Non-Invasive Sampling for Population Genetics of Wild Terrestrial Mammals (2015–2025): A Systematic Review
Abstract
1. Introduction
2. Methodology
2.1. Literature Search
2.2. Inclusion and Exclusion Criteria
3. Invasive and Non-Invasive Genetic Sampling in Terrestrial Mammals: Techniques, Challenges, and Conservation Applications
4. Genetic Variability in the Order Carnivora
4.1. Main Species and Their Heterozygosity Values
4.2. Population Balance and Inbreeding Coefficients
5. Genetic Variability in the Order Artiodactyla
5.1. Main Species and Their Heterozygosity Values
5.2. Population Balance and Inbreeding Coefficients
| Species | Main Threats | Conservation Strategy | Genetic Sampling | Expected and Observed Heterocigosity | Reference |
|---|---|---|---|---|---|
| Ammotragus lervia | Illegal hunting | Conservation and translocation management units | Faeces, hair, bone and tissue | He = 0.49 Ho = 0.43 | [56] |
| Ammotragus lervia | Poaching and habitat loss | None | Blood and tissue | He = 0.48 Ho = 0.46 | [57] |
| Alces americanus americanus | Physical and anthropogenic barriers | Connectivity between towns | Hair and fabric | He = 0.32 Ho = 0.35 | [25] |
| Sus scrofa | Habitat reduction | Urban colonisation | Muscle and tissue | He = 0.59 Ho = 0.56 | [55] |
| Naemorhedus griseus | Consanguinity | Species in captivity for reintroduction | Blood | He = 0.45 Ho = 0.19 | [58] |
| Lama glama | Species utilisation | Species in captivity for reintroduction | Blood | He = 0.76 Ho = 0.71 | [59] |
| Camelus dromedarius | None | Research only | Blood | He = 0.72 Ho = 0.67 | [60] |
| Ozotoceros bezoarticus | Agriculture and urbanisation | Population genetic variability studies | Faeces | He = 0.75 Ho = 0.72 | [26] |
| Tragelaphus eurycerus ssp. isaaci | Human activities | Species in captivity for reintroduction | Faeces and tissue | He = 0.42 Ho = 0.71 | [61] |
| Odocoileus hemionus fuliginatus | Fragmentation of habitat and urbanisation | Natural protected areas and biological corridors | Faeces | He = 0.56 Ho = 0.56 | [27] |
| Cervus elaphus hanglu | Habitat reduction | Protected natural areas | Hair | He = 0.66 Ho = 0.44 | [28] |
6. Genetic Variability in the Orders Proboscidea, Primates and Rodentia
6.1. Main Species and Their Expected and Observed Heterozygosity Values
6.2. Population Balance and Inbreeding Coefficients
7. 2015–2025: Methodological Shifts and Best Practices in Non-Invasive Population Genomics
8. Future Orientations
9. Limitations of the Study
10. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Beedanagari, S.; Vulimiri, S.V.; Bhatia, S.; Mahadevan, B. Genotoxicity biomarkers: Molecular basis of genetic variability and susceptibility. In Biomarkers in Toxicology; Elsevier: Amsterdam, The Netherlands, 2014; ISBN 978-0-12-404630-6. [Google Scholar]
- Rivera-Ortiz, F.A.; Aguilar, R.; Arizmendi, M.D.; Quesada, M.; Oyama, K. Habitat fragmentation and genetic variability of tetrapod populations. Anim. Conserv. 2015, 18, 249–258. [Google Scholar] [CrossRef]
- Bolaji, U.-F.O.; Ajasa, A.A.; Ahmed, R.O.; Bello, S.F.; Ositanwosu, O.E. Cattle conservation in the 21st century: A mini review. Open J. Anim. Sci. 2021, 11, 304–332. [Google Scholar] [CrossRef]
- Salgotra, R.K.; Chauhan, B.S. Genetic diversity, conservation, and utilization of plant genetic resources. Genes 2023, 14, 174. [Google Scholar] [CrossRef]
- Zemanova, M.A. Towards more compassionate wildlife research through the 3Rs principles: Moving from invasive to non-invasive methods. Wildl. Biol. 2020, 1, wlb.00607. [Google Scholar] [CrossRef]
- Schilling, A.-K.; Mazzamuto, M.V.; Romeo, C. A review of non-invasive sampling in wildlife disease and health research: What’s new? Animals 2022, 12, 1719. [Google Scholar] [CrossRef] [PubMed]
- Nishikawa-Pacher, A. Research questions with PICO: A universal mnemonic. Publications 2022, 10, 21. [Google Scholar] [CrossRef]
- Page, M.J.; McKenzie, J.E.; Bossuyt, P.M.; Boutron, I.; Hoffmann, T.C.; Mulrow, C.D.; Shamseer, L.; Tetzlaff, J.M.; Akl, E.A.; Brennan, S.E.; et al. The PRISMA 2020 statement: An updated guideline for reporting systematic reviews. BMJ 2021, 372, n71. [Google Scholar] [CrossRef]
- Byrne, M.S.; Peralta, D.M.; Ibañez, E.A.; Nardelli, M.; Túnez, J.I. Non-invasive sampling techniques applied to conservation genetic studies in mammals. In Molecular Ecology and Conservation Genetics of Neotropical Mammals; Nardelli, M., Túnez, J.I., Eds.; Springer International Publishing: Cham, Switzerland, 2021; pp. 63–83. [Google Scholar] [CrossRef]
- Carroll, E.L.; Bruford, M.W.; DeWoody, J.A.; Leroy, G.; Strand, A.; Waits, L.; Wang, J. Genetic and genomic monitoring with minimally invasive sampling methods. Evol. Appl. 2018, 11, 1094–1119. [Google Scholar] [CrossRef] [PubMed]
- Von Thaden, A.; Nowak, C.; Tiesmeyer, A.; Reiners, T.E.; Alves, P.C.; Lyons, L.A.; Mattucci, F.; Randi, E.; Cragnolini, M.; Galián, J.; et al. Applying genomic data in wildlife monitoring: Development guidelines for genotyping degraded samples with reduced single nucleotide polymorphism panels. Mol. Ecol. Resour. 2020, 20, 662–680. [Google Scholar] [CrossRef]
- Robinson, J.J.; Crichlow, A.D.; Hacker, C.E.; Munkhtsog, B.; Zhang, Y.; Swanson, W.F.; Lyons, L.A.; Janecka, J.E. Genetic variation in the Pallas’s cat (Otocolobus manul) in zoo-managed and wild populations. Diversity 2024, 16, 228. [Google Scholar] [CrossRef]
- Barbora, G.; Belotti, E.; Bufka, L.; Volfová, J.; Wölfl, S.; Mináriková, T.; Hollerbach, L.; Duľa, M.; Kleven, O.; Kutal, M.; et al. Long-term genetic monitoring of a reintroduced Eurasian lynx population does not indicate an ongoing loss of genetic diversity. Glob. Ecol. Conserv. 2023, 42, e02399. [Google Scholar] [CrossRef]
- Wultsch, C.; Zeller, K.A.; Welfelt, L.S.; Beausoleil, R.A. Genetic diversity, gene flow, and source-sink dynamics of cougars in the Pacific Northwest. Conserv. Genet. 2023, 24, 793–806. [Google Scholar] [CrossRef]
- Buono, V.; Burgio, S.; Macrì, N.; Catania, G.; Hauffe, H.C.; Mucci, N.; Davoli, F. Microsatellite characterization and panel selection for brown bear (Ursus arctos) population assessment. Genes 2022, 13, 2164. [Google Scholar] [CrossRef]
- Balestrieri, A.; Gariano, P.; Grandinetti, M.; Verduci, F.; Gianfranceschi, L.; Gatti, E.; Mucci, C.; Mengoni, C.; Tremolada, P. Faecal DNA-based genetic survey of a relict Eurasian otter (Lutra lutra) population (Sila Massif, S Italy). Conserv. Genet. Res. 2022, 14, 453–461. [Google Scholar] [CrossRef]
- Korablev, M.P.; Poyarkov, A.D.; Karnaukhov, A.S.; Zvychaynaya, E.Y.; Kuksin, A.N.; Malykh, S.V.; Istomov, S.V.; Spitsyn, S.V.; Aleksandrov, D.Y.; Hernández-Blanco, J.A.; et al. Large-scale and fine-grain population structure and genetic diversity of snow leopards (Panthera uncia Schreber, 1776) from the northern and western parts of the range with an emphasis on the Russian population. Conserv. Genet. 2021, 22, 397–410. [Google Scholar] [CrossRef]
- Bou, N.; Soutullo, Á.; Hernández, D.; Mannise, N.; González, S.; Bartesaghi, L.; Pereira, J.; Merino, M.; Espinosa, C.; Trigo, T.C.; et al. Population structure and gene flow of Geoffroy’s cat (Leopardus geoffroyi) in the Uruguayan Savanna ecoregion. J. Mammal. 2021, 102, 879–890. [Google Scholar] [CrossRef]
- Van Tuijl, C.; van Bochove, K.; de Visser, M. Genetic structure of badger populations in a fragmented landscape: How do barriers affect populations on a genetic level? Lutra 2019, 62, 65–76. [Google Scholar]
- Menchaca, A.; Rossi, N.A.; Froidevaux, J.; Dias-Freedman, I.; Caragiulo, A.; Wultsch, C.; Harmsen, B.; Foster, R.; de la Torre, J.A.; Medellín, R.A.; et al. Population genetic structure and habitat connectivity for jaguar (Panthera onca) conservation in Central Belize. BMC Genet. 2019, 20, 100. [Google Scholar] [CrossRef] [PubMed]
- Kolipakam, V.; Singh, S.; Pant, B.; Qureshi, Q.; Jhala, Y.V. Genetic structure of tigers (Panthera tigris tigris) in India and its implications for conservation. Glob. Ecol. Conserv. 2019, 20, e00710. [Google Scholar] [CrossRef]
- Amaike, Y.; Murakami, T.; Masuda, R. Low genetic diversity in an isolated red fox (Vulpes vulpes) population on Mt. Hakodate, Japan, revealed by microsatellite analyses of fecal samples. Mammal Study 2018, 43, 141–152. [Google Scholar] [CrossRef]
- Mattucci, F.; Galaverni, M.; Pertoldi, C.; Fabbri, E.; Sliwa, A.; Caniglia, R. How to spot a black-footed cat? Successful application of cross-species markers to identify captive-bred individuals from non-invasive genetic sampling. Mammal Res. 2018, 64, 133–145. [Google Scholar] [CrossRef]
- Singh, S.K.; Aspi, J.; Kvist, L.; Sharma, R.; Pandey, P.; Mishra, S.; Singh, R.; Agrawal, M.; Goyal, S.P. Fine-scale population genetic structure of the Bengal tiger (Panthera tigris tigris) in a human-dominated western Terai Arc Landscape, India. PLoS ONE 2017, 12, e0174371. [Google Scholar] [CrossRef]
- Rosenblatt, E.; Gieder, K.; Donovan, T.; Murdoch, J.; Smith, T.P.L.; Heaton, M.P.; Kalbfleisch, T.S.; Murdoch, B.M.; Bhattarai, S.; Pacht, E.; et al. Genetic diversity and connectivity of moose (Alces americanus americanus) in eastern North America. Conserv. Genet. 2023, 24, 235–248. [Google Scholar] [CrossRef]
- Mantellatto, A.M.B.; Caparroz, R.; Christofoletti, M.D.; Piovezan, U.; Duarte, J.M.B. Genetic diversity of the pampas deer (Ozotoceros bezoarticus) population in the Brazilian Pantanal assessed by combining fresh fecal DNA analysis and a set of heterologous microsatellite loci. Genet. Mol. Biol. 2017, 40, 774–780. [Google Scholar] [CrossRef]
- Mitelberg, A.; Vandergast, A.G. Non-Invasive Genetic Sampling of Southern Mule Deer (Odocoileus hemionus fuliginatus) Reveals Limited Movement Across California State Route 67 in San Diego County. West. Wildl. 2016, 3, 8–18. [Google Scholar]
- Mukesh, K.V.P.; Sharma, L.K.; Shukla, M.; Sathyakumar, S. Pragmatic Perspective on Conservation Genetics and Demographic History of the Last Surviving Population of Kashmir Red Deer (Cervus elaphus hanglu) in India. PLoS ONE 2015, 10, e0117069. [Google Scholar] [CrossRef]
- Budd, K.; Sampson, C.; Leimgruber, P.; Tonkyn, D.; Storey, K.; Garrett, M.; Eggert, L.S. Population structure and demography of Myanmar’s conflict elephants. Glob. Ecol. Conserv. 2021, 31, e01828. [Google Scholar] [CrossRef]
- Oklander, L.I.; Miño, C.I.; Fernández, G.; Caputo, M.; Corach, D. Genetic structure in the southernmost populations of black-and-gold howler monkeys (Alouatta caraya) and its conservation implications. PLoS ONE 2017, 12, e0185867. [Google Scholar] [CrossRef]
- DeMarco, C.; Cooper, D.S.; Torres, E.; Muchlinski, A.; Aguilar, A. Effects of urbanization on population genetic structure of western gray squirrels. Conserv. Genet. 2021, 22, 67–81. [Google Scholar] [CrossRef]
- Byrne, M.S.; Quintana, R.D.; Bolkovic, M.L.; Túnez, J.I. Population genetics of the capybara, Hydrochoerus hydrochaeris, in the Chaco-pampean region. Mamm. Biol. 2019, 96, 14–22. [Google Scholar] [CrossRef]
- de Flamingh, A.; Ishida, Y.; Pečnerová, P.; Vilchis, S.; Siegismund, H.R.; van Aarde, R.J.; Malhi, R.S.; Roca, A.L. Combining methods for non-invasive fecal DNA enables whole genome and metagenomic analyses in wildlife biology. Front. Genet. 2023, 13, 1021004. [Google Scholar] [CrossRef]
- Cueva, D.F.; Gutierrez, B.; Bruque, G.; Molina, S.; Torres, M.L. Mitochondrial DNA reveals low genetic diversity in Ecuadorian Andean bears. Ursus 2018, 29, 43–50. [Google Scholar] [CrossRef]
- De Barba, M.; Miquel, C.; Lobréaux, S.; Quenette, P.Y.; Swenson, J.E.; Taberlet, P. High-throughput microsatellite genotyping in ecology: Improved accuracy, efficiency, standardization and success with low-quantity and degraded DNA. Mol. Ecol. Resour. 2017, 17, 492–507. [Google Scholar] [CrossRef] [PubMed]
- Beng, K.C.; Corlett, R.T. Applications of environmental DNA (eDNA) in ecology and conservation: Opportunities, challenges and prospects. Biodivers. Conserv. 2020, 29, 2089–2121. [Google Scholar] [CrossRef]
- Banerjee, P.; Stewart, K.A.; Antognazza, C.M.; Bunholi, I.V.; Deiner, K.; Barnes, M.A.; Saha, S.; Verdier, H.; Doi, H.; Maity, J.P.; et al. Plant–animal interactions in the era of environmental DNA (eDNA)—A review. Environ. DNA 2022, 4, 987–999. [Google Scholar] [CrossRef]
- Gutiérrez, E.E.; Helgen, K.M.; McDonough, M.M.; Bauer, F.; Hawkins, M.T.R.; Escobedo-Morales, L.A.; Patterson, B.D.; Maldonado, J.E. A gene-tree test of the traditional taxonomy of American deer: The importance of voucher specimens, geographic data, and dense sampling. ZooKeys 2017, 697, 87–131. [Google Scholar] [CrossRef]
- Asier, R.L.; Guitián, P. Intraspecific variation in fruit size and shape in Corema album (Ericaceae) along a latitudinal gradient: From fruits to populations. Biol. J. Linn. Soc. 2016, 118, 940–950. [Google Scholar] [CrossRef]
- Bidon, T.; Randi, E.; Hailer, F.; Frosch, C.; Hofreiter, M. Phylogenetic and phylogeographic analyses of the brown bear (Ursus arctos) in Europe and Asia. Mol. Ecol. 2018, 27, 1–14. [Google Scholar] [CrossRef]
- Wang, J. Estimating genotyping errors from genotype and reconstructed pedigree data. Methods Ecol. Evol. 2018, 9, 109–120. [Google Scholar] [CrossRef]
- Groen, K.; Trimbos, K.B.; Hein, S.; Blaauw, A.I.; van Bodegom, P.M.; Hahne, J.; Jacob, J. Establishment of a faecal DNA quantification technique for rare and cryptic diet constituents in small mammals. Mol. Ecol. Resour. 2022, 22, 2220–2231. [Google Scholar] [CrossRef]
- Andrews, K.R.; De Barba, M.; Russello, M.A.; Waits, L.P. Advances in Using Non-invasive, Archival, and Environmental Samples for Population Genomic Studies. In Population Genomics: Wildlife; Hohenlohe, P.A., Rajora, O.P., Eds.; Springer: Cham, Switzerland, 2018; pp. 63–99. [Google Scholar] [CrossRef]
- Kamra, D.N.; Singh, B. Rumen Microbiome and Plant Secondary Metabolites (PSM): Inhibition of Methanogenesis and Improving Nutrient Utilization. In Advancing Frontiers in Mycology & Mycotechnology; Satyanarayana, T., Deshmukh, S., Deshpande, M., Eds.; Springer: Singapore, 2019; pp. 325–345. [Google Scholar] [CrossRef]
- Lear, G.; Dickie, I.; Banks, J.; Boyer, S.; Buckley, H.L.; Buckley, T.R.; Cruickshank, R.; Dopheide, A.; Handley, K.M.; Hermans, S.; et al. Methods for the Extraction, Storage, Amplification and Sequencing of DNA from Environmental Samples. N. Zealand J. Ecol. 2018, 42, 10. [Google Scholar] [CrossRef]
- Zhang, Y.; Chi, X.; Feng, L.; Wu, X.; Qi, X. Improvement of Multiplex Semi-Nested PCR System for Screening of Rare Mutations by High-Throughput Sequencing. BioTechniques 2019, 67, 294–298. [Google Scholar] [CrossRef] [PubMed]
- Salcido, C.J.; Polly, P.D. The relationship between form and function of the carnivore mandible. Anat. Rec. 2025, 308, 1–20. [Google Scholar] [CrossRef]
- Meeus, M.P.; Lescroart, J.; Svardal, H. Genomic diversity in felids correlates with range and density, not census size. Conserv. Genet. 2025, 26, 873–890. [Google Scholar] [CrossRef]
- Azizan, A.; Anile, S.; Nielsen, C.K.; Paradis, E.; Devillard, S. Population density and genetic diversity are positively correlated in wild felids globally. Glob. Ecol. Biogeogr. 2023, 32, 1858–1869. [Google Scholar] [CrossRef]
- Willi, Y.; Sgrò, C.M.; Weeks, A.R.; Ørsted, M.; Hoffmann, A.A. Conservation genetics as a management tool: The five best-supported paradigms to assist the management of threatened species. Proc. Natl. Acad. Sci. USA 2022, 119, e2105076119. [Google Scholar] [CrossRef] [PubMed]
- Morandin, C.; Loveridge, A.J.; Segelbacher, G.; Elliot, N.; Madzikanda, H.; Macdonald, D.W.; Höglund, J. Gene flow and immigration: Genetic diversity and population structure of lions (Panthera leo) in Hwange National Park, Zimbabwe. Conserv. Genet. 2014, 15, 697–706. [Google Scholar] [CrossRef]
- Orliac, M.J.; Maugoust, J.; Balcarcel, A.; Gilissen, E. Paleoneurology of Artiodactyla, an Overview of the Evolution of the Artiodactyl Brain. In Paleoneurology of Amniotes; Dozo, M.T., Paulina-Carabajal, A., Macrini, T.E., Walsh, S., Eds.; Springer: Cham, Switzerland, 2023; pp. 507–555. [Google Scholar]
- Jeon, J.; Black, A.; Heenkenda, E.; Mularo, A.; Lamka, G.; Janjua, S.; Brüniche-Olsen, A.; Bickham, J.; Willoughby, J.; DeWoody, J. Genomic Diversity as a Key Conservation Criterion: Proof-of-Concept From Mammalian Whole-Genome Resequencing Data. Evol. Appl. 2024, 17, e70000. [Google Scholar] [CrossRef]
- Hoban, S.; Archer, F.I.; Bertola, L.D.; Bragg, J.G.; Breed, M.F.; Bruford, M.W.; Coleman, M.A.; Ekblom, R.; Funk, W.C.; Grueber, C.E.; et al. Global genetic diversity status and trends: Towards a suite of Essential Biodiversity Variables (EBVs) for genetic composition. Biol. Rev. 2022, 97, 1511–1538. [Google Scholar] [CrossRef]
- Hagemann, J.; Conejero, C.; Stillfried, M.; Mentaberre, G.; Castillo-Contreras, R.; Fickel, J.; López-Olvera, J.R. Genetic population structure defines wild boar as an urban exploiter species in Barcelona, Spain. Sci. Total Environ. 2022, 833, 155126. [Google Scholar] [CrossRef]
- Pizzigalli, C.; Silva, T.L.; Abáigar, T.; Bertorelle, G.; Cassinello, J.; Brito, J.C. Assessment of population structure and genetic diversity of wild and captive populations of Ammotragus lervia provide insights for conservation management. Conserv. Genet. 2024, 25, 59–73. [Google Scholar] [CrossRef]
- Stipoljev, S.; Safner, T.; Gančević, P.; Galov, A.; Stuhne, T.; Svetličić, I.; Grignolio, S.; Cassinello, J.; Šprem, N. Population structure and genetic diversity of non-native aoudad populations. Sci. Rep. 2021, 11, 12300. [Google Scholar] [CrossRef]
- Jangtarwan, K.; Kamsongkram, P.; Subpayakom, N.; Sillapaprayoon, S.; Muangmai, N.; Kongphoemph, A.; Wongsodchuen, A.; Intapan, S.; Chamchumroon, W.; Safoowong, M.; et al. Predictive genetic plan for a captive population of the Chinese goral (Naemorhedus griseus) and prescriptive action for ex situ and in situ conservation management in Thailand. PLoS ONE 2020, 15, e0234064. [Google Scholar] [CrossRef] [PubMed]
- Paredes, G.F.; Yalta-Macedo, C.E.; Gutierrez, G.A.; Veli-Rivera, E.A. Genetic Diversity and Population Structure of Llamas (Lama glama) from the Camelid Germplasm Bank—Quimsachata. Genes 2020, 11, 541. [Google Scholar] [CrossRef] [PubMed]
- Piro, M.; Mabsoute, F.E.; El Khattaby, N.; Laghouaouta, H.; Boujenane, I. Genetic variability of dromedary camel populations based on microsatellite markers. Animal 2020, 14, 2452–2462. [Google Scholar] [CrossRef]
- Svengren, H.; Prettejohn, M.; Bunge, D.; Fundi, P.; Björklund, M. Relatedness and genetic variation in wild and captive populations of Mountain Bongo in Kenya obtained from genome-wide single-nucleotide polymorphism (SNP) data. Glob. Ecol. Conserv. 2017, 11, 196–206. [Google Scholar] [CrossRef]
- Kratzer, K. The Genomic History of Elephants Supports the Existence of a Third Modern Species and has Important Implications for Conservation. Proc. Man. Undergrad. Sci. Eng. Res. 2018, 4, 87–89. [Google Scholar]
- McGraw, W.S. Primates Defined. In A Companion to Biological Anthropology; Wiley: Hoboken, NJ, USA, 2023; pp. 277–299. [Google Scholar] [CrossRef]
- D’Elía, G.; Pierre-Henri, F.; Lessa, E.P. Rodent systematics in an age of discovery: Recent advances and prospects. J. Mammal. 2019, 100, 852–871. [Google Scholar] [CrossRef]
- Fernandez-Fournier, P.; Lewthwaite, J.M.M.; Mooers, A.Ø. Do we need to identify adaptive genetic variation when prioritizing populations for conservation? Conserv. Genet. 2021, 22, 205–216. [Google Scholar] [CrossRef]
- Maurer, G.; Dubois, M.P.; Oo, Z.M.; Chanthavong, V.; Mulot, B.; Gimenez, O.; Kjellberg, F. Genetic structure and diversity of semi-captive populations: The anomalous case of the Asian elephant. Conserv. Genet. 2024, 25, 973–984. [Google Scholar] [CrossRef]


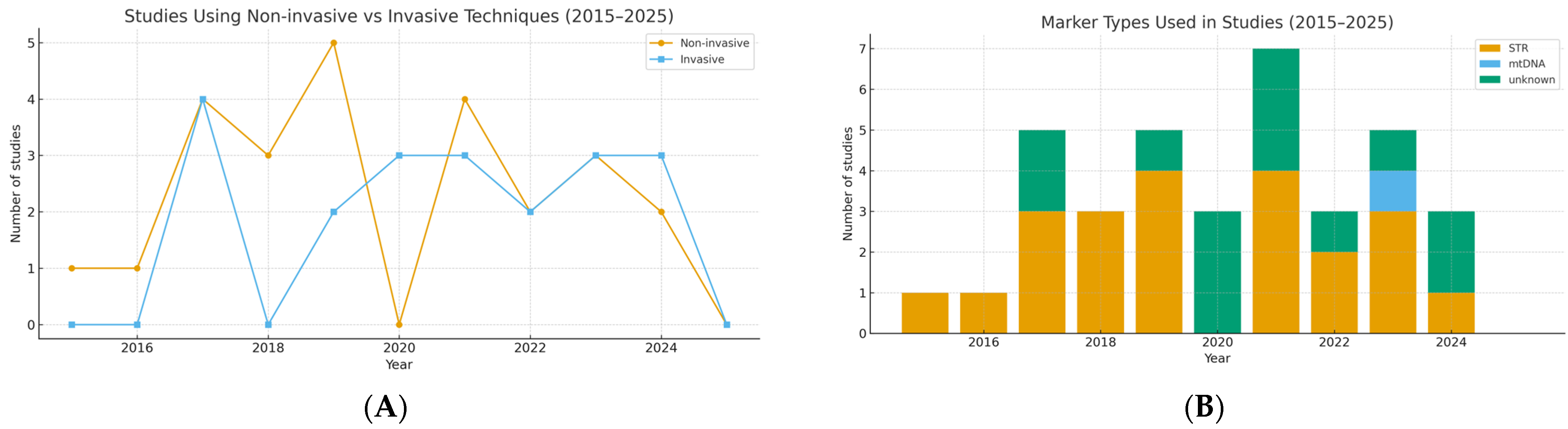
| Species | N | n of Non-Invasive Samples | n of Invasive Samples | Type of Marker | Reference |
|---|---|---|---|---|---|
| Otocolobus manul | 16 | 6 | 10 | STR,12S ribosomal | [12] |
| Lynx lynx | 6 | 5 | 1 | STR | [13] |
| Puma concolor | 3355 | 0 | 3355 | STR | [14] |
| Ursus arctos | 44 | 27 | 17 | STR | [15] |
| Lutra lutra | 47 | 47 | 0 | STR, mitochondrial cytochrome b gene | [16] |
| Panthera uncia | 213 | 213 | 0 | STR, mitochondrial cytochrome b gene | [17] |
| Leopardus geoffroyi | 172 | 163 | 9 | STR | [18] |
| Mellivora capensis | 194 | 113 | 81 | STR | [19] |
| Panthera onca | 536 | 536 | 0 | STR | [20] |
| Panthera tigris | 718 | 718 | 0 | STR | [21] |
| Vulpes vulpes | 150 | 150 | 0 | STR | [22] |
| Felis nigripes | 23 | 23 | 0 | STR, mithochondrial DNA ND5 | [23] |
| Panthera tigris tigris | 71 | 12 | 59 | STR | [24] |
| Alces alces | 1058 | 529 | 529 | STR | [25] |
| Ozotoceros bezoarticus | 155 | 142 | 13 | STR, mithochondrial DNA | [26] |
| Odocoileus hemionus fuliginatus | 238 | 238 | 0 | STR | [27] |
| Cervus hanglu hanglu | 160 | 160 | 0 | STR, mitochondrial D-loop | [28] |
| Elephas maximus | 252 | 252 | 0 | STR, mitochondrial DNA | [29] |
| Alouatta caraya | 138 | 132 | 6 | STR, mitochondrial DNA | [30] |
| Sciurus griseus | 117 | 114 | 3 | STR, mitochondrial DNA D-loop | [31] |
| Hydrochoerus hydrochaeris | 54 | 27 | 27 | STR, mitochondrial DNA | [32] |
| Loxodonta africana | 37 | 37 | 0 | Mitochondrial genomes | [33] |
| Tremarctos ornatus | 38 | 38 | 0 | STR, mitochondrial D-loop | [34] |
| Species | Main Threats | Conservation Strategy | Type of Sample | Expected and Observed Heterocigosity * | Reference |
|---|---|---|---|---|---|
| Otocolobus manul | Habitat degradation and climate change | Poaching mitigation, creation of NPAs and Captive Areas | Faeces and blood. | He = 0.62 Ho = 0.57 | [12] |
| Lynx lynx | Poaching and poor habitat quality | Reintroduction | Faeces, blood and tissue | - | [13] |
| Puma concolor | Urbanisation and habitat loss and fragmentation | Protected natural areas | Skin and muscle | He = 0.59 Ho = 0.52 | [14] |
| Ursus arctos arctos | Human activities | Reintroduction | Blood, tissue, excreta and hair | He = 0.61 Ho = 0.64 | [15] |
| Ursus arctos marsicanus | Human activities | Reintroduction | Blood, tissue, excreta and hair | He = 0.40 Ho = 0.39 | [15] |
| Lutra lutra | Habitat fragmentation | Reintroduction | Faeces | Ho = 0.37 | [16] |
| Panthera uncia | Habitat fragmentation | Translocation | Faeces | He = 0.57 Ho = 0.54 | [17] |
| Leopardus geoffroyi | Habitat fragmentation and land use change | Biological corridors | Tissue and blood | He = 0.77 Ho = 0.70 | [18] |
| Meles meles | Urbanisation | Biological corridors | Fresh, dead tissue and faeces | He = 0.45 Ho = 0.42 | [19] |
| Panthera onca | Habitat fragmentation | Biological corridors and protected areas | Faeces | He = 0.61 Ho = 0.55 | [20] |
| Panthera tigris tigris | Poaching and habitat loss | Biological corridors | Faeces | He = 0.77 Ho = 0.68 | [21] |
| Vulpes vulpes | Urbanisation and population isolation | - | Faeces | He = 0.55 Ho = 0.52 | [22] |
| Felis nigripes | Overgrazing and extensive agriculture | Captive areas | Hair | He = 0.62 Ho = 0.68 | [23] |
| Panthera tigris tigris | Poaching and habitat loss | Biological corridors | Blood and excreta | He = 0.64 Ho = 0.50 | [24] |
| Species | Main Threats | Conservation Strategy | Genetic Sampling | Expected and Observed Heterocigosity | Reference |
|---|---|---|---|---|---|
| Elephas maximus | Habitat fragmentation and reduced gene flow | Semi-captive species | Blood | He = 0.64 Ho = 0.63 | [66] |
| Elephas maximus | Illegal hunting | Semi-captive species | Faeces | He = 0.67 Ho = 0.57 | [29] |
| Alouatta caraya | Deforestation, agriculture and livestock | Reintroduction and management units | Faeces and tissue | He = 0.44 Ho = 0.46 | [30] |
| Sciurus griseus * | Habitat loss and fragmentation due to urbanization | - | Hair | He = 0.49 Ho = 0.45 | [31] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ramírez-García, J.G.; Maciel-Torres, S.P.; Hernández-Rodríguez, M.; Arenas-Báez, P.; Orzuna-Orzuna, J.F.; Granados-Rivera, L.D. Non-Invasive Sampling for Population Genetics of Wild Terrestrial Mammals (2015–2025): A Systematic Review. Diversity 2025, 17, 760. https://doi.org/10.3390/d17110760
Ramírez-García JG, Maciel-Torres SP, Hernández-Rodríguez M, Arenas-Báez P, Orzuna-Orzuna JF, Granados-Rivera LD. Non-Invasive Sampling for Population Genetics of Wild Terrestrial Mammals (2015–2025): A Systematic Review. Diversity. 2025; 17(11):760. https://doi.org/10.3390/d17110760
Chicago/Turabian StyleRamírez-García, Jesús Gabriel, Sandra Patricia Maciel-Torres, Martha Hernández-Rodríguez, Pablo Arenas-Báez, José Felipe Orzuna-Orzuna, and Lorenzo Danilo Granados-Rivera. 2025. "Non-Invasive Sampling for Population Genetics of Wild Terrestrial Mammals (2015–2025): A Systematic Review" Diversity 17, no. 11: 760. https://doi.org/10.3390/d17110760
APA StyleRamírez-García, J. G., Maciel-Torres, S. P., Hernández-Rodríguez, M., Arenas-Báez, P., Orzuna-Orzuna, J. F., & Granados-Rivera, L. D. (2025). Non-Invasive Sampling for Population Genetics of Wild Terrestrial Mammals (2015–2025): A Systematic Review. Diversity, 17(11), 760. https://doi.org/10.3390/d17110760







