β-Glucosidases: In Silico Analysis of Physicochemical Properties and Domain Architecture Diversity Revealed by Metagenomic Technology
Abstract
1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Phylogenetic Analysis of β-Glucosidase Sequences
2.3. In Silico Analysis of Physicochemical Properties and Domain Architectures of β-Glucosidases
2.4. Taxonomic Assignment
2.5. Purification and Characterization of β-Glucosidase GH3-31
2.6. Statistical Analysis
3. Results
3.1. Diversity of β-Glucosidase Sequences from Goat Rumen, Wood Humus, and Termite Gut
3.2. Physicochemical Properties of Bacterial β-Glucosidases from Wood Humus, Termite Gut and Goat Rumen
3.3. Domain Architectures of β-Glucosidases from Goat Rumen, Wood Humus and Termite Gut
3.4. Diversity of Bacteria Producing β-Glucosidase
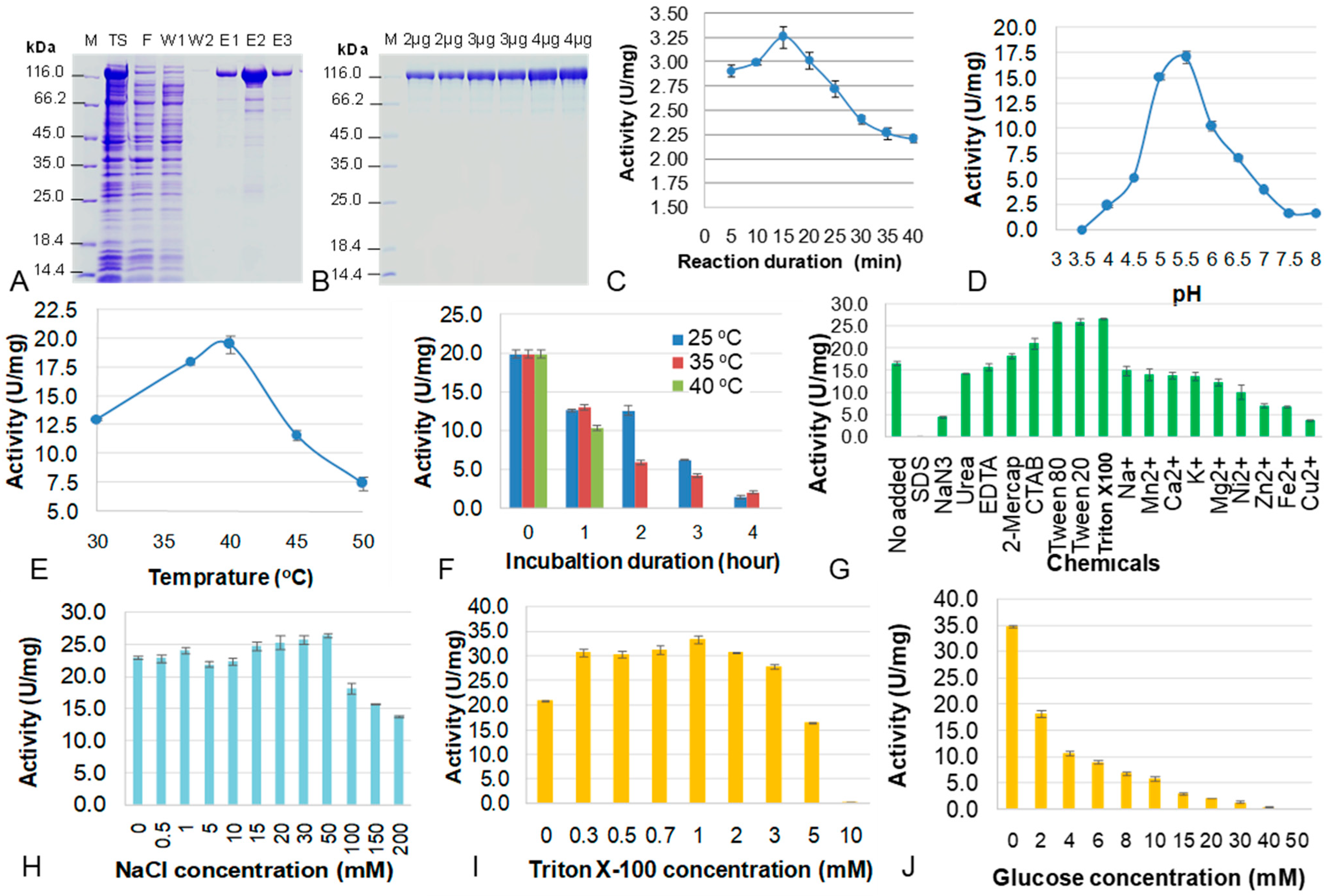
3.5. Purification and Characterization of β-Glucosidase GH3-31
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| GH | Glycosyl hydrolase |
| CAZy | Carbohydrate-Active enZYmes |
| KEGG | Kyoto Encyclopedia of Genes and Genomes |
| FN3 | fibronectin type III |
| CBM | carbohydrate-binding module |
| pI | isoelectric point |
| Tm | melting temperature |
| MEGAN | The Metagenome Analyzer program |
| NR | NCBI non-redundant protein |
| BGC-GH3-31 | Beta-glucosidase carried domains GH3 and GH31 |
References
- Godse, R.; Fernandes, J.M.; Kulkarni, R. Characterization of β-Glucosidase Activity of a Lactiplantibacillus plantarum 6-Phospho-β-Glucosidase. Appl. Microbiol. Biotechnol. 2025, 109, 86. [Google Scholar] [CrossRef]
- Paventi, G.; Di Martino, C.; Coppola, F.; Iorizzo, M. β-Glucosidase Activity of Lactiplantibacillus plantarum: A Key Player in Food Fermentation and Human Health. Foods 2025, 14, 1451. [Google Scholar] [CrossRef]
- Ketudat Cairns, J.R.; Esen, A. β-Glucosidases. Cell Mol. Life Sci. 2010, 67, 3389–3405. [Google Scholar] [CrossRef] [PubMed]
- Muradova, M.; Proskura, A.; Canon, F.; Aleksandrova, I.; Schwartz, M.; Heydel, J.M.; Baranenko, D.; Nadtochii, L.; Neiers, F. Unlocking Flavor Potential Using Microbial β-Glucosidases in Food Processing. Foods 2023, 12, 4484. [Google Scholar] [CrossRef]
- Cao, H.; Zhang, Y.; Shi, P.; Ma, R.; Yang, H.; Xia, W.; Cui, Y.; Luo, H.; Bai, Y.; Yao, B. A Highly Glucose-Tolerant GH1 β-Glucosidase with Greater Conversion Rate of Soybean Isoflavones in Monogastric Animals. J. Ind. Microbiol. Biotechnol. 2018, 45, 369–378. [Google Scholar] [CrossRef]
- Tran, T.N.A.; Son, J.S.; Awais, M.; Ko, J.H.; Yang, D.C.; Jung, S.K. β-Glucosidase and Its Application in Bioconversion of Ginsenosides in Panax ginseng. Bioengineering 2023, 10, 484. [Google Scholar] [CrossRef]
- Magwaza, B.; Amobonye, A.; Pillai, S. Microbial β-Glucosidases: Recent Advances and Applications. Biochimie 2024, 225, 49–67. [Google Scholar] [CrossRef]
- Ashwini, M. Beta Glucosidase Market Report 2025 (Global Edition). 2025. Available online: https://www.cognitivemarketresearch.com/beta-glucosidase-market-report (accessed on 9 November 2025).
- Singh, N.; Sithole, B.; Kumar, A.; Govinden, R. A Glucose Tolerant β-Glucosidase from a Newly Isolated Neofusicoccum parvum Strain F7: Production, Purification, and Characterization. Sci. Rep. 2023, 13, 5134. [Google Scholar] [CrossRef]
- Ouyang, B.; Wang, G.; Zhang, N.; Zuo, J.; Huang, Y.; Zhao, X. Recent Advances in β-Glucosidase Sequence and Structure Engineering: A Brief Review. Molecules 2023, 28, 4990. [Google Scholar] [CrossRef] [PubMed]
- Solden, L.; Lloyd, K.; Wrighton, K. The Bright Side of Microbial Dark Matter: Lessons Learned from the Uncultivated Majority. Curr. Opin. Microbiol. 2016, 31, 217–226. [Google Scholar] [CrossRef] [PubMed]
- Salgado, J.C.S.; Meleiro, L.P.; Carli, S.; Ward, R.J. Glucose Tolerant and Glucose Stimulated β-Glucosidases—A Review. Bioresour. Technol. 2018, 267, 704–713. [Google Scholar] [CrossRef]
- Kaçıran, A.; Şahinkaya, M.; Çolak, D.N.; Zada, N.S.; Kaçağan, M.; Güler, H.İ.; Saygın, H.; Beldüz, A.O. Biochemical Characterization of a Novel, Glucose-Tolerant β-Glucosidase from Jiangella ureilytica KC603, and Determination of Resveratrol Production Capacity from Polydatin. Appl. Biochem. Biotechnol. 2025, 197, 5104–5130. [Google Scholar] [CrossRef]
- Tiwari, R.; Kumar, K.; Singh, S.; Nain, L.; Shukla, P. Molecular Detection and Environment-Specific Diversity of Glycosyl Hydrolase Family 1 β-Glucosidase in Different Habitats. Front. Microbiol. 2016, 7, 1597. [Google Scholar] [CrossRef] [PubMed]
- Mao, G.; Song, M.; Li, H.; Lin, J.; Wang, K.; Liu, Q.; Su, Z.; Zhang, H.; Su, L.; Xie, H.; et al. Biochemical and Structural Characterization of a Highly Glucose-Tolerant β-Glucosidase from the Termite Reticulitermes perilucifugus. Int. J. Mol. Sci. 2025, 26, 3118. [Google Scholar] [CrossRef]
- Dong, S.; Liu, Y.J.; Zhou, H.; Xiao, Y.; Xu, J.; Cui, Q.; Wang, X.; Feng, Y. Structural Insight into a GH1 β-Glucosidase from the Oleaginous Microalga, Nannochloropsis Oceanica. Int. J. Biol. Macromol. 2021, 170, 196–206. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, K.H.V.; Dao, T.K.; Nguyen, H.D.; Nguyen, K.H.; Nguyen, T.Q.; Nguyen, T.T.; Nguyen, T.M.P.; Truong, N.H.; Do, T.H. Some Characters of Bacterial Cellulases in Goats’ Rumen Elucidated by Metagenomic DNA Analysis and the Role of Fibronectin 3 Module for Endoglucanase Function. Anim. Biosci. 2021, 34, 867–879. [Google Scholar] [CrossRef]
- Sidar, A.; Voshol, G.P.; Arentshorst, M.; Ram, A.F.J.; Vijgenboom, E.; Punt, P.J. Deciphering Domain Structures of Aspergillus and Streptomyces GH3-β-Glucosidases: A Screening System for Enzyme Engineering and Biotechnological Applications. BMC Res. Notes 2024, 17, 257. [Google Scholar] [CrossRef]
- He, Y.; Wang, C.; Jiao, R.; Ni, Q.; Wang, Y.; Gao, Q.; Zhang, Y.; Xu, G. Biochemical Characterization of a Novel Glucose-Tolerant GH3 β-Glucosidase (Bgl1973) from Leifsonia Sp. ZF2019. Appl. Microbiol. Biotechnol. 2022, 106, 5063–5079. [Google Scholar] [CrossRef]
- Georgakis, N.; Premetis, G.E.; Pantiora, P.; Varotsou, C.; Bodourian, C.S.; Labrou, N.E. The Impact of Metagenomic Analysis on the Discovery of Novel Endolysins. Appl. Microbiol. Biotechnol. 2025, 109, 126. [Google Scholar] [CrossRef]
- Jeilu, O.; Alexandersson, E.; Johansson, E.; Simachew, A.; Gessesse, A. A Novel GH3-β-Glucosidase from Soda Lake Metagenomic Libraries with Desirable Properties for Biomass Degradation. Sci. Rep. 2024, 14, 10012. [Google Scholar] [CrossRef] [PubMed]
- Ariaeenejad, S.; Nooshi-Nedamani, S.; Rahban, M.; Kavousi, K.; Pirbalooti, A.G.; Mirghaderi, S.; Mohammadi, M.; Mirzaei, M.; Salekdeh, G.H. A Novel High Glucose-Tolerant β-Glucosidase: Targeted Computational Approach for Metagenomic Screening. Front. Bioeng. Biotechnol. 2020, 8, 813. [Google Scholar] [CrossRef]
- Kaushal, G.; Rai, A.K.; Singh, S.P. A Novel β-Glucosidase from a Hot-Spring Metagenome Shows Elevated Thermal Stability and Tolerance to Glucose and Ethanol. Enzym. Microb. Technol. 2021, 145, 109764. [Google Scholar] [CrossRef]
- Mai, Z.; Su, H.; Zhang, S. Characterization of a Metagenome-Derived β-Glucosidase and Its Application in Conversion of Polydatin to Resveratrol. Catalysts 2016, 6, 35. [Google Scholar] [CrossRef]
- Matsuzawa, T.; Watanabe, M.; Nakamichi, Y.; Akita, H.; Yaoi, K. Crystal Structure of Metagenomic β-Glycosidase MeBglD2 in Complex with Various Saccharides. Appl. Microbiol. Biotechnol. 2022, 106, 4539–4551. [Google Scholar] [CrossRef]
- Do, T.H.; Nguyen, T.T.; Nguyen, T.N.; Le, Q.G.; Nguyen, C.; Kimura, K.; Truong, N.H. Mining Biomass-Degrading Genes through Illumina-Based de Novo Sequencing and Metagenomic Analysis of Free-Living Bacteria in the Gut of the Lower Termite Coptotermes gestroi Harvested in Vietnam. J. Biosci. Bioeng. 2014, 118, 665–671. [Google Scholar] [CrossRef]
- Do, T.H.; Dao, T.K.; Nguyen, K.H.V.; Le, N.G.; Nguyen, T.M.P.; Le, T.L.; Phung, T.N.; van Straalen, N.M.; Roelofs, D.; Truong, N.H. Metagenomic Analysis of Bacterial Community Structure and Diversity of Lignocellulolytic Bacteria in Vietnamese Native Goat Rumen. Asian-Australas. J. Anim. Sci. 2018, 31, 738–747. [Google Scholar] [CrossRef]
- Do, T.H.; Le, N.G.; Dao, T.K.; Nguyen, T.M.P.; Le, T.L.; Luu, H.L.; Nguyen, K.H.V.; Nguyen, V.L.; Le, L.A.; Phung, T.N.; et al. Metagenomic Insights into Lignocellulose-Degrading Genes through Illumina-Based de Novo Sequencing of the Microbiome in Vietnamese Native Goats’ Rumen. J. Gen. Appl. Microbiol. 2018, 64, 108–116. [Google Scholar] [CrossRef] [PubMed]
- Dao, T.K.; Do, T.H.; Le, N.G.; Nguyen, H.D.; Nguyen, T.Q.; Le, T.T.H.; Truong, N.H. Understanding the Role of Prevotella Genus in the Digestion of Lignocellulose and Other Substrates in Vietnamese Native Goats’ Rumen by Metagenomic Deep Sequencing. Animals 2021, 11, 3257. [Google Scholar] [CrossRef] [PubMed]
- Le, T.T.H.; Nguyen, T.B.; Nguyen, H.D.; Nguyen, H.D.; Le, N.G.; Dao, T.K.; Nguyen, T.Q.; Do, T.H.; Truong, N.H. De Novo Metagenomic Analysis of Microbial Community Contributing in Lignocellulose Degradation in Humus Samplesharvested from Cuc Phuong Tropical Forest in Vietnam. Diversity 2022, 14, 220. [Google Scholar] [CrossRef]
- Lin, H.; Chen, W.; Ding, H. AcalPred: A Sequence-Based Tool for Discriminating between Acidic and Alkaline Enzymes. PLoS ONE 2013, 8, e75726. [Google Scholar] [CrossRef]
- Nguyen, K.; Nguyen, T.; Truong, N.; Do, T. Application of Bioinformatic Tools for Prediction of Active pH and Temperature Stability of Endoglucanases Based on Coding Sequences from Metagenomic DNA Data. Biol. Forum-Int. J. 2019, 11, 14–20. [Google Scholar]
- Lim, S.; Seo, J.; Choi, H.; Yoon, D.; Nam, J.; Kim, H.; Cho, S.; Chang, J. Metagenome Analysis of Protein Domain Collocation within Cellulase Genes of Goat Rumen Microbes. Asian-Australas. J. Anim. Sci. 2013, 26, 1144–1151. [Google Scholar] [CrossRef]
- Lombard, V.; Henrissat, B.; Garron, M.-L. CAZac: An Activity Descriptor for Carbohydrate-Active Enzymes. Nucleic Acids Res. 2025, 53, D625–D633. [Google Scholar] [CrossRef]
- Huson, D.H.; Auch, A.F.; Qi, J.; Schuster, S.C. MEGAN Analysis of Metagenomic Data. Genome Res. 2007, 17, 377–386. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, T.Q.; Do, T.H.; Nguyen, T.K.L.; Nguyen, H.D.; Truong, N.H. Expression of Beta-Glucosidase Mined from Metagenomic DNA Data of Bacteria in Vietnamese Goats’ Rumen in Eschrichia coli System. Acad. J. Biol. 2022, 44, 43–52. [Google Scholar] [CrossRef]
- Binh, N.T.; Quy, N.T.; Huyen, D.T.; Hong, L.T.T.; Hai, T.N. Selection of Optimal Culture Conditions for Expression of Recombinant Beta-Glucosidase in Escherichia coli. Vietnam J. Biotechnol. 2022, 20, 425–433. [Google Scholar] [CrossRef]
- Binh, N.T.; Quy, N.T.; Hong, L.T.T.; Hai, T.N. Purification and Characterization of a Recombinant Beta-Glucosidase in Escherichia coli. Vietnam J. Biotechnol. 2022, 20, 599–607. [Google Scholar] [CrossRef]
- Bradford, M.M. A Rapid and Sensitive Method for the Quantitation of Microgram Quantities of Protein Utilizing the Principle of Protein-Dye Binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Fang, S.; Chang, J.; Lee, Y.S.; Guo, W.; Choi, Y.L.; Zhou, Y. Cloning and Characterization of a New Broad specific β-Glucosidase from Lactococcus sp. FSJ4. World J. Microbiol. Biotechnol. 2014, 30, 213–223. [Google Scholar] [CrossRef] [PubMed]
- Qi, J.; Zhang, M.; Chen, C.; Feng, Y.; Xuan, J. Cellulosome Systems in the Digestive Tract: Underexplored Enzymatic Machine for Lignocellulose Bioconversion. Catalysts 2025, 15, 387. [Google Scholar] [CrossRef]
- Bule, P.; Pires, V.M.R.; Alves, V.D.; Carvalho, A.L.; Prates, J.A.M.; Ferreira, L.M.A.; Smith, S.P.; Gilbert, H.J.; Noach, I.; Bayer, E.A.; et al. Higher Order Scaffoldin Assembly in Ruminococcus Flavefaciens Cellulosome Is Coordinated by a Discrete Cohesin-Dockerin Interaction. Sci. Rep. 2018, 8, 6987. [Google Scholar] [CrossRef]
- Alahuhta, M.; Xu, Q.; Bomble, Y.J.; Brunecky, R.; Adney, W.S.; Ding, S.Y.; Himmel, M.E.; Lunin, V.V. The Unique Binding Mode of Cellulosomal CBM4 from Clostridium thermocellum Cellobiohydrolase A. J. Mol. Biol. 2010, 402, 374–387. [Google Scholar] [CrossRef]
- Huang, Y.; Busk, P.K.; Grell, M.N.; Zhao, H.; Lange, L. Identification of a β-Glucosidase from the Mucor Circinelloides Genome by Peptide Pattern Recognition. Enzym. Microb. Technol. 2014, 67, 47–52. [Google Scholar] [CrossRef]
- Yang, Y.; Zhang, X.; Yin, Q.; Fang, W.; Fang, Z.; Wang, X.; Zhang, X.; Xiao, Y. A Mechanism of Glucose Tolerance and Stimulation of GH1 β-Glucosidases. Sci. Rep. 2015, 5, 17296. [Google Scholar] [CrossRef] [PubMed]
- Correia, M.A.S.; Pires, V.M.R.; Gilbert, H.J.; Bolam, D.N.; Fernandes, V.O.; Alves, V.D.; Prates, J.A.M.; Ferreira, L.M.A.; Fontes, C.M.G.A. Family 6 Carbohydrate-Binding Modules Display Multiple Beta1,3-Linked Glucan-Specific Binding Interfaces. FEMS Microbiol. Lett. 2009, 300, 48–57. [Google Scholar] [CrossRef] [PubMed]
- Rigden, D.J.; Mello, L.V.; Galperin, M.Y. The PA14 Domain, a Conserved All-β Domain in Bacterial Toxins, Enzymes, Adhesins and Signaling Molecules. Trends Biochem. Sci. 2004, 29, 335–339. [Google Scholar] [CrossRef] [PubMed]
- Castro-Costa, A.; Salama, A.A.K.; Moll, X.; Aguiló, J.; Caja, G. Using Wireless Rumen Sensors for Evaluating the Effects of Diet and Ambient Temperature in Nonlactating Dairy Goats. J. Dairy. Sci. 2015, 98, 4646–4658. [Google Scholar] [CrossRef]
- Mamuad, L.L.; Lee, S.S.; Lee, S.S. Recent Insight and Future Techniques to Enhance Rumen Fermentation in Dairy Goats. Asian-Australas. J. Anim. Sci. 2019, 32, 1321–1330. [Google Scholar] [CrossRef]
- Huang, Y.Y.; Lv, Z.H.; Zheng, H.Z.; Zhu, Q.; Liu, M.T.; Sang, P.; Wang, F.; Zhu, D.; Xian, W.D.; Yin, Y.R. Characterization of a Thermophilic and Glucose-Tolerant GH1 β-Glucosidase from Hot Springs and Its Prospective Application in Corn Stover Degradation. Front. Microbiol. 2023, 14, 1286682. [Google Scholar] [CrossRef]
- Zhu, Q.; Huang, Y.; Yang, Z.; Wu, X.; Zhu, Q.; Zheng, H.; Zhu, D.; Lv, Z.; Yin, Y. A Recombinant Thermophilic and Glucose-Tolerant GH1 β-Glucosidase Derived from Hehua Hot Spring. Molecules 2024, 29, 1017. [Google Scholar] [CrossRef]
- Talley, K.; Alexov, E. On the pH-Optimum of Activity and Stability of Proteins. Proteins 2010, 78, 2699–2706. [Google Scholar] [CrossRef] [PubMed]
- Krisch, J.; Takó, M.; Papp, T.; Vágvölgyi, C. Characteristics and Potential Use of β-Glucosidases from Zygomycetes. In Current Research; Technology and Education Topics in Applied Microbiology and Microbial Biotechnology; Formatex Research Center: Badajoz, Spain, 2010; pp. 891–896. ISBN 978-84-614-6195-0. [Google Scholar]
- Hong, M.R.; Kim, Y.S.; Park, C.S.; Lee, J.K.; Kim, Y.S.; Oh, D.K. Characterization of a Recombinant Beta-Glucosidase from the Thermophilic Bacterium Caldicellulosiruptor Saccharolyticus. J. Biosci. Bioeng. 2009, 108, 36–40. [Google Scholar] [CrossRef] [PubMed]



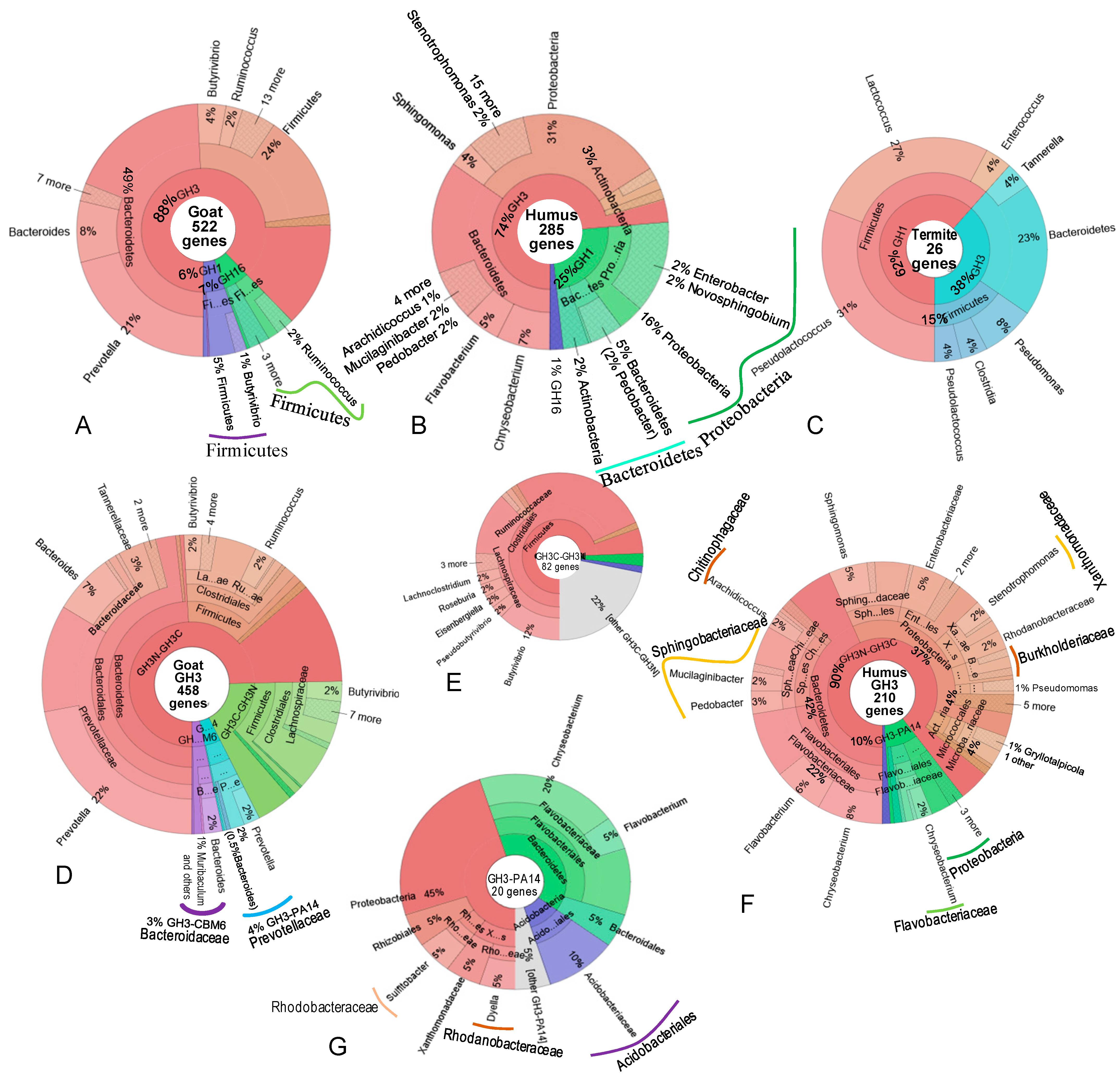
| No | Modular Structure | Goat | Humus | Termite | Total |
|---|---|---|---|---|---|
| Total β-glucosidase GH1 | 29 | 70 | 16 | 115 | |
| 1 | GH1 | 29 | 57 | 16 | 102 |
| 2 | Sig-GH1 | 13 | 13 | ||
| Total β-glucosidase GH16 | 35 | 5 | 0 | 40 | |
| 1 | GH16 | 6 | 1 | 0 | 7 |
| 2 | SigP-GH16 | 9 | 3 | 12 | |
| 3 | GH16-CBM4 | 6 | 6 | ||
| 4 | SigP-GH16-CBM4 | 13 | 13 | ||
| 5 | SigP-Dockerin-GH16_CBM4-CBM4 | 1 | 1 | ||
| 6 | GH16-CBM32-Por_sec_tail | 1 | 1 | ||
| Total β-glucosidase GH3 | 458 | 210 | 10 | 678 | |
| 1 | SigP-GH3C-FN3-GH3N | 7 | 7 | ||
| 2 | GH3C-FN3-GH3N | 72 | 2 | 74 | |
| 3 | GH3C-GH3N | 3 | 3 | ||
| 4 | GH3N-GH3C-FN3-Lactamase | 1 | 1 | ||
| 5 | GH3N-GH3C-FN3 | 174 | 69 | 2 | 245 |
| 6 | GH3N-GH3C | 25 | 3 | 28 | |
| 7 | GH3N-GH3C-ExopC | 2 | 2 | ||
| 8 | SigP-GH3N-GH3C-FN3-GH5 | 1 | 1 | ||
| 9 | SigP-GH3N-GH3C-FN3-GH31 | 1 | 1 | ||
| 10 | SigP-GH3N-GH3C-FN3 | 121 | 110 | 4 | 235 |
| 11 | SigP-GH3N-GH3C | 19 | 4 | 1 | 24 |
| 12 | GH3N-GH3C:PA14:GH3C-FN3 | 8 | 7 | 3 | 18 |
| 13 | GH3N-GH3C-FN3-PA14 | 1 | 1 | ||
| 14 | SigP-GH3N-GH3C:PA14:GH3C-FN3 | 8 | 13 | 21 | |
| 15 | SigP-DUF-GH3N-GH3C:PA14:GH3C-FN3 | 1 | 1 | ||
| 16 | GH3N-GH3C:PA14:GH3C | 1 | 1 | ||
| 17 | GH3N-GH3C:CBM6:GH3C-FN3-CE8 | 1 | 1 | ||
| 18 | GH3N-GH3C:CBM6:GH3C-FN3-Big2-GH43 | 1 | 1 | ||
| 19 | SigP-GH3N-GH3C:CBM6:GH3C-FN3-CE8 | 1 | 1 | ||
| 20 | SigP-GH3N-GH3C:CBM6:GH3C-FN3-Big2-GH43 | 1 | 1 | ||
| 21 | SigP-GH3N-GH3C:CBM6:GH3C-FN3-GH43 | 2 | 2 | ||
| 22 | SigP-GH3N-GH3C:CBM6:GH3C-FN3 | 9 | 9 | ||
| Total | 522 | 285 | 26 | 833 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nguyen, T.Q.; Do, T.H.; Le, N.G.; Nguyen, H.D.; Dao, T.K.; Dinh, N.T.; Truong, N.H. β-Glucosidases: In Silico Analysis of Physicochemical Properties and Domain Architecture Diversity Revealed by Metagenomic Technology. Diversity 2025, 17, 804. https://doi.org/10.3390/d17110804
Nguyen TQ, Do TH, Le NG, Nguyen HD, Dao TK, Dinh NT, Truong NH. β-Glucosidases: In Silico Analysis of Physicochemical Properties and Domain Architecture Diversity Revealed by Metagenomic Technology. Diversity. 2025; 17(11):804. https://doi.org/10.3390/d17110804
Chicago/Turabian StyleNguyen, Thi Quy, Thi Huyen Do, Ngoc Giang Le, Hong Duong Nguyen, Trong Khoa Dao, Nho Thai Dinh, and Nam Hai Truong. 2025. "β-Glucosidases: In Silico Analysis of Physicochemical Properties and Domain Architecture Diversity Revealed by Metagenomic Technology" Diversity 17, no. 11: 804. https://doi.org/10.3390/d17110804
APA StyleNguyen, T. Q., Do, T. H., Le, N. G., Nguyen, H. D., Dao, T. K., Dinh, N. T., & Truong, N. H. (2025). β-Glucosidases: In Silico Analysis of Physicochemical Properties and Domain Architecture Diversity Revealed by Metagenomic Technology. Diversity, 17(11), 804. https://doi.org/10.3390/d17110804






