Mapping Global Biodiversity and Habitat Distribution of Lactobacillaceae Using NCBI Sequence Metadata
Abstract
1. Introduction
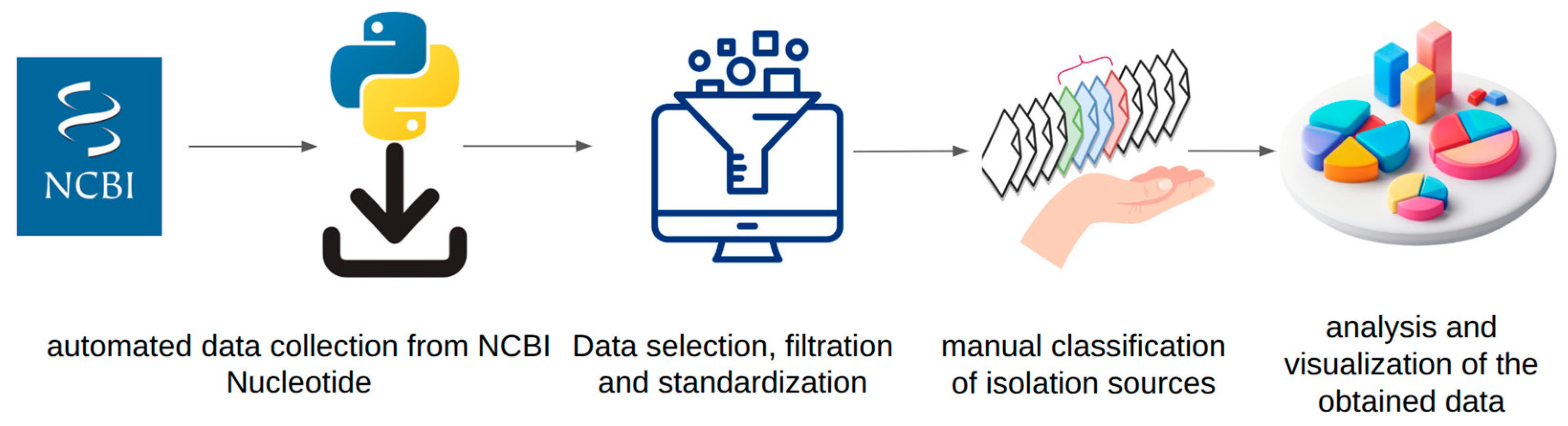
2. Materials and Methods
2.1. Data Acquisition and Primary Database Construction
2.2. Database Curation and Verification
2.3. Habitat Classification and Dictionary Development
2.4. Data Analysis and Visualization
3. Results
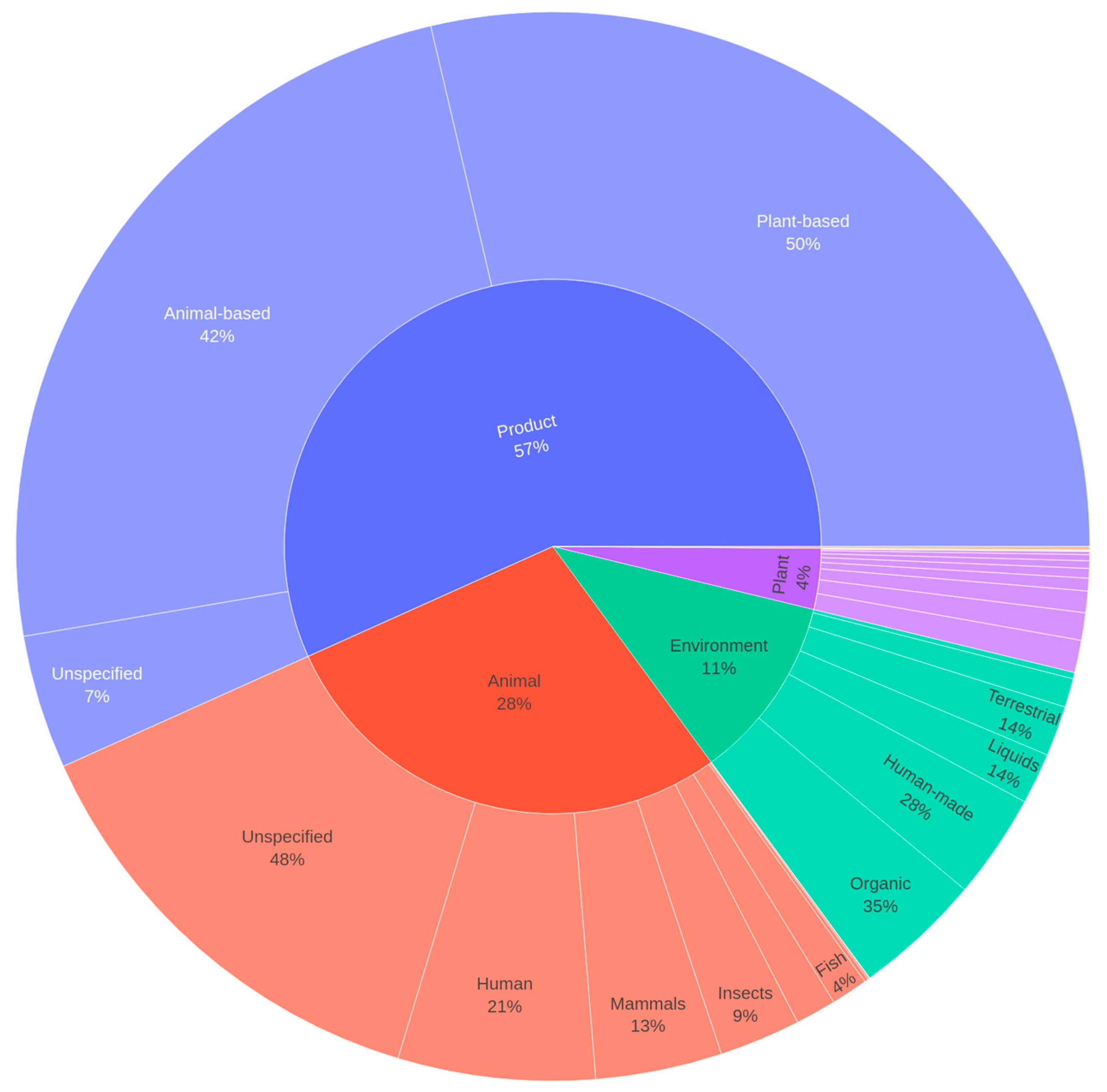
3.1. General Habitat Distribution of Lactobacillaceae
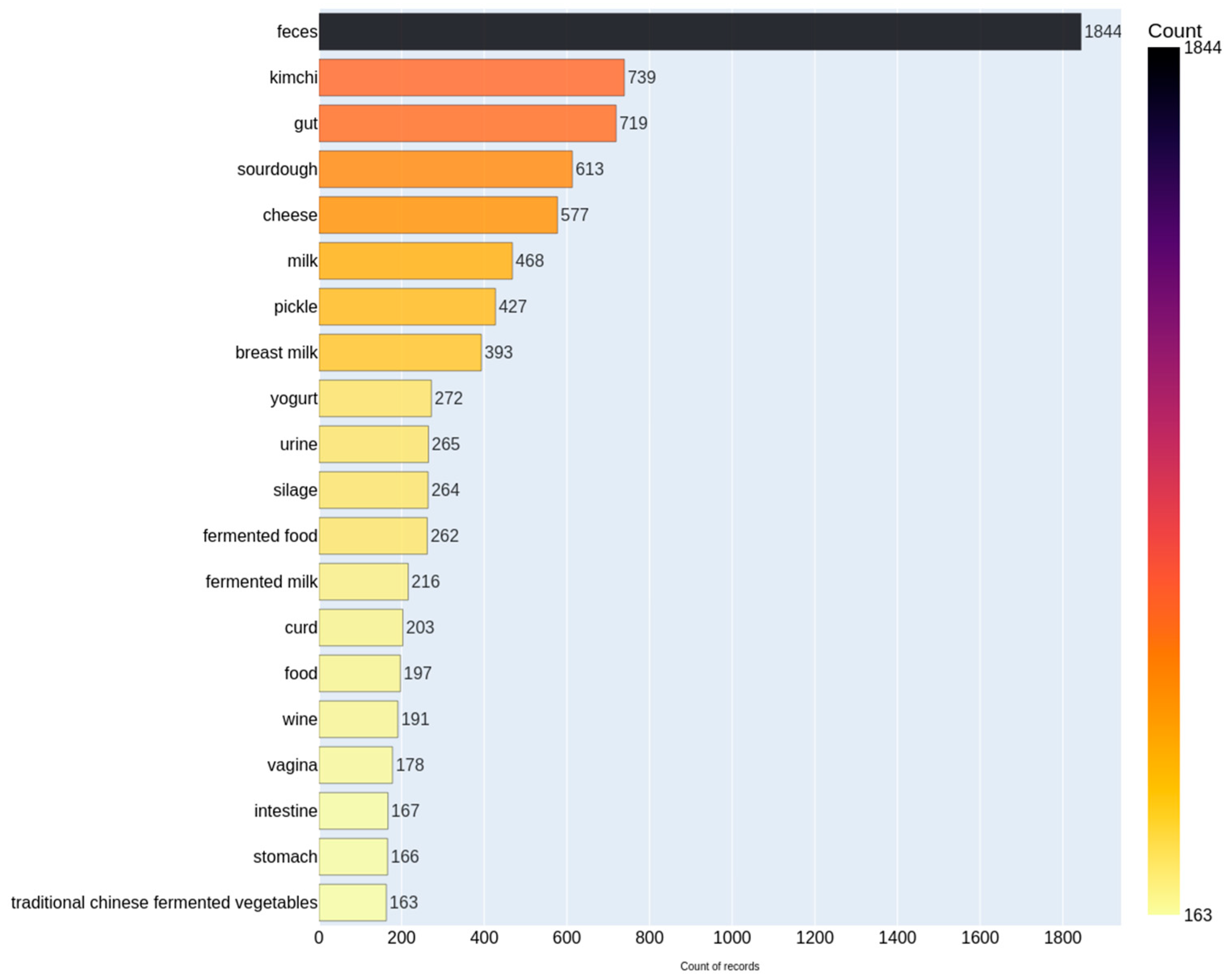
3.2. Distribution of Top Isolation Sources
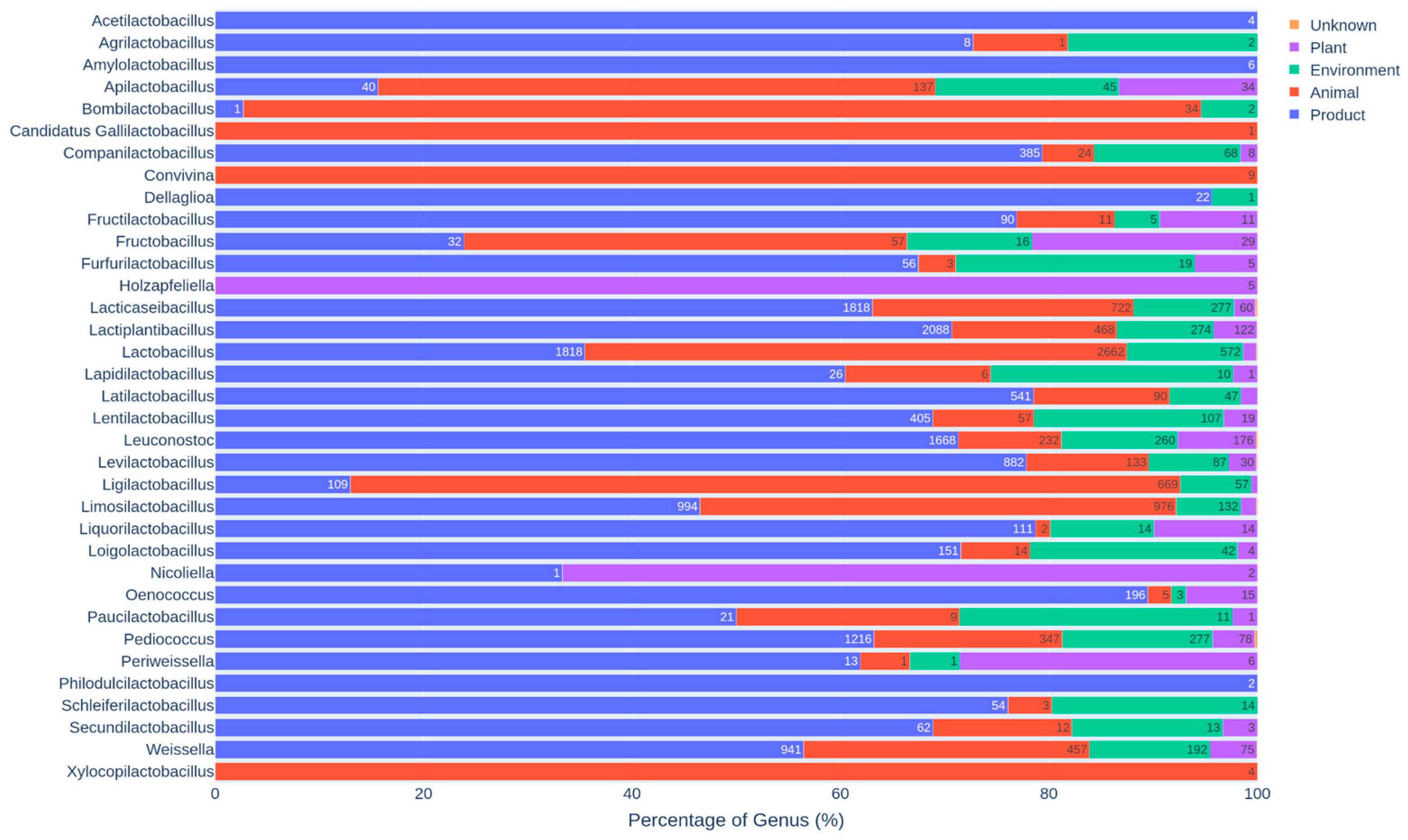
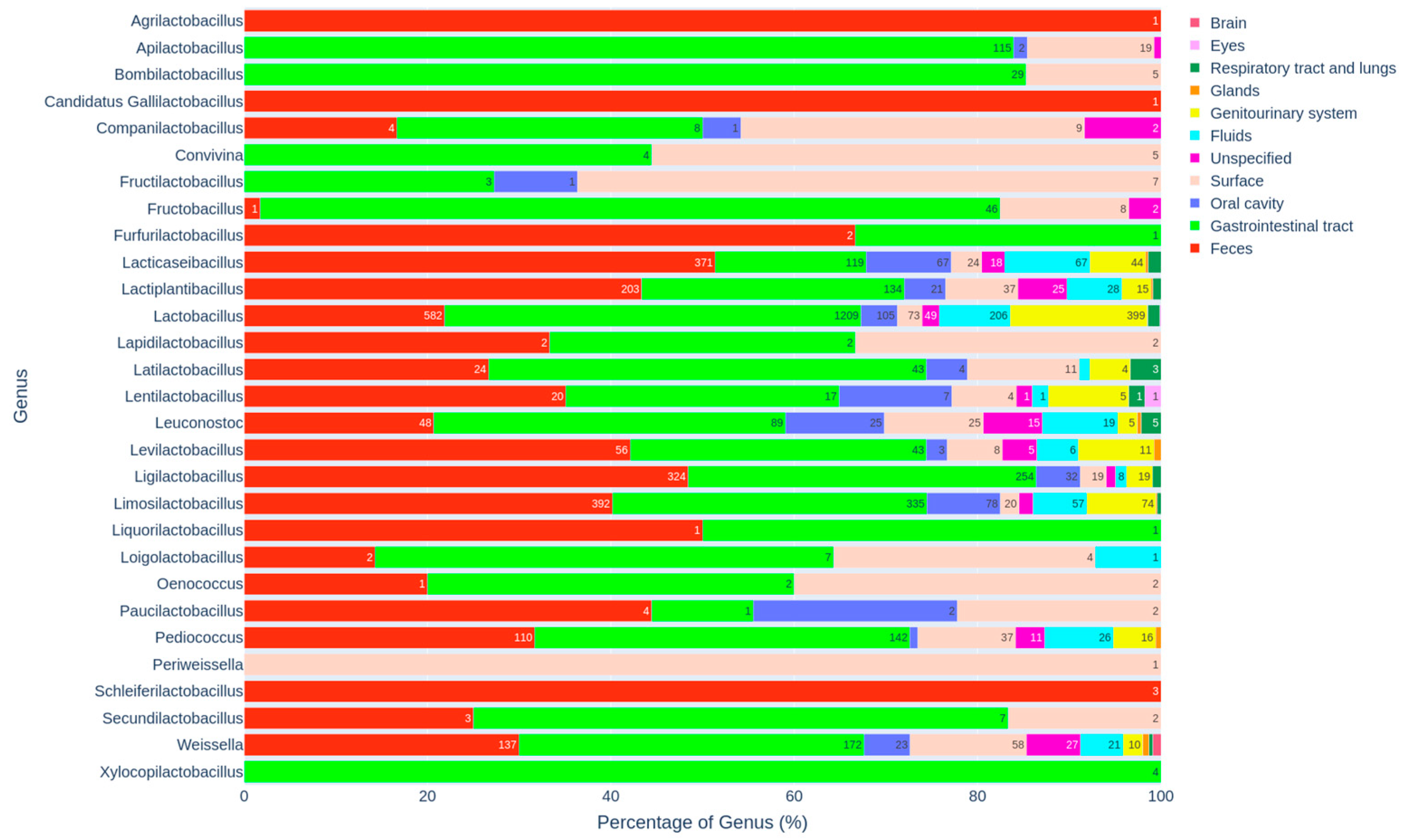
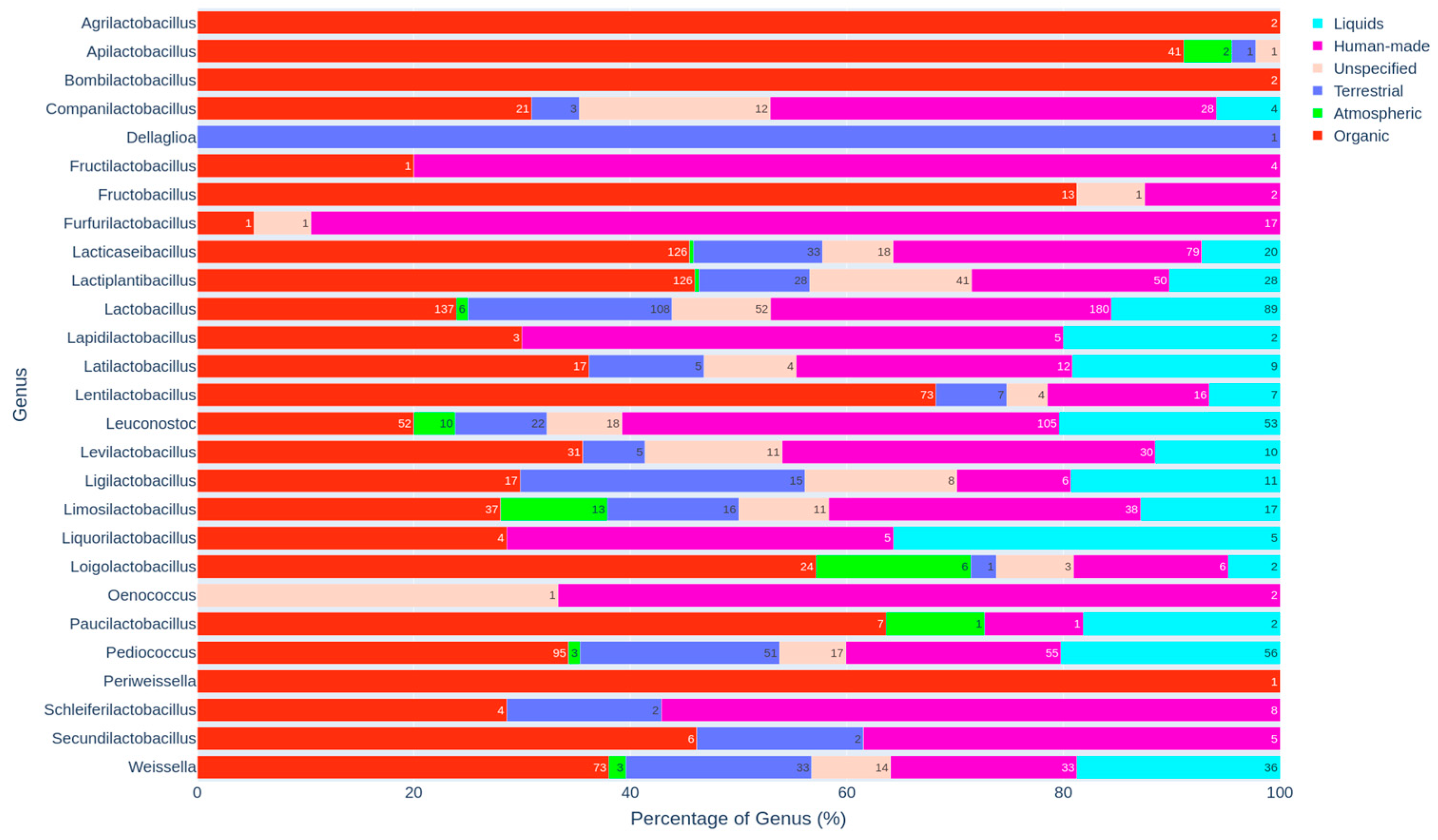
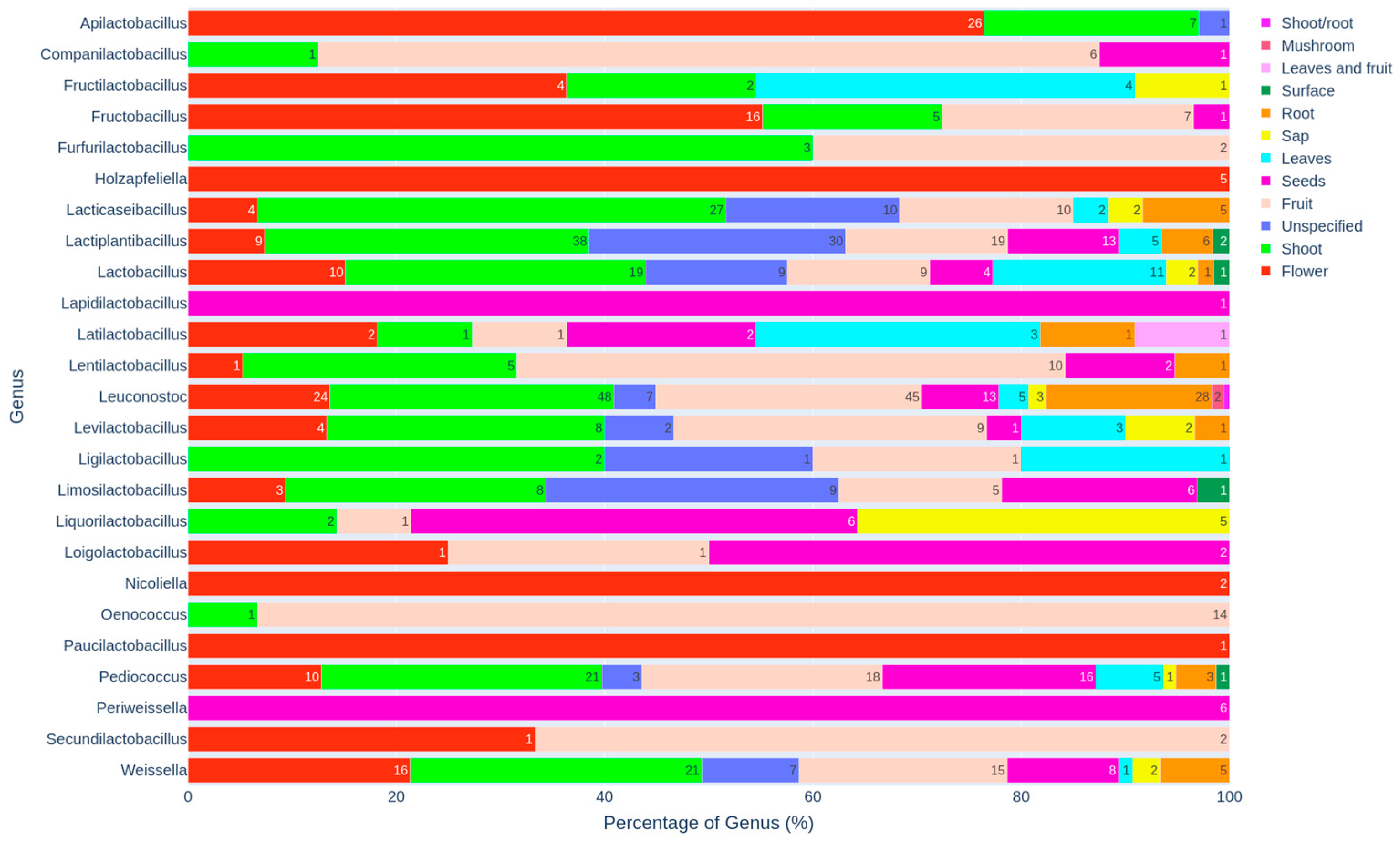
3.3. Distribution of Genera and Species Across Specific Habitats
3.3.1. Genus-Level Distribution and Habitat Preferences
3.3.2. Specialization Within Food-Associated Habitats
3.3.3. Distribution Within Animal-Associated Habitats and Host Specificity
3.3.4. Ecological Associations in Soil, Silage, Water, and Organic Wastes
3.3.5. Plant-Associated Habitats
3.3.6. Synthesis: A Spectrum from Habitat Generalists to Extreme Specialists
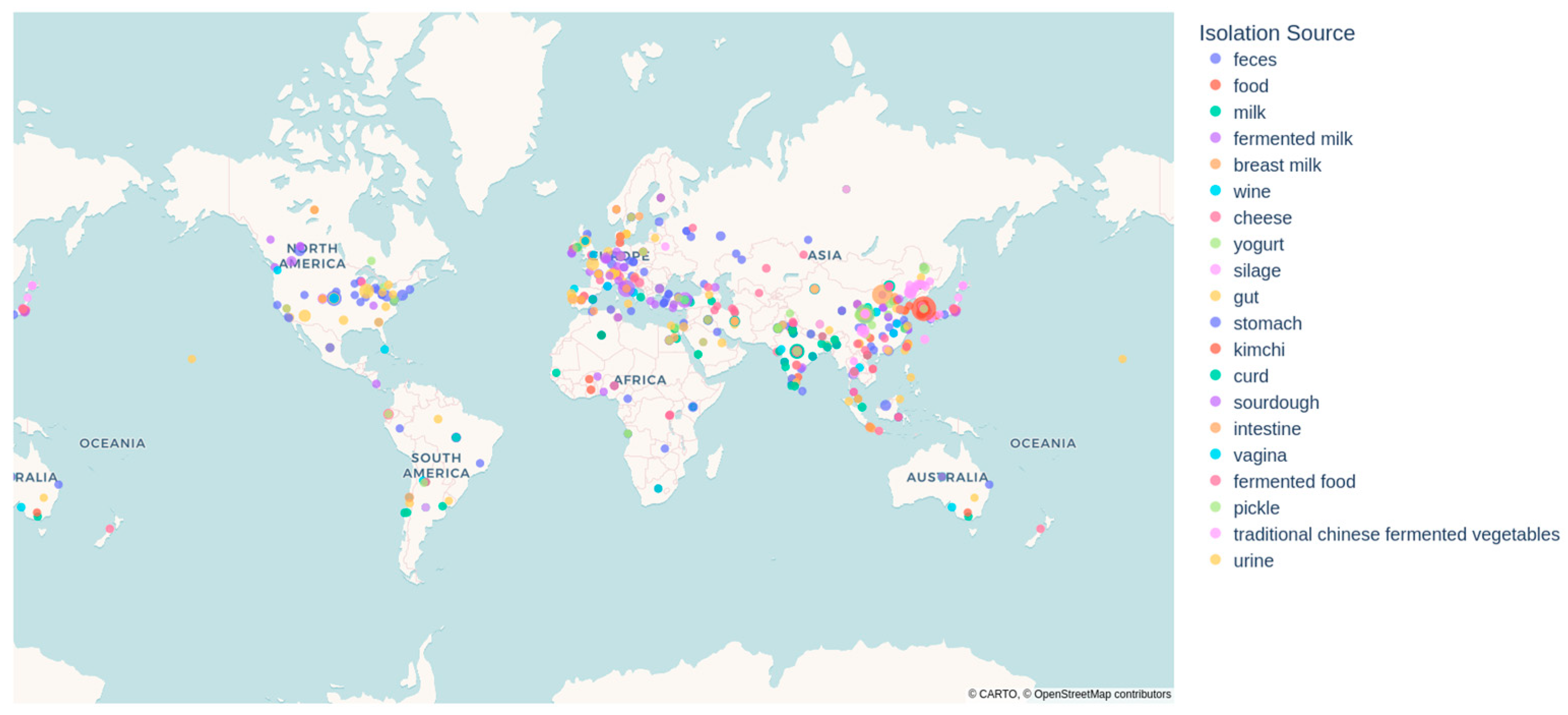
3.4. Geographical Trends and Sampling Biases in Lactobacillaceae Research
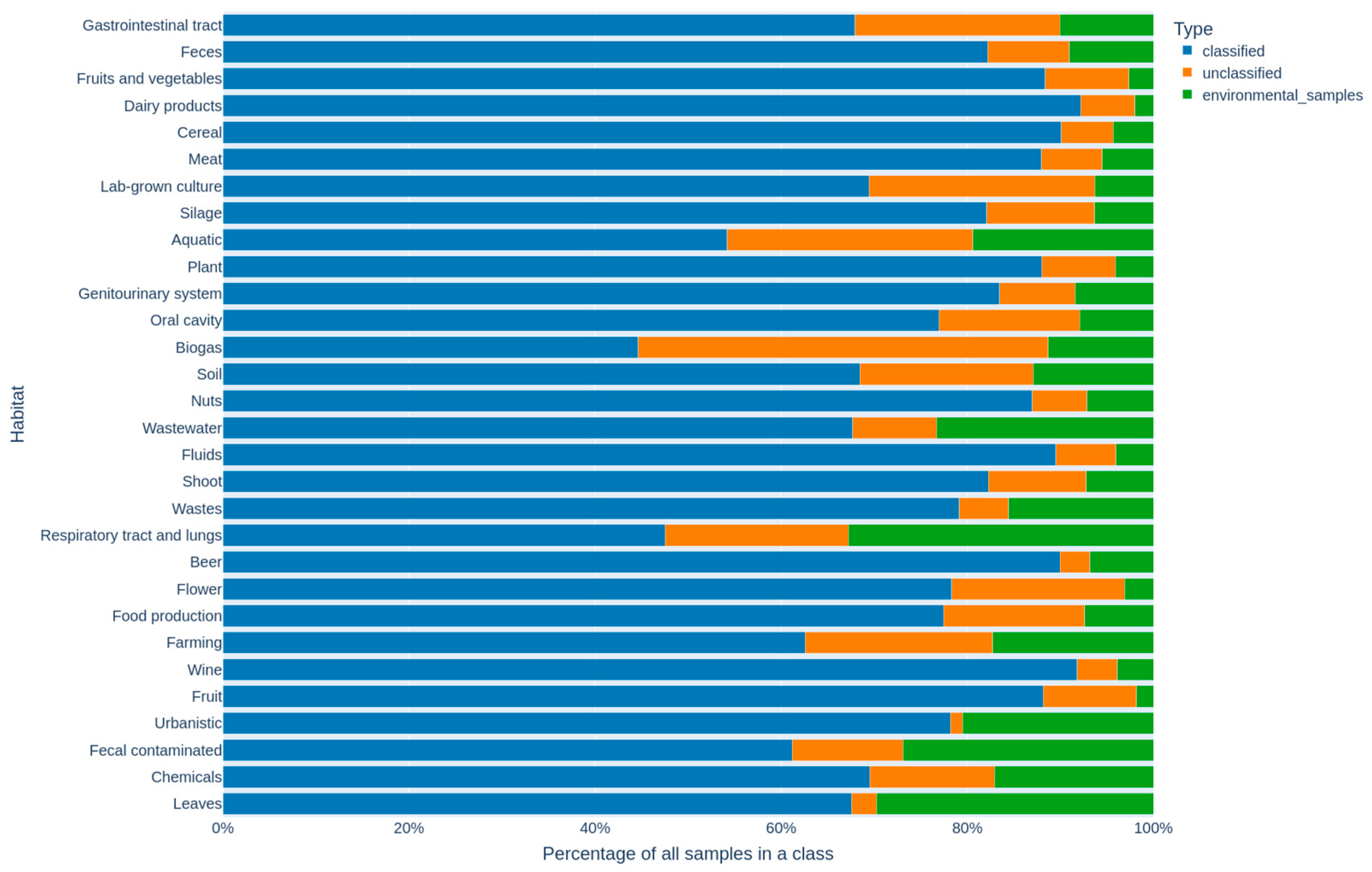
3.5. Analysis of Unclassified and Environmental Samples Taxids
4. Discussion
4.1. The Necessity of Data Standardization and Comprehensive Metadata for Ecological Analysis
4.2. Correlation Between Taxonomy and Habitat
4.3. Hotspots for Novel Taxa Discovery
4.4. Ecological Significance and Future Directions
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| NCBI | National Center for Biotechnology Information |
| LAB | Lactic acid bacteria |
| TSV | Tab-separated value |
| MIxS | Minimum Information about any (x) Sequence |
References
- Davray, D.; Deo, D.; Kulkarni, R. Plasmids encode niche-specific traits in Lactobacillaceae. Microb. Genom. 2021, 7, 472. [Google Scholar] [CrossRef]
- George, F.; Daniel, C.; Thomas, M.; Singer, E.; Guilbaud, A.; Tessier, F.J.; Revol-Junelles, A.-M.; Borges, F.; Foligné, B. Occurrence and Dynamism of Lactic Acid Bacteria in Distinct Ecological Niches: A Multifaceted Functional Health Perspective. Front. Microbiol. 2018, 9, 2899. [Google Scholar] [CrossRef] [PubMed]
- Akpoghelie, P.O.; Edo, G.I.; Ali, A.B.; Yousif, E.; Zainulabdeen, K.; Owheruo, J.O.; Isoje, E.F.; Igbuku, U.A.; Essaghah, A.E.A.; Makia, R.S.; et al. Lactic acid bacteria: Nature, characterization, mode of action, products and applications. Process Biochem. 2025, 152, 1–28. [Google Scholar] [CrossRef]
- Anumudu, C.K.; Miri, T.; Onyeaka, H. Multifunctional Applications of Lactic Acid Bacteria: Enhancing Safety, Quality, and Nutritional Value in Foods and Fermented Beverages. Foods 2024, 13, 3714. [Google Scholar] [CrossRef] [PubMed]
- Yadav, M.K.; Song, J.H.; Vasquez, R.; Lee, J.S.; Kim, I.H.; Kang, D.-K. Methods for Detection, Extraction, Purification, and Characterization of Exopolysaccharides of Lactic Acid Bacteria—A Systematic Review. Foods 2024, 13, 3687. [Google Scholar] [CrossRef]
- Zoghi, A.; Todorov, S.D.; Khosravi-Darani, K. Bio-detoxification of mycotoxin-contaminated feedstuffs: Using lactic acid bacteria and yeast. Appl. Environ. Biotechnol. 2024, 9, 62–75. [Google Scholar] [CrossRef]
- Štyriaková, D.; Hajnal-Jafari, T.; Žunić, V.; Šuba, J.; Prekopová, M.; Yetik, A.K.; Štyriaková, L. Influence of biostimulant applications on vegetative growth and yield of strawberry under full and reduced fertilization. DYSONA-Appl. Sci. 2026, 7, 41–49. [Google Scholar] [CrossRef]
- Ayed, L.; M’hir, S.; Nuzzolese, D.; Di Cagno, R.; Filannino, P. Harnessing the Health and Techno-Functional Potential of Lactic Acid Bacteria: A Comprehensive Review. Foods 2024, 13, 1538. [Google Scholar] [CrossRef]
- Novikova, V.A.; Bondarenko, K.D.; Sazonov, A.E.; Rozanov, A.S. The Effect of Probiotic Lactic Acid Bacteria on the Symptoms of Mental Disorders. Nanobiotechnol. Rep. 2024, 19, 645–666. [Google Scholar] [CrossRef]
- Garzone, S.; Charitos, I.A.; Mandorino, M.; Maggiore, M.E.; Capozzi, L.; Cakani, B.; Lopes, G.C.D.; Bocchio-Chiavetto, L.; Colella, M. Can We Modulate Our Second Brain and Its Metabolites to Change Our Mood? A Systematic Review on Efficacy, Mechanisms, and Future Directions of “Psychobiotics”. Int. J. Mol. Sci. 2025, 26, 1972. [Google Scholar] [CrossRef] [PubMed]
- Qiao, N.; Wittouck, S.; Mattarelli, P.; Zheng, J.; Lebeer, S.; Felis, G.E.; Gänzle, M.G. After the storm—Perspectives on the taxonomy of Lactobacillaceae. JDS Commun. 2022, 3, 222–227. [Google Scholar] [CrossRef]
- Walter, J.; O’Toole, P.W. Microbe Profile: The Lactobacillaceae. Microbiology 2023, 169, 1414. [Google Scholar] [CrossRef] [PubMed]
- Zheng, J.; Wittouck, S.; Salvetti, E.; Franz, C.M.A.P.; Harris, H.M.B.; Mattarelli, P.; O’Toole, P.W.; Pot, B.; Vandamme, P.; Walter, J.; et al. A taxonomic note on the genus Lactobacillus: Description of 23 novel genera, emended description of the genus Lactobacillus Beijerinck 1901, and union of Lactobacillaceae and Leuconostocaceae. Int. J. Syst. Evol. Microbiol. 2020, 70, 2782–2858. [Google Scholar] [CrossRef] [PubMed]
- Yu, A.O.; Leveau, J.H.J.; Marco, M.L. Abundance, Diversity and Plant—Specific Adaptations of Plant—Associated Lactic Acid Bacteria. Environ. Microbiol. Rep. 2020, 12, 16–29. [Google Scholar] [CrossRef]
- Cock, P.J.A.; Antao, T.; Chang, J.T.; Chapman, B.A.; Cox, C.J.; Dalke, A.; Friedberg, I.; Hamelryck, T.; Kauff, F.; Wilczynski, B.; et al. Biopython: Freely available Python tools for computational molecular biology and bioinformatics. Bioinformatics 2009, 25, 1422–1423. [Google Scholar] [CrossRef]
- Huerta-Cepas, J.; Serra, F.; Bork, P. ETE 3: Reconstruction, Analysis, and Visualization of Phylogenomic Data. Mol. Biol. Evol. 2016, 33, 1635–1638. [Google Scholar] [CrossRef]
- Data Apps for Production|Plotly. Available online: https://plotly.com/ (accessed on 24 September 2025).
- GeoPy’s Documentation. 2025. Available online: https://geopy.readthedocs.io/en/stable/ (accessed on 24 September 2025).
- Freire, M.O.d.L.; Neto, J.P.R.C.; Lemos, D.E.d.A.; de Albuquerque, T.M.R.; Garcia, E.F.; de Souza, E.L.; Alves, J.L.d.B. Limosilactobacillus fermentum Strains as Novel Probiotic Candidates to Promote Host Health Benefits and Development of Biotherapeutics: A Comprehensive Review. Probiotics Antimicrob. Proteins 2024, 16, 1483–1498. [Google Scholar] [CrossRef]
- Schneider, E.; Balasubramanian, R.; Ferri, A.; Cotter, P.D.; Clarke, G.; Cryan, J.F. Fibre & fermented foods: Differential effects on the microbiota-gut-brain axis. Proc. Nutr. Soc. 2024, 1–16. [Google Scholar] [CrossRef]
- Yilmaz, B.; Bangar, S.P.; Echegaray, N.; Suri, S.; Tomasevic, I.; Lorenzo, J.M.; Melekoglu, E.; Rocha, J.M.; Ozogul, F. The Impacts of Lactiplantibacillus plantarum on the Functional Properties of Fermented Foods: A Review of Current Knowledge. Microorganisms 2022, 10, 826. [Google Scholar] [CrossRef] [PubMed]
- Jiang, D.; Li, B.; Zheng, M.; Niu, D.; Zuo, S.; Xu, C. Effects of Pediococcus pentosaceus on fermentation, aerobic stability and microbial communities during ensiling and aerobic spoilage of total mixed ration silage containing alfalfa (Medicago sativa L.). Grassl. Sci. 2020, 66, 215–224. [Google Scholar] [CrossRef]
- Benkerroum, N.; Misbah, M.; Sandine, W.E.; Elaraki, A.T. Development and use of a selective medium for isolation of Leuconostoc spp. from vegetables and dairy products. Appl. Environ. Microbiol. 1993, 59, 607–609. [Google Scholar] [CrossRef]
- Stefanovic, E.; Kilcawley, K.; Rea, M.; Fitzgerald, G.; McAuliffe, O. Genetic, enzymatic and metabolite profiling of the Lactobacilluscasei group reveals strain biodiversity and potential applications for flavour diversification. J. Appl. Microbiol. 2017, 122, 1245–1261. [Google Scholar] [CrossRef]
- Moser, A.; Schafroth, K.; Meile, L.; Egger, L.; Badertscher, R.; Irmler, S. Population Dynamics of Lactobacillus helveticus in Swiss Gruyère-Type Cheese Manufactured With Natural Whey Cultures. Front. Microbiol. 2018, 9, 637. [Google Scholar] [CrossRef] [PubMed]
- Marasco, R.; Gazzillo, M.; Campolattano, N.; Sacco, M.; Muscariello, L. Isolation and Identification of Lactic Acid Bacteria from Natural Whey Cultures of Buffalo and Cow Milk. Foods 2022, 11, 233. [Google Scholar] [CrossRef] [PubMed]
- Özcan, E.; Selvi, S.S.; Nikerel, E.; Teusink, B.; Öner, E.T.; Çakır, T. A genome-scale metabolic network of the aroma bacterium Leuconostoc mesenteroides subsp. cremoris. Appl. Microbiol. Biotechnol. 2019, 103, 3153–3165. [Google Scholar] [CrossRef]
- Fusco, V.; Quero, G.M.; Cho, G.S.; Kabisch, J.; Meske, D.; Neve, H.; Bockelmann, W.; Franz, C.M.A.P. The genus Weissella: Taxonomy, ecology and biotechnological potential. Front. Microbiol. 2015, 6, 155. [Google Scholar] [CrossRef]
- He, Y.; Zhu, L.; Chen, J.; Tang, X.; Pan, M.; Yuan, W.; Wang, H. Efficacy of Probiotic Compounds in Relieving Constipation and Their Colonization in Gut Microbiota. Molecules 2022, 27, 666. [Google Scholar] [CrossRef]
- Zheng, N.; Guo, R.; Wang, J.; Zhou, W.; Ling, Z. Contribution of Lactobacillus iners to Vaginal Health and Diseases: A Systematic Review. Front. Cell. Infect. Microbiol. 2021, 11, 792787. [Google Scholar] [CrossRef]
- Vergalito, F.; Testa, B.; Cozzolino, A.; Letizia, F.; Succi, M.; Lombardi, S.J.; Tremonte, P.; Pannella, G.; Di Marco, R.; Sorrentino, E.; et al. Potential Application of Apilactobacillus kunkeei for Human Use: Evaluation of Probiotic and Functional Properties. Foods 2020, 9, 1535. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.-S.; Yanagida, F.; Shinohara, T. Isolation and identification of lactic acid bacteria from soil using an enrichment procedure. Lett. Appl. Microbiol. 2005, 40, 195–200. [Google Scholar] [CrossRef]
- Rabelo, C.H.S.; Basso, F.C.; Lara, E.C.; Jorge, L.G.O.; Härter, C.J.; Mesquita, L.G.; Silva, L.F.P.; Reis, R.A. Effects of Lactobacillus buchneri as a silage inoculant and as a probiotic on feed intake, apparent digestibility and ruminal fermentation and microbiology in wethers fed low-dry-matter whole-crop maize silage. Grass Forage Sci. 2017, 73, 67–77. [Google Scholar] [CrossRef]
- Chmielewski, R.A.N.; Frank, J.F. Biofilm Formation and Control in Food Processing Facilities. Compr. Rev. Food Sci. Food Saf. 2003, 2, 22–32. [Google Scholar] [CrossRef]
- Yilmaz, P.; Kottmann, R.; Field, D.; Knight, R.; Cole, J.R.; Amaral-Zettler, L.; Gilbert, J.A.; Karsch-Mizrachi, I.; Johnston, A.; Cochrane, G.; et al. Minimum information about a marker gene sequence (MIMARKS) and minimum information about any (x) sequence (MIxS) specifications. Nat. Biotechnol. 2011, 29, 415–420. [Google Scholar] [CrossRef] [PubMed]
- Sarton-Lohéac, G.; da Silva, C.G.N.; Mazel, F.; Baud, G.; de Bakker, V.; Das, S.; El Chazli, Y.; Ellegaard, K.; Garcia-Garcera, M.; Glover, N.; et al. Deep Divergence and Genomic Diversification of Gut Symbionts of Neotropical Stingless Bees. mBio 2023, 14, e0353822. [Google Scholar] [CrossRef] [PubMed]
- Duar, R.M.; Lin, X.B.; Zheng, J.; Martino, M.E.; Grenier, T.; Pérez-Muñoz, M.E.; Leulier, F.; Gänzle, M.; Walter, J. Lifestyles in transition: Evolution and natural history of the genus Lactobacillus. FEMS Microbiol. Rev. 2017, 41, S27–S48. [Google Scholar] [CrossRef] [PubMed]
- Sun, Z.; Harris, H.M.B.; McCann, A.; Guo, C.; Argimón, S.; Zhang, W.; Yang, X.; Jeffery, I.B.; Cooney, J.C.; Kagawa, T.F.; et al. Expanding the biotechnology potential of lactobacilli through comparative genomics of 213 strains and associated genera. Nat. Commun. 2015, 6, 8322. [Google Scholar] [CrossRef]





Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sokolova, T.S.; Namsaraev, Z.B.; Wolf, E.R.; Kulyashov, M.A.; Akberdin, I.R.; Sazonov, A.E. Mapping Global Biodiversity and Habitat Distribution of Lactobacillaceae Using NCBI Sequence Metadata. Diversity 2025, 17, 776. https://doi.org/10.3390/d17110776
Sokolova TS, Namsaraev ZB, Wolf ER, Kulyashov MA, Akberdin IR, Sazonov AE. Mapping Global Biodiversity and Habitat Distribution of Lactobacillaceae Using NCBI Sequence Metadata. Diversity. 2025; 17(11):776. https://doi.org/10.3390/d17110776
Chicago/Turabian StyleSokolova, Tatiana S., Zorigto B. Namsaraev, Ekaterina R. Wolf, Mikhail A. Kulyashov, Ilya R. Akberdin, and Aleksey E. Sazonov. 2025. "Mapping Global Biodiversity and Habitat Distribution of Lactobacillaceae Using NCBI Sequence Metadata" Diversity 17, no. 11: 776. https://doi.org/10.3390/d17110776
APA StyleSokolova, T. S., Namsaraev, Z. B., Wolf, E. R., Kulyashov, M. A., Akberdin, I. R., & Sazonov, A. E. (2025). Mapping Global Biodiversity and Habitat Distribution of Lactobacillaceae Using NCBI Sequence Metadata. Diversity, 17(11), 776. https://doi.org/10.3390/d17110776








