Reassessment of the Taxonomic Borders Within Pipistrellus (Chiroptera, Vespertilionidae, Pipistrellini)
Abstract
1. Introduction
2. Materials and Methods
2.1. Molecular Genetic Analysis
2.2. Morphological Analysis
3. Results
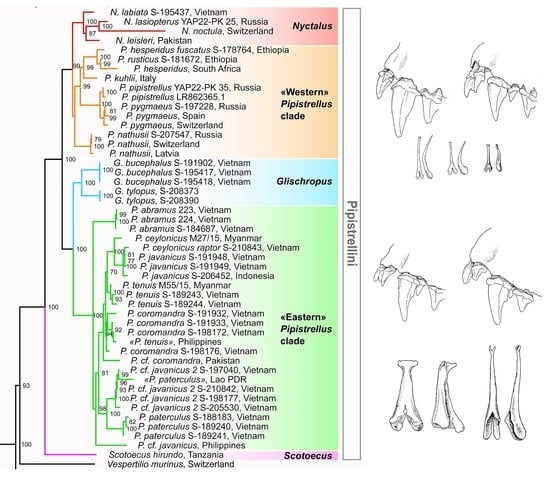
3.1. Molecular Genetic Results
3.1.1. Mitochondrial Data
3.1.2. Nuclear Data
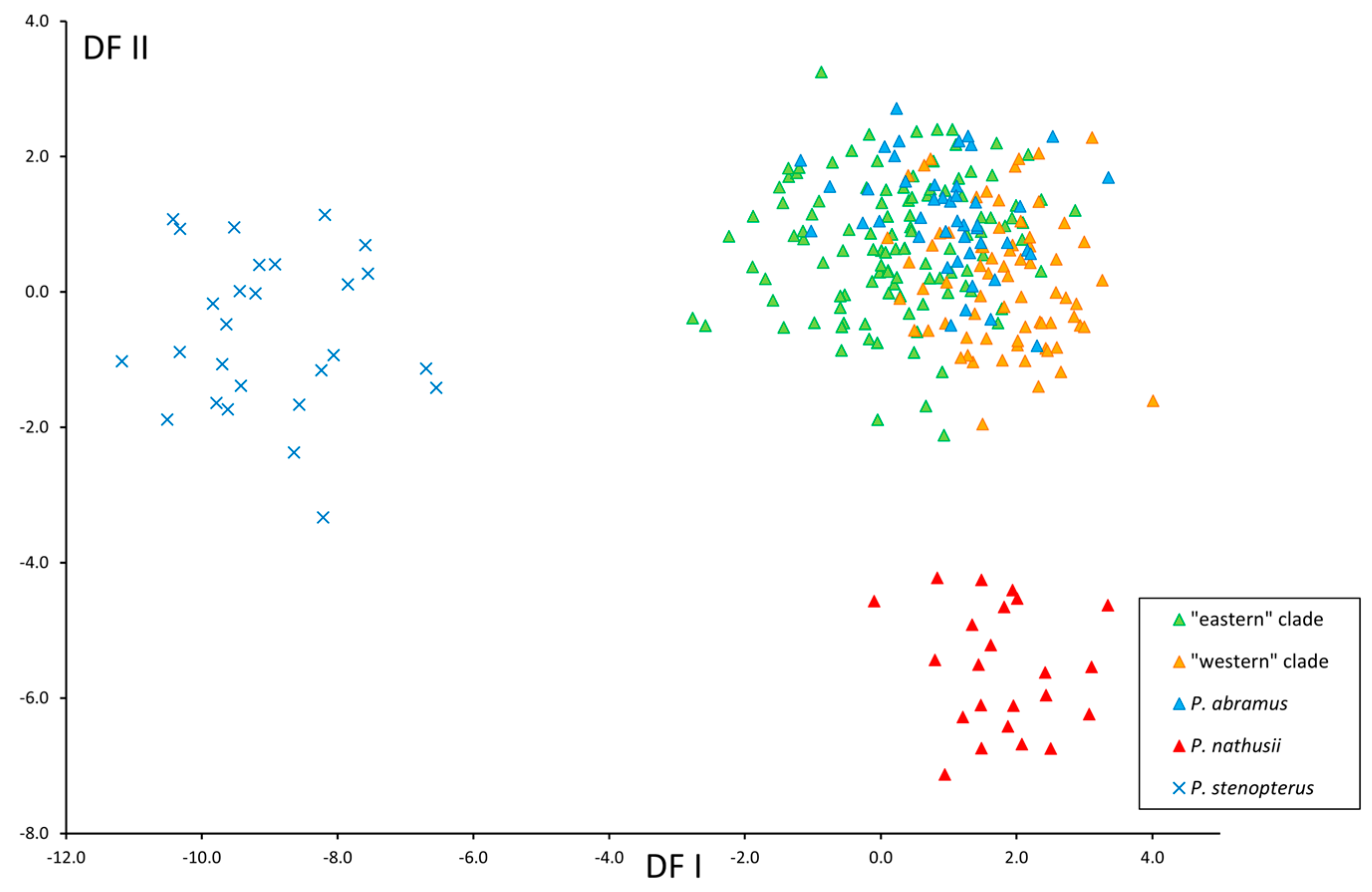
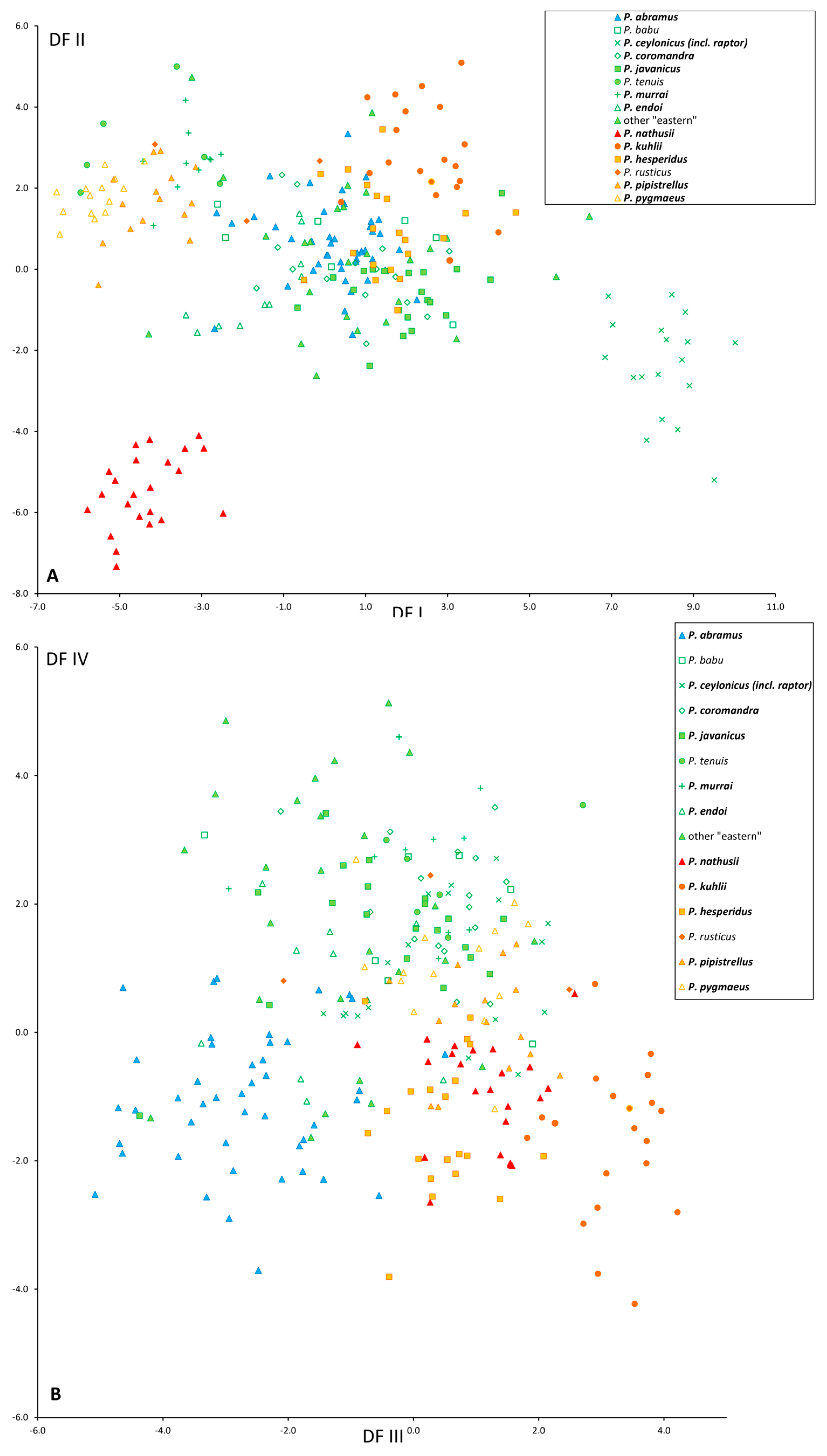
3.2. Morphological Results
3.2.1. Cranial Structure and Proportions
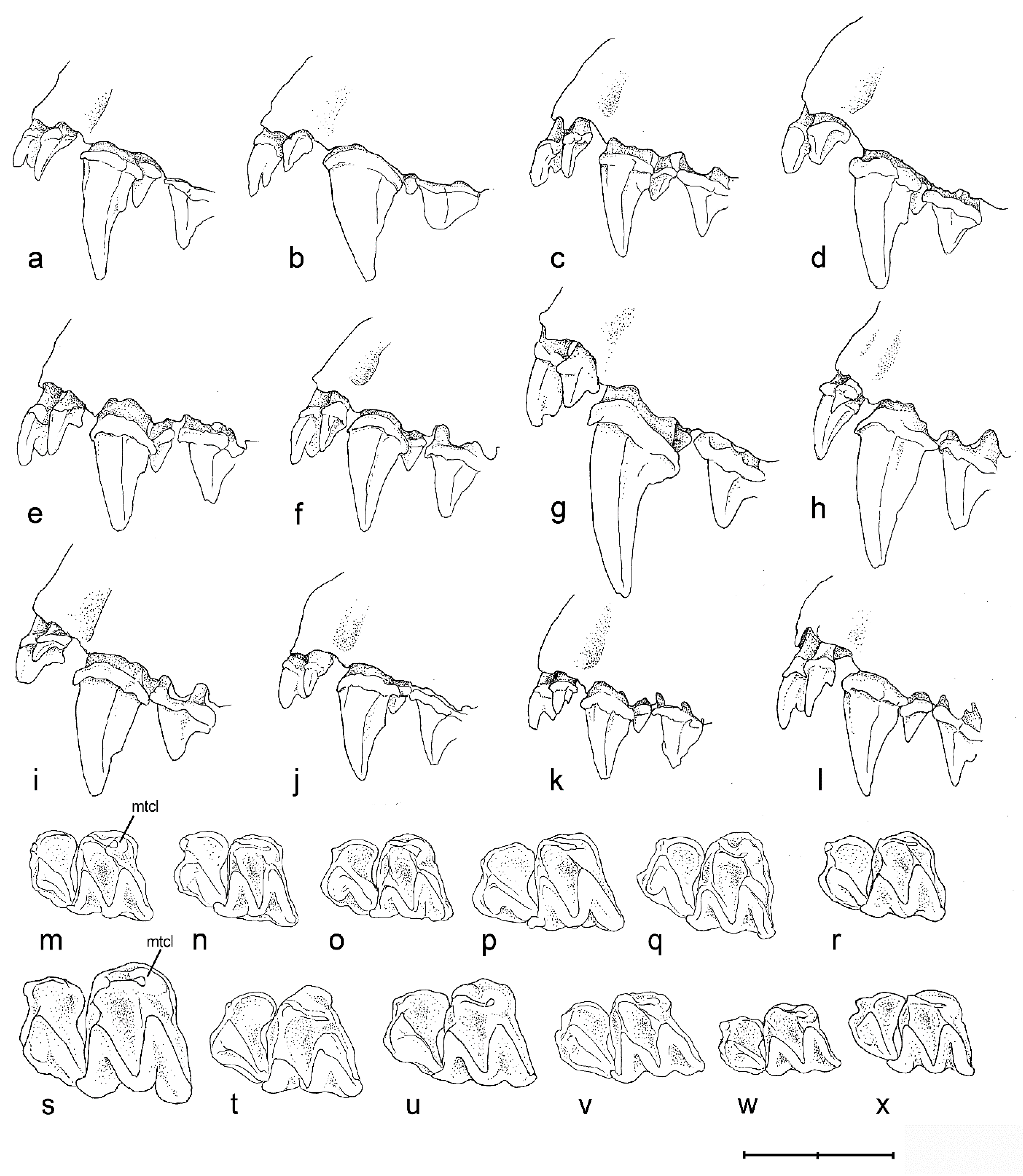
3.2.2. Dental Structure
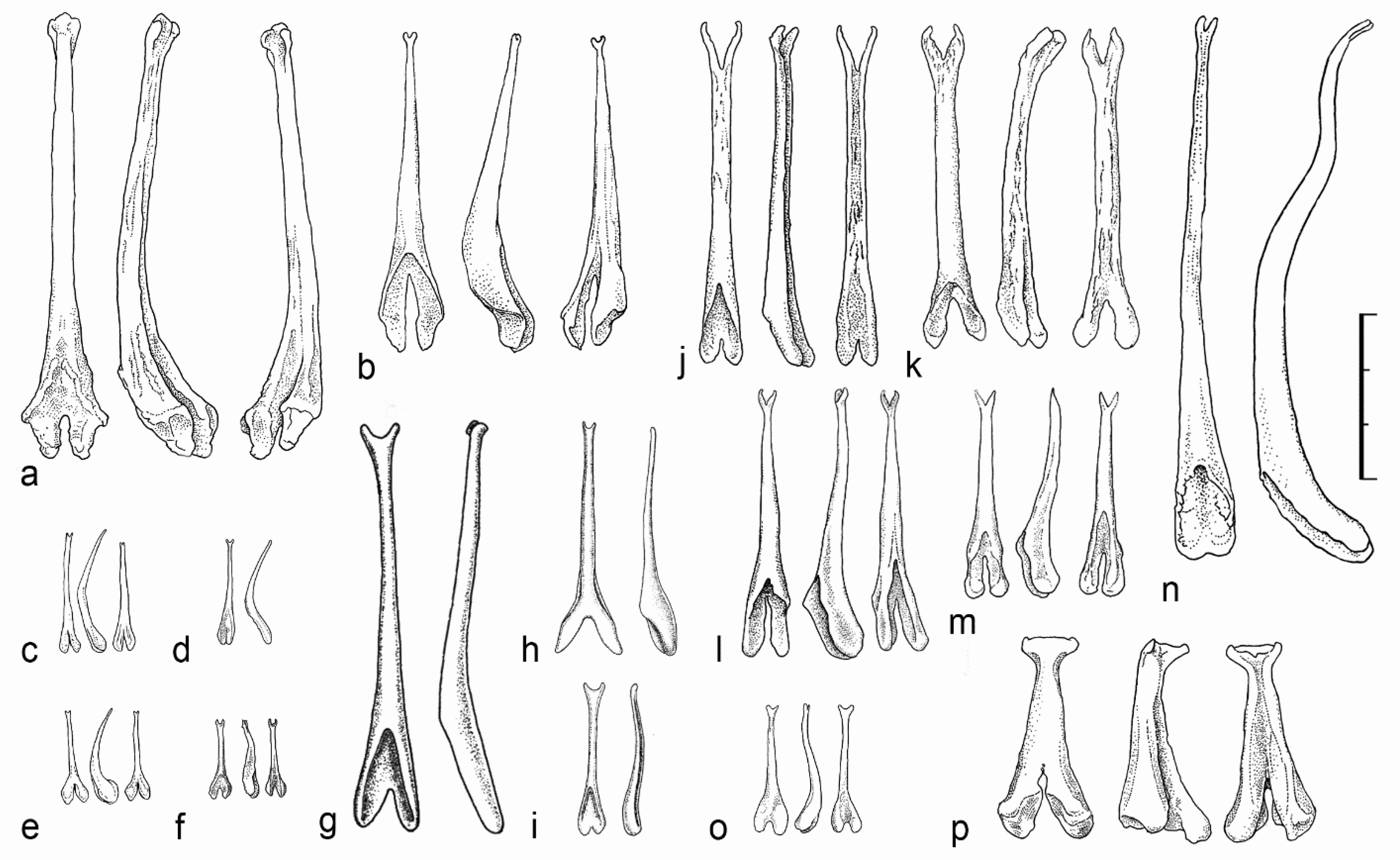
3.2.3. Baculum
3.3. Taxonomic Part
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A
References
- Dobson, G.E. Descriptions of new or little-known species of bats of the genus Vesperugo. Proc. Zool. Soc. Lond. 1875, 470–474. [Google Scholar]
- Miller, G.S. The Families and Genera of Bats; Government Printing Office: Washington, DC, USA, 1907; pp. 1–282. [Google Scholar]
- Ognev, S.I. Mammals of the Eastern Europe and Northern Asia; Glavnauka: Moscow, Rusia, 1928; Volume 1, pp. 1–631. (In Russian) [Google Scholar]
- Tate, G.H.H. Results of the Archbold Expeditions 47. Review of the vespertilionine bats, with special attention to genera and species of the Archbold Expeditions. Bull. Am. Mus. Nat. Hist. 1942, 80, 221–297. [Google Scholar]
- Kuzyakin, A.P. Bats (Systematics, Life History and Utility in Agriculture and Forestry); Sovetskaya Nauka: Moscow, Rusia, 1950; pp. 1–444. (In Russian) [Google Scholar]
- Ellerman, J.R.; Morrison-Scott, T.C.S. Checklist of Palaearctic and Indian Mammals 1758 to 1946; British Museum (Nat. Hist.): London, UK, 1951; pp. 1–810. [Google Scholar]
- Corbet, G.B. The Mammals of the Palaearctic Region: A Taxonomic Review; Cornell University Press: London, UK; Ithaca, NY, USA, 1978; pp. 1–314. [Google Scholar]
- Hill, J.E.; Harrison, D.L. The baculum in Vespertilioninae (Chiroptera: Vespertilionidae) with a systematic review, a synopsis of Pipistrellus and Eptesicus, and the description of a new genus and subgenus. Bull. Br. Mus. Zool. 1987, 52, 225–305. [Google Scholar] [CrossRef]
- Corbet, G.B.; Hill, J.E. The Mammals of the Indomalayan Region; Oxford University Press: Oxford, UK, 1992; pp. 1–488. [Google Scholar]
- Koopman, K.F. Chiroptera: Systematics. Handbook of Zoology. Mammalia; Walter de Gruyter: Berlin, Germany, 1994; Volume 8, Part 60; pp. 1–217. [Google Scholar]
- Heller, K.-G.; Volleth, M. Taxonomic position of ‘Pipistrellus sociestatis’ Hill, 1972 and the karyological characteristics of the genus Eptesicus (Chiroptera: Vespertilionidae). Z. Zool. Syst. Evol. 1984, 22, 65–77. [Google Scholar] [CrossRef]
- Horacek, I.; Hanak, V. Generic status of Pipistrellus savii and comments on classification of the genus Pipistrellus (Chiroptera, Vespertilionidae). Myotis 1986, 23–24, 9–16. [Google Scholar]
- Menu, H. Morphotypes dentaires actuels et fossiles des chiropteres. 2eme partie: Implications systematiques et phylogeniques. Palaeovertebrata 1987, 17, 77–150. [Google Scholar]
- Volleth, M.; Tidemann, C.R. The origin of the Australian Vespertilioninae bats, as indicated by chromosomal studies. Z. Saugetierkd. 1991, 56, 321–330. [Google Scholar]
- Volleth, M.; Heller, K.-G. Phylogenetic relationships of vespertilionid genera (Mammalia: Chiroptera) as revealed by karyological analysis. J. Zool. Syst. Evol. Res. 1994, 32, 11–34. [Google Scholar] [CrossRef]
- Kearney, T.C.; Volleth, M.; Contrafatto, G.; Taylor, P.J. Systematic implications of chromosome GTG-band and bacula morphology for Southern African Eptesicus and Pipistrellus and several other species of Vespertilioninae (Chiroptera: Vespertilionidae). Acta Chiropterologica 2002, 4, 55–76. [Google Scholar] [CrossRef]
- Hoofer, S.R.; Van Den Bussche, R.A. Molecular phylogenetics of the chiropteran family Vespertilionidae. Acta Chiropterologica 2003, 5, 1–63. [Google Scholar] [CrossRef]
- Koubínová, D.; Irwin, N.; Hulva, P.; Koubek, P.; Zima, J. Hidden diversity in Senegalese bats and associated findings in the systematics of the family Vespertilionidae. Front. Zool. 2013, 10, 48. [Google Scholar] [CrossRef]
- Roehrs, Z.P.; Lack, J.B.; Van Den Bussche, R.A. Tribal phylogenetic relationships within Vespertilioninae (Chiroptera: Vespertilionidae) based on mitochondrial and nuclear sequence data. J. Mammal. 2010, 91, 1073–1092. [Google Scholar] [CrossRef]
- Thorn, E.; Kock, D.; Cuisin, J. 2007 Status of the African bats Vesperugo grandidieri Dobson 1876 and Vesperugo flavescens Seabra 1900 (Chiroptera, Vespertilionidae), with description of a new subgenus. Mammalia 2007, 71, 70–79. [Google Scholar] [CrossRef]
- Monadjem, A.; Demos, T.C.; Dalton, D.L.; Webala, P.W.; Musila, S.; Kerbis Peterhans, J.C.; Patterson, B.D. A revision of pipistrelle-like bats (Mammalia: Chiroptera: Vespertilionidae) in East Africa with the description of new genera and species. Zool. J. Linnean Soc. 2021, 191, 1114–1146. [Google Scholar] [CrossRef]
- Benda, P.; Uvizl, M.; Šklíba, J.; Mazoch, V.; Červený, J. African bats in the collection of the National Museum, Prague (Chiroptera). I. Bats from Zambia. Lynx Ser. Nova 2022, 53, 291–332. [Google Scholar] [CrossRef]
- Demos, T.C.; Webala, P.W.; Monadjem, A.; Patterson, B.D. Nuclear Introns Support the Subtribe Laephotina and Recently Proposed Genera of African Vespertilionidae. Acta Chiropterologica 2024, 26, 143–152. [Google Scholar] [CrossRef]
- Kruskop, S.V.; Solovyeva, E.N.; Kaznadzey, A.D. Unusual Pipistrelle: Taxonomic position of the Malayan Noctule (Pipistrellus stenopterus; Vespertilionidae; Chiroptera). Zool. Stud. 2018, 57, 1–15. [Google Scholar] [CrossRef]
- Moratelli, R.; Burgin, C.; Cláudio, V.; Novaes, R.; López-Baucells, A.; Haslauer, R. Family Vespertilionidae (Vesper Bats). In Handbook of the Mammals of the World, Volume 9: Bats; Wilson, D.E., Mittermeier, R.A., Eds.; Lynx Edicions: Barcelona, Spain, 2019; pp. 716–981. [Google Scholar]
- Zhukova, S.S.; Solovyeva, E.N.; Artyushin, I.V.; Kruskop, S.V. Paraphyly of the Pipistrelles (Pipistrellus; Vespertilionidae) is confirmed by the analysis of the nuclear gene markers. Doklady Biochem. Biophys. 2022, 507, 302–306. [Google Scholar] [CrossRef]
- Mayer, F.; Dietz, C.; Kiefer, A. Molecular species identification boosts bat diversity. Front. Zool. 2007, 4, 4. [Google Scholar] [CrossRef]
- Zhukova, S.S.; Speranskaya, A.S.; Lisenkova, A.A.; Kruskop, S.V. The complete mitochondrial genome of Glischropus bucephalus (Vespertilionidae; Chiroptera) provides new evidence for Pipistrellus paraphyly. Diversity 2023, 15, 1085. [Google Scholar] [CrossRef]
- Dool, S.E.; Puechmaille, S.J. Challenges in the Vespertilionidae phylogeny: Resolving Pipistrellus nathusii placement and affirming generic status for Asian pipistrelles. J. Mammal. 2025, 106, 458–468. [Google Scholar] [CrossRef]
- Zhukova, S.S.; Kruskop, S.V. Is Nathusius’ pipistrelle Pipistrellus nathusii a special phylogenetic lineage? In Theriofauna of Belarus and Neighboring Regions, Proceedings of the International Scientific and Practical Conference Dedicated to the 90th Anniversary of the Birth of Professor P. G. Kozlo, Minsk, Belarus, 24–26 September 2024; Belarusskaya Nauka: Minsk, Belarus, 2024; pp. 343–345. (In Russian). [Google Scholar]
- Sambrook, J.; Fritsch, E.F.; Maniatis, T. Molecular Cloning: A Laboratory Manual, 2nd ed.; Cold Spring Harbour Lab Press: New York, NY, USA, 1989; pp. 1–1659. [Google Scholar]
- Eick, G.N.; Jacobs, D.S.; Matthee, C.A. A nuclear DNA phylogenetic perspective on the evolution of echolocation and historical biogeography of extant bats (Chiroptera). Mol. Biol. Evol. 2005, 22, 1869–1886. [Google Scholar] [CrossRef] [PubMed]
- Artyushin, I.V.; Bannikova, A.A.; Lebedev, V.S.; Kruskop, S.V. Mitochondrial DNA relationships among North Palaearctic Eptesicus (Vespertilionidae, Chiroptera) and past hybridization between Common Serotine and Northern Bat. Zootaxa 2009, 2262, 40–52. [Google Scholar] [CrossRef]
- Artyushin, I.V.; Kruskop, S.V.; Lebedev, V.S.; Bannikova, A.A. Molecular phylogeny of Serotines (Mammalia, Chiroptera, Eptesicus): Evolutionary and taxonomical aspects of the E. serotinus species group. Biol. Bull. 2018, 45, 469–477. [Google Scholar] [CrossRef]
- Kruskop, S.V.; Artyushin, I.V.; Yusefovich, A.P.; Undrakhbayar, E.; Speranskaya, A.S.; Lisenkova, A.A.; Bannikova, A.A.; Lebedev, V.S. Genetic diversity of Mongolian long-eared bats (Plecotus; Vespertilionidae; Chiroptera). Acta Chiropterologica 2020, 22, 243–255. [Google Scholar] [CrossRef]
- Burland, T.G. DNASTAR’s Lasergene sequence analysis software. Methods Mol. Biol. 2000, 132, 71–91. [Google Scholar]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular evolutionary genetics analysis version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef]
- Nguyen, L.-T.; Schmidt, H.A.; von Haeseler, A.; Minh, B.Q. IQ-TREE: A fast and effective stochastic algorithm for estimating maximum likelihood phylogenies. Mol. Biol. Evol. 2015, 32, 268–274. [Google Scholar] [CrossRef]
- Kalyaanamoorthy, S.; Minh, B.Q.; Wong, T.K.F.; von Haeseler, A.; Jermiin, L.S. ModelFinder: Fast model selection for accurate phylogenetic estimates. Nat. Methods 2017, 14, 587–589. [Google Scholar] [CrossRef]
- Minh, B.Q.; Nguyen, M.A.T.; von Haeseler, A. Ultrafast approximation for phylogenetic boot-strap. Mol. Biol. Evol. 2013, 30, 1188–1195. [Google Scholar] [CrossRef]
- Fracasso, M.P.A.; de Oliveira Salles, L.; Perini, F.A. Upper molar morphology and relationships among higher taxa in bats. J. Mammal. 2011, 92, 421–432. [Google Scholar] [CrossRef]
- Herdina, A.N.; Kelly, D.A.; Jahelková, H.; Lina, P.H.C.; Horáček, I.; Metscher, B.D. Testing hypotheses of bat baculum function with 3D models derived from microCT. J. Anat. 2015, 226, 229–235. [Google Scholar] [CrossRef]
- Herdina, A.N.; Plenk, H., Jr.; Benda, P.; Lina, P.H.; Herzig-Straschil, B.; Hilgers, H.; Metscher, B.D. Correlative 3D-imaging of Pipistrellus penis micromorphology: Validating quantitative microCT images with undecalcified serial ground section histomorphology. J. Morphol. 2015, 276, 695–706. [Google Scholar] [CrossRef]
- Kruskop, S.V. The bacula of bats from Indochina: Serotines and pipistrelles (Chiroptera; Vespertilionidae: Vespertilioninae). Plecotus 2019, 22, 88–103. (In Russian) [Google Scholar]
- Srinivasulu, B.; Kaur, H.; Shah, T.A.; Devender, G.; Gopi, A.; Raman, S.; Srinivasulu, C. A review of the bacular morphology of some Indian bats (Mammalia: Chiroptera). J. Threat. Taxa 2020, 12, 15985–16005. [Google Scholar] [CrossRef]
- Bates, P.J.J.; Ratrimomanarivo, F.H.; Harrison, D.L.; Goodman, S.M. A description of a new species of Pipistrellus (Chiroptera: Vespertilionidae) from Madagascar with a review of related Vespertilioninae from the island. Acta Chiropterologica 2006, 8, 299–324. [Google Scholar] [CrossRef]
- Palmer, T.S. Index Generum Mammalium: A List of the Genera and Families of Mammals; US Government Printing Office: Washington, DC, USA, 1904; pp. 1–984. [Google Scholar]
- Pavlinov, I.Y.; Rossolimo, O.L. Systematics of Mammals of USSR; Moscow State University Publishing: Moscow, Rusia, 1987; pp. 1–253. (In Russian) [Google Scholar]
- Volleth, M.; Bronner, G.; Göpfert, M.C.; Heller, K.-G.; von Helversen, O.; Yong, H.-S. Karyotype comparison and phylogenetic relationships of Pipistrellus-like bats (Vespertilionidae; Chiroptera; Mammalia). Chromosome Res. 2001, 9, 25–46. [Google Scholar] [CrossRef]
- Zima, J.; Horáček, I. Synopsis of karyotypes of vespertilionid bats (Mammalia: Chiroptera). Acta Univ. Carol. Biol. 1985, 1981, 311–329. [Google Scholar]
- Hulva, P.; Fornůsková, A.; Chudárková, A.; Evin, A.; Allegrini, B.; Benda, P.; Bryja, J. Mechanisms of radiation in a bat group from the genus Pipistrellus inferred by phylogeography, demography and population genetics. Mol. Ecol. 2010, 19, 5417–5431. [Google Scholar] [CrossRef]
- Fedyk, S.; Ruprecht, A.L. Karyotypes of Pipistrellus pipistrellus (Schreber 1774) and P. nathusii (Keyserling and Blasius 1839) (Chiroptera: Vespertilionidae). Caryologia 1976, 29, 283–289. [Google Scholar] [CrossRef]
- Zima, J. Chromosomal homology in the complements of bats of the family Vespertilionidae. II. G-band karyotypes of some Myotis, Eptesicus and Pipistrellus species. Folia Zool. 1982, 31, 31–36. [Google Scholar]
- Volleth, M. Differences in the location of nucleolus organizer regions in European vespertilionid bats. Cytogenet. Cell Genet. 1987, 44, 186–197. [Google Scholar] [CrossRef]
- Harada, M. Chromosomes of nine Chiropteran species in Japan (Chiroptera). La Kromosomo 1973, 91, 2885–2895. [Google Scholar]
- Dulic, B. Chromosomes of three species of Indianin the genus Pipistrellus may be facilitated by use of the Microchiroptera. Myotis 1981, 1819, 76–82. [Google Scholar]
- Ando, K.; Harada, M.; Uchida, T.A. A karyological study on five Japanese species of Myotis and Pipistrellus, with special attention to composition of their C-band materials. J. Mammal. Soc. Jpn. 1987, 12, 25–29. [Google Scholar]
- Morales, C.; Ballinger, W.; Bickham, W.; Greenbaum, F.; Schlitter, A. Genetic relationships among eight species of Eptesicus and Pipistrellus (Chiroptera: Vespertilionidae). J. Mammlogy 1991, 72, 286–291. [Google Scholar] [CrossRef]
- Sreepada, K.S.; Naldu, K.N.; Gururaj, M.E. Karyotypíc studies of four species of Pipistrellus (Mammalia: Chiroptera) from India. Mammalia 1996, 60, 407–416. [Google Scholar] [CrossRef]
- Lin, L.K.; Motokawa, M.; Harada, M. Karyological study of the house bat Pipistrellus abramus (Mammalia: Chiroptera) from Taiwan with comments on its taxonomic status. Raffles Bull. of Zool. 2002, 50, 507–510. [Google Scholar]
- Wu, Y.; Motokawa, M.; Li, Y.C.; Harada, M.; Chen, Z.; Lin, L.K. Karyology of eight species of bats (Mammalia: Chiroptera) from Hainan Island, China. Int. J. Biol. Sci. 2009, 5, 659–666. [Google Scholar] [CrossRef]
- Flannery, T.F. Mammals of New Guinea (Rev. and Upd. Ed.); Cornell University Press: Ithaca, NY, USA, 1995; pp. 1–568. [Google Scholar]
- Volleth, M.; Mayer, F.; Heller, K.-G.; Müller, S.; Fahr, J. Karyotype comparison of five African Vespertilionini species with comments on phylogenetic relationships and proposal of a new subtribe. Acta Chiropterologica 2023, 25, 35–52. [Google Scholar] [CrossRef]
- Monadjem, A.; Richards, L.R.; Decher, J.; Hutterer, R.; Mamba, M.L.; JGuyton, J.; Naskrecki, P.; Markotter, W.; Wipfler, B.; Kropff, A.S.; et al. A phylogeny for African Pipistrellus species with the description of a new species from West Africa (Mammalia: Chiroptera). Zool. J. Linnean Soc. 2021, 191, 548–574. [Google Scholar] [CrossRef]
- Cláudio, V.C.; Novaes, R.L.; Gardner, A.L.; Nogueira, M.R.; Wilson, D.E.; Maldonado, J.E.; Oliveira, J.A.; Moratelli, R. Taxonomic reevaluation of New World Eptesicus and Histiotus (Chiroptera: Vespertilionidae), with the description of a new genus. Zoologia 2023, 40, e22029. [Google Scholar] [CrossRef]
- Yi, X.; Latch, E.K. 2022 Systematics of the New World bats Eptesicus and Histiotus suggest trans-marine dispersal followed by Neotropical cryptic diversification. Mol. Phyl. Evol. 2022, 175, 107582. [Google Scholar] [CrossRef] [PubMed]
- Yi, X.; Latch, E.K.; Lim, B.K.; Koroiva, R.; Da Rocha, P.A.; Feijó, A. UCE-derived mitochondrial phylogeny reveals pervasive mito-nuclear discordances in serotine bats (genus Eptesicus) and complex evolutionary history in Eptesicus (Histiotus). Mamm. Biol. 2024, 104, 417–430. [Google Scholar] [CrossRef]
- Lebedev, V.S.; Shenbrot, G.I.; Krystufek, B.; Mahmoudi, A.; Melnikova, M.N.; Solovyeva, E.N.; Lisenkova, A.A.; Undrakhbayar, E.; Rogovin, K.A.; Surov, A.V.; et al. Phylogenetic relations and range history of jerboas of the Allactaginae subfamily (Dipodidae, Rodentia). Sci. Rep. 2022, 12, 842. [Google Scholar] [CrossRef]
- Mein, P. The Late Miocene small mammal succession from France, with emphasis on the Rhône valley localities. In The Evolution of Neogene Terrestrial Ecosystem in Europe. Hominoid Evolution and Climatic Change in Europe; Agustí, J., Rook, L., Andrews, P., Eds.; Cambridge University Press: Cambridge, UK, 1999; pp. 140–164. [Google Scholar]
- Rosina, V.V.; Sinitsa, M.V. Bats (Chiroptera, Mammalia) from the Turolian of the Ukraine: Phylogenetic and biostratigraphic considerations. N. Jb. Geol. Paläont. Abh. 2014, 272, 147–166. [Google Scholar] [CrossRef]
- Horáček, I. On the early history of vespertilionid bats in Europe: The Lower Miocene record from the Bohemian Massif. Lynx 2001, 32, 123–154. [Google Scholar]
- Ruedi, M.; Eger, J.L.; Lim, B.K.; Csorba, G. A new genus and species of vespertilionid bat from the Indomalayan region. J. Mammal. 2018, 99, 209–222. [Google Scholar] [CrossRef]
- Görföl, T.; Kruskop, S.V.; Vuong, T.T.; Estók, P.; Nguyen, T.S.; Csorba, G. A new genus of Vespertilionid bat: The end of a long journey for Joffre’s pipistrelle (Chiroptera: Vespertilionidae). J. Mammal. 2020, 101, 331–348. [Google Scholar] [CrossRef]

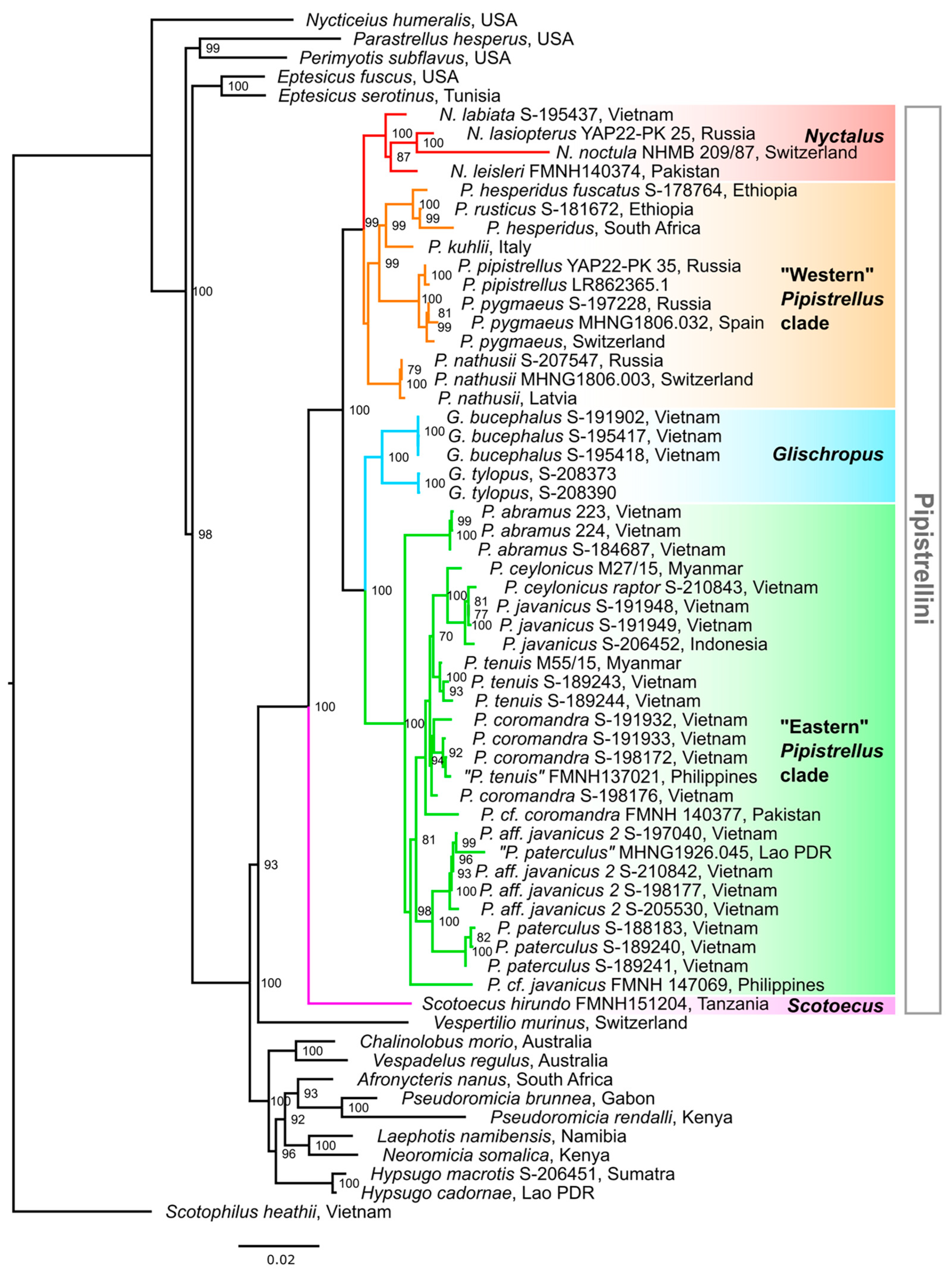

| Group | Scotoecus | Nyctalus | Glischropus | “Eastern” Pipistrellus Clade | “Western” Pipistrellus Clade |
|---|---|---|---|---|---|
| Scotoecus | 0.0569 | 0.0543 | 0.0591 | 0.0523 | |
| Nyctalus | 0.185 | 0.0399 | 0.0429 | 0.0314 | |
| Glischropus | 0.188 | 0.175 | 0.0324 | 0.0344 | |
| “Eastern” Pipistrellus clade | 0.193 | 0.184 | 0.169 | 0.0395 | |
| “Western” Pipistrellus clade | 0.187 | 0.172 | 0.171 | 0.174 | |
| Vansonia rueppellii | 0.216 | 0.191 | 0.194 | 0.197 | 0.187 |
| Measurements | Factor 1 | Factor 2 |
|---|---|---|
| TL | 0.65909 | 0.714665 |
| CCL | 0.71694 | 0.669831 |
| CBL | 0.67911 | 0.697700 |
| MW | 0.61110 | 0.768663 |
| BCW | 0.47121 | 0.860871 |
| OH | 0.59325 | 0.742683 |
| ZW | 0.62708 | 0.732749 |
| POC | 0.40049 | 0.883035 |
| IOW | 0.62428 | 0.744754 |
| RW | 0.63857 | 0.741287 |
| RL | 0.53120 | 0.701985 |
| CC | 0.65365 | 0.668459 |
| MM | 0.72146 | 0.659743 |
| CM | 0.83234 | 0.538769 |
| PM | 0.86454 | 0.485116 |
| C | 0.77851 | 0.556921 |
| M3W | 0.82139 | 0.498778 |
| M3L | 0.73807 | 0.584250 |
| Pal | 0.83859 | 0.442444 |
| cm | 0.82380 | 0.545565 |
| MdL | 0.72582 | 0.672085 |
| MdH | 0.73123 | 0.598691 |
| Eigenvalue | 19.92583 | 0.57624 |
| % Total variance | 90.57198 | 2.61927 |
| Training Set | ||||||
|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | ||
| 1 | Pipistrellus “western” | 37.9343 | 12.3037 | 8.26871 | 118.6221 | |
| 2 | Pipistrellus nathusii | 0.000 | 49.7333 | 44.01163 | 144.8407 | |
| 3 | Pipistrellus abramus | 0.000 | 0.000 | 10.24641 | 108.5053 | |
| 4 | Pipistrellus “eastern” | 0.000 | 0.000 | 0.000 | 89.6127 | |
| 5 | Pipistrellus stenopterus | 0.000 | 0.000 | 0.000 | 0.000 |
| Training Set | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | ||
| 1 | Pipistrellus abramus | 89.114 | 23.113 | 16.816 | 16.240 | 23.909 | 45.937 | 46.136 | 73.882 | 40.508 | 54.531 | |
| 2 | Pipistrellus ceylonicus | 0.000 | 69.046 | 112.343 | 66.346 | 52.153 | 79.267 | 175.165 | 176.022 | 174.851 | 211.266 | |
| 3 | Pipistrellus coromandra | 0.000 | 0.000 | 23.229 | 20.240 | 16.033 | 37.731 | 34.689 | 69.197 | 37.417 | 52.244 | |
| 4 | Pipistrellus endoi | 0.000 | 0.000 | 0.000 | 31.474 | 27.623 | 59.913 | 36.481 | 49.492 | 28.653 | 33.603 | |
| 5 | Pipistrellus hesperidus | 0.000 | 0.000 | 0.000 | 0.000 | 21.968 | 19.987 | 54.548 | 81.408 | 42.232 | 65.583 | |
| 6 | Pipistrellus javanicus | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 41.419 | 55.336 | 78.148 | 56.643 | 72.393 | |
| 7 | Pipistrellus kuhlii | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 70.561 | 120.742 | 61.243 | 86.181 | |
| 8 | Pipistrellus murrai | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 87.510 | 26.889 | 29.772 | |
| 9 | Pipistrellus nathusii | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 56.466 | 64.457 | |
| 10 | Pipistrellus pipistrellus | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 7.058 | |
| 11 | Pipistrellus pygmaeus | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.005 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhukova, S.S.; Yuzefovich, A.P.; Lebedev, V.S.; Kruskop, S.V. Reassessment of the Taxonomic Borders Within Pipistrellus (Chiroptera, Vespertilionidae, Pipistrellini). Diversity 2025, 17, 317. https://doi.org/10.3390/d17050317
Zhukova SS, Yuzefovich AP, Lebedev VS, Kruskop SV. Reassessment of the Taxonomic Borders Within Pipistrellus (Chiroptera, Vespertilionidae, Pipistrellini). Diversity. 2025; 17(5):317. https://doi.org/10.3390/d17050317
Chicago/Turabian StyleZhukova, Svetlana S., Alexander P. Yuzefovich, Vladimir S. Lebedev, and Sergei V. Kruskop. 2025. "Reassessment of the Taxonomic Borders Within Pipistrellus (Chiroptera, Vespertilionidae, Pipistrellini)" Diversity 17, no. 5: 317. https://doi.org/10.3390/d17050317
APA StyleZhukova, S. S., Yuzefovich, A. P., Lebedev, V. S., & Kruskop, S. V. (2025). Reassessment of the Taxonomic Borders Within Pipistrellus (Chiroptera, Vespertilionidae, Pipistrellini). Diversity, 17(5), 317. https://doi.org/10.3390/d17050317