, G.; Yan, L.; Chen, J.; Zhao, J.; Wang, J. Environmental DNA for Assessing Population and Spatial Distribution of Spinibarbus caldwelli in the Liuxi River. Diversity 2025, 17, 320. https://doi.org/10.3390/d17050320
Wang J, Lin H, Xiao J, Tan G, Yan L, Chen J, Zhao J, Wang J. Environmental DNA for Assessing Population and Spatial Distribution of Spinibarbus caldwelli in the Liuxi River. Diversity. 2025; 17(5):320. https://doi.org/10.3390/d17050320
Chicago/Turabian StyleWang, Jujing, Haimei Lin, Jinsheng Xiao, Guiyu Tan, Luobin Yan, Jiabo Chen, Jun Zhao, and Junjie Wang. 2025. "Environmental DNA for Assessing Population and Spatial Distribution of Spinibarbus caldwelli in the Liuxi River" Diversity 17, no. 5: 320. https://doi.org/10.3390/d17050320
APA StyleWang, J., Lin, H., Xiao, J., Tan, G., Yan, L., Chen, J., Zhao, J., & Wang, J. (2025). Environmental DNA for Assessing Population and Spatial Distribution of Spinibarbus caldwelli in the Liuxi River. Diversity, 17(5), 320. https://doi.org/10.3390/d17050320






