Abstract
The assumption of neutrality for mitochondrial DNA has been widely questioned due to the functional importance of the encoded proteins. Mitochondrial genes involved in the oxidative phosphorylation (OXPHOS) pathway can be subjected to selection due to changes in some environmental conditions or in the living habits of specimens in order to adapt to their environment. In this study, We search for evidence of positive selection in COI, Cytb, and ND2 mitochondrial OXPHOS genes in 100 specimens of Xiphias gladius belonging to Atlantic, Indian, and Mediterranean stocks through codon models for inference of site-specific positive selection. For comparison, the same analysis was conducted on sequences of the same genes belonging to the closely related species Istiophorus platypterus, which are present in GenBank. The ND2 sequence analysis identified a non-synonymous transversion (T to A) at the nucleotide position 293, resulting in a codon change in the Greek population of X. gladius. Selection tests showed diversifying positive selection for only the ND2 dataset in both species, at 324 codon positions by MEME and FUBAR in X. gladius and only by the FUBAR test at 19 codon positions in I. platypterus.
1. Introduction
Migratory fish species that undertake long-distance movements and exhibit high energy consumption, necessary to sustain the metabolic capacities involved in crossing highly variable environments [1,2]. This high energy expenditure, resulting both from maintaining stable body temperature and from their prolonged migratory behavior, is directly related to intensified mitochondrial activity [3].
Mitochondria, often described as the cell’s “powerhouses”, are responsible for more than 95% of total ATP (adenosine triphosphate) production through oxidative phosphorylation (OXPHOS), carried out by multimeric enzyme complexes [4]. These complexes depend on the coordinated action of proteins encoded by both 13 genes present in the mitochondrial genome and nuclear genes [5]. Among the five complexes involved in OXPHOS, only the components of complex II are exclusively encoded by nuclear genes. Complex I (NADH ubiquinone reductase) is the largest complex and transfers electrons from NADH to coenzyme Q; complex III (quinol-cytochrome c reductase) mediates the transfer of electrons to cytochrome c; complex IV (cytochrome c oxidase) controls the oxidation of cytochrome c and the reduction of oxygen to water; and finally, complex V (ATP synthase) synthesizes ATP using the energy generated by the proton flow across the mitochondrial membrane promoted by the previous complexes [6].
These OXPHOS system components and their functions are highly conserved across all organisms, as they constitute the primary energy source for cells, but at the same time, these components may be subject to selective pressures to meet the specific energetic adaptive demands of organisms [3,7,8]. For this reason, although mitochondrial DNA was long considered to have evolved under neutral selection, more recent evidence shows that it is not modified solely by random genetic drift [9]. Evidence of positive selection in mitochondrial DNA has been detected, for example, in fish that have increased their capacity to adapt to changes in various environmental parameters or changes in their lifestyle. Genes such as ND5, ND6, and ATP6 show signatures of positive selection in regionally endothermic teleosts, such as tunas, needlefish, and butterfly mackerels, likely to optimize ATP production and increase heat production and metabolic efficiency of these organisms [10].
Similar studies on the opah (Lampris guttatus), a whole-body endothermic fish that inhabits deep marine environments, revealed that the ATP8 and COXIII genes had undergone positive selection. Specifically, amino acid substitutions in these genes, which may be associated with enhanced metabolic performance [11], have likely evolved in tandem with endothermy [12]. In this context, the billfish species such as the swordfish, Xiphias gladius Linnaeus 1758 and the sailfish Istiophorus platypterus (Shaw and Nodder 1792), stand out. Swordfish is a large pelagic and migratory fish widely distributed in open waters of tropical and temperate regions, found in all oceans, as well as in the Mediterranean, Marmara, Black, and Azov seas [13,14]. Sailfish, is a highly migratory, circumtropical epipelagic species mainly distributed in subtropical and tropical oceanic waters; it is considered occasional in the Mediterranean Sea [15]. Both species are classified as regionally endothermic fish because they have highly specialized extraocular muscles that act as a thermogenic organ, warming the brain and eyes [16]. Swordfish are of great commercial value especially in the Mediterranean Sea, where catch levels are relatively high, although this region represents less than 10% of the species’ global distribution area [17]. The sailfish is currently the most commonly caught species of the Istiophoridae family in its distribution area. The swordfish’s migratory behavior is complex and multidirectional [13]. Seasonally, adult swordfish migrate to spawning areas for reproductive purposes, but they also exhibit a daily pattern of vertical migration, staying in deep waters during the day (between 300 and 600 m) and rising at night to shallower layers to feed [18,19]. These migratory behaviors require high energy expenditure and expose individuals to significant thermal variations, representing an important physiological challenge, especially in terms of energy metabolism and thermoregulation.
Based on all the above considerations, this study examines the sequences of the Cytochrome Oxidase I (COI), Cytochrome b (Cytb), and NADH dehydrogenase 2 (ND2) genes from several populations of Xiphias gladius sampled in the Mediterranean Sea, Atlantic, and Indian Oceans and, for comparison, the sequences retrieved from GenBank of the same genes belonging to the closely related species Istiophorus platypterus, were also examined. The objective is to determine whether these three mitochondrial genes, encoding subunits involved in the OXPHOS pathway, show signs of positive selection, which could be interpreted as molecular adaptations related to energy metabolism and regional endothermy.
2. Materials and Methods
Sampling sites
A total of 100 certified origin samples of swordfish were obtained from import/export fishery companies and directly from fishermen. Swordfish samples came from the Atlantic (N = 25), Indian (N = 20), and Mediterranean area (N = 55; 15 of which from Spain, 10 from Greece, and 30 not specified).
DNA Extraction
Total genomic DNA was extracted from muscle tissue (25–30 mg) and fin clips using the DNeasy Blood and Tissue Kit (QIAGEN, Hilden, Germany) following the manufacturer’s instructions. The quality of the extracted DNA was evaluated on 0.8% agarose gel and then the isolated DNA was kept in storage at −20 °C for further studies.
Amplification and Sequencing
Three different primers pairs were used to amplify three mitochondrial genes belonging to different complexes of the OXPHOS chain: a portion of 1002 bp for ND2 (NADH: ubiquinone oxidoreductase core subunit 2) gene for respiratory complex I, 801 bp for Cytb (cytochrome b) for respiratory complex III gene, and a portion of 681 bp for COI (Cytochrome c Oxidase I) gene for respiratory complex IV (Table 1).

Table 1.
Primers used in this study to amplify the three target genes.
PCR amplifications were performed in 50 µL of the total reaction mixtures using Platinum Taq DNA Polymerase (Thermofisher Scientific, Walthman, MA, USA) and using different thermal profiles. In particular, for COI amplification, the thermal profile consisted of an initial denaturation at 94 °C for 2 min, followed by denaturation at 94 °C (30 s), annealing at 52 °C (40 s), extension at 72 °C (1 min) repeated for 35 cycles, and a final extension at 72 °C for 10 min; for Cytb amplification, the steps were an initial denaturation at 95 °C for 7 min, followed by denaturation at 94 °C (30 s), annealing at 55 °C (35 s), extension at 72 °C (45 s) repeated for 38 cycles, and a final extension at 72 °C for 7 min; for ND2 amplification the thermal profile had an initial denaturation at 95 °C for 4 min, followed by denaturation at 95 °C (30 s), annealing at 55 °C (30 s), extension at 72 °C (1 min) repeated for 30 cycles, and a final extension at 72 °C for 5 min. Every PCR run included negative controls to ensure there was no cross-contamination. In order to check the obtained double-stranded products, 0.8% agarose gel electrophoresis was conducted, and the amplicons were visualized through a Safe Imager TM 2.0 Blue Light Transilluminator (Thermo Fisher, Waltham, MA, USA). All amplicons were purified using the QIAquick PCR purification kit (Qiagen, Hilden, Germany) and were then bidirectionally sequenced using an ABI 3730 automated sequencing machine at Eurofins Genomics Italy (https://eurofinsgenomics.eu, accessed on 3 September 2023), and, in particular, only for the COI gene were the M13 sequencing primers were used.
Data analysis
The sequence chromatograms were checked visually and assembled. Multiple-sequence alignment was carried out using the online version of MAFFT v.7 [24]. Ambiguous sequences were trimmed and primer sequences were cut. Finally, the sequences were submitted to the GenBank database. Phylogenetic relationships were inferred through maximum likelihood (ML) criterion and carried out in RAxML-NG v.1.2.0. [25]. The multisequence alignments from the three mitochondrial genes (COI: 669 bp; Cytb: 801 bp; and ND2: 1002 bp, respectively) were concatenated into a single 2472 bp sequence dataset. The nucleotide substitution model was set to a General Time Reversible (GTR) substitution model with a Gamma distribution (G) to account for rate heterogeneity across sites, as suggested by the JModelTest2 [26]. The initial tree search comprised 25 trees based on parsimony and 25 random starting trees. This approach aimed to comprehensively sample tree space and, therefore, increase the likelihood of identifying the global maximum likelihood tree. To assess the support for individual clades, we conducted nonparametric bootstrap analyses with 10,000 replicates. The analysis was conducted with a random number of seeds set to 12,345 to ensure the reproducibility of the results.
dN/dS ratios-based selection tests
Selection tests were performed on the X. gladius COI, Cytb, and ND2 datasets, and for comparison on a dataset of 63 sequences of each gene from the closely related species I. platypterus, retrieved from GenBank (accession numbers AB470306, KU315124, OP404092–OP404151, and NC012676).
To test the presence of mitochondrial recombinants for each sequenced portion, we used Genetic Algorithms for Recombination Detection (GARD) (HyPhy package, accessed on 28 February 2025 at www.datamonkey.org). To test the presence of selection on targeted mitochondrial genes, a preliminary one-tailed Z test [27] was performed in MEGA X [28]. Furthermore, three codon models were used to estimate codons under positive or purifying selections: FEL (Fixed Effects Likelihood) [29], FUBAR (Fast, Unconstrained Bayesian Approximation for Inferring Selection) [30], and MEME (Mixed effects model of evolution) [31].
Sites with p-values below 0.05 for FEL and MEME, as well as sites with a posterior probability higher than 0.9 for FUBAR, were all considered as being under selection [32].
To visualize the position of positively selected sites in a 3-dimensional space, we used the SWISS-MODEL server (https://swissmodel.expasy.org/, accessed on 17 March 2024) with default parameters [33]. The spatial orientation of the molecules was assessed by submitting the downloaded pdb files into the OPM (Orientation of Proteins in Membranes) database [34]. SIFT [35] was used to predict whether or not amino acid substitution (AAS) was deleterious or tolerable and PyMol [36] was used to assess any possible change in the structure of the ND2 proteins of both species resulting from the AAS.
3. Results
Xiphias gladius OXPHOS gene sequence analysis
Unambiguous alignments were obtained for 669 bp of COI, 801 bp of Cytb, and 1002 bp of ND2 from 100 X. gladius tissue samples. All sequences are available in GenBank (accession numbers in Tables S1–S3). No insertions, deletions, or stop codons were observed. The absence of stop codons indicates that all amplified sequences are functional mitochondrial sequences, as they were all the same length. This suggests that NUMTs (nuclear DNA sequences originating from mitochondrial DNA sequences) were not sequenced, as vertebrate NUMTs are generally smaller than 600 bp. The single-gene alignments were then concatenated into a 2472 bp-long dataset that was subject to phylogenetic inference analysis in RAxML-NG.
The resulting tree indicates that samples from the Mediterranean Sea and Indian Ocean (OTUs represented by blue and yellow dots, respectively, in Figure 1) are more inclined to form standalone clusters of sequences sampled from the same area, while samples from the Atlantic Ocean (OTUs represented by red dots in Figure 1) tend to be more scattered across the tree.
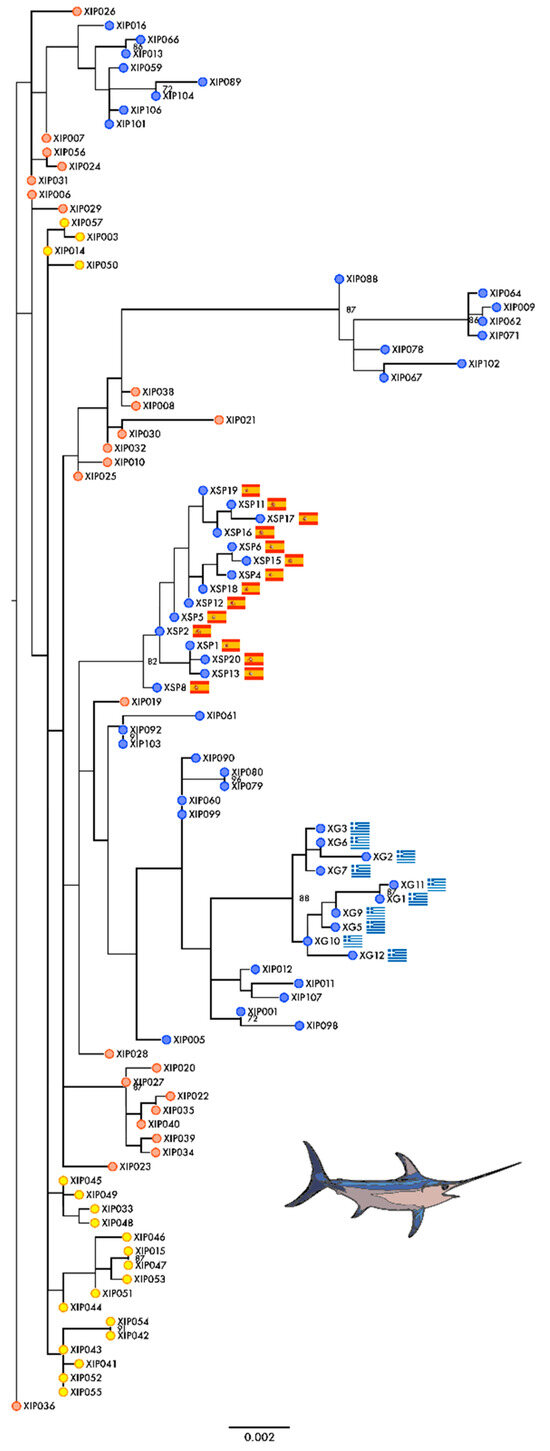
Figure 1.
Maximum likelihood unrooted tree representing phylogenetic relationships among Xiphias gladius concatenated mitochondrial sequences. Maximum likelihood tree showing the relationships among X. gladius COI, ND2, and CytB concatenated sequences. Bootstrap support values at nodes are only shown when above 70. Atlantic samples are displayed with red dots. Samples from the Indian Ocean are represented by yellow dots. Mediterranean samples are shown with blue dots. Among the Mediterranean samples, Spanish and Greek samples additionally featured their respective flags to represent their defined sampling location.
dN/dS ratios-based selection tests
No evidence for recombination was found in all gene datasets with GARD. The null hypothesis of strict neutrality was rejected by the Z test in favor of the alternative hypothesis of positive selection in the three analyzed genes for both species.
Xiphias gladius datasets
For the COI gene, seven codons were found to be under purifying selection, and no positive selection signals were identified. For the Cytb gene, nine codons were under purifying selection, and none under positive selection. For the ND2 gene, six codons were under purifying selection, and only codon 324 resulted in positive selection with the FUBAR and MEME methods (Table 2).

Table 2.
Positively and negatively selected sites in Xiphias gladius mitochondrial genes estimated by the FUBAR (# = bpp ≥ 0.9; ## = bpp ≥ 0.95; ### = bpp ≥ 0.99), FEL, and MEME models (* = p < 0.05; ** = p < 0.01; *** = p < 0.001).
By aligning the amino acid sequences of ND2, it was possible to establish that, in some individuals, proline (small apolar amino acid) is replaced by arginine (positively charged amino acid) at codon 324 in Xiphias gladius, and that codon 19 experienced positive selection in Istiophorus platypterus, where the dominant amino acid valine (which features a hydrophobic side chain) was replaced by either isoleucine or leucine, which both share the same nature (Figure 2).
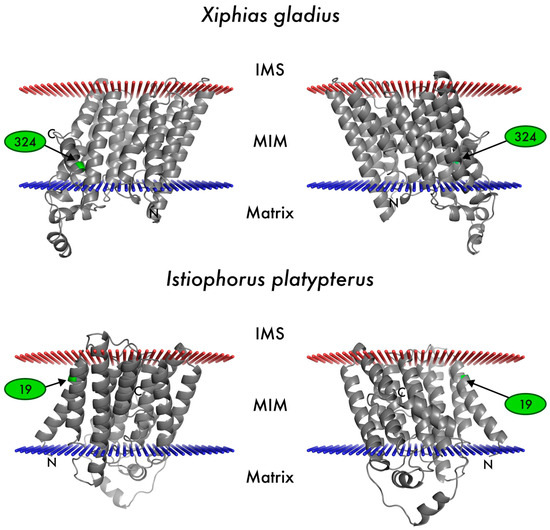
Figure 2.
Mapping of the positively selected amino acid residues (indicated in green) in the 3-dimensional obtained models for swordfish Xiphias gladius and Istiophorus platypterus ND2 protein. MIM, mitochondrial inner membrane; IMS, intermembrane space; N, amino-terminal tail; C, carboxy-terminal tail. Numbers and arrows indicate codon positions.
Furthermore, the ND2 sequence analysis identified a non-synonymous transversion (T to A) at the nucleotidic position 293, resulting in a codon change that is not subject to selection. All ND2 sequences of Greece had an asparagine (polar amino acid) in position 98 of the translated sequence, while an isoleucine (apolar amino acid) was present in the same position in the rest of the samples.
SIFT described the I98N change and P324R as tolerable, having a score of 0.53 and 0.74, respectively (values with a score < 0.05 indicate a probably harmful effect). ChimeraX 1.10 confirmed that no change in the structure of the ND2 proteins results from amino acid substitution.
Istiophorus platypterus datasets and interspecific comparison
For the COI gene, eight codons were under purifying selection, and no positive selection signals were detected. For the Cytb gene, 21 codons were under purifying selection, and none were under positive selection. For the ND2 gene, one codon was under purifying selection, and only codon 19 resulted in positive selection with the FUBAR method (Table 3). The amino acid sequence analysis indicates that in this position, a substitution between three aliphatic amino acids, valine, isoleucine, and leucine, were present, with valine being the most represented.

Table 3.
Positively and negatively selected sites in Istiophorus platypterus mitochondrial genes estimated by the FUBAR (# = bpp ≥ 0.9; ## = bpp ≥ 0.95; ### = bpp ≥ 0.99), FEL, and MEME models (* = p < 0.05).
Comparing the amino acid sequences at position 324 in the two species reveals that proline is consistently present in I. platypterus, whereas isoleucine is consistently present at position 19 in X. gladius (Supplementary Figure S1).
4. Discussion
The sequence analysis of the three mitochondrial genes investigated, COI, Cytb, and ND2, highlighted signs of selection in these OXPHOS genes in X. gladius, using the FUBAR, MEME, and FEL methods. Furthermore, a notable feature of our results is that sequences from the Spanish and Greek populations of swordfish are gathered together in the ML tree to form exclusive and well-supported clusters (bootstrap support values of 82 and 88, respectively), indicating unique genetic features that isolate them from the other sequences in this study. It is also notable that the Spanish samples branch basally from the rest of the clade in the larger Mediterranean cluster, which comprises mostly Mediterranean samples only interspersed with the Atlantic specimen XIP019. Their proximity to Atlantic samples suggests possible introgression from the Atlantic Ocean, which is foreseeable, given Spain’s geographic location at the entrance to the Mediterranean Sea via the Strait of Gibraltar. Additionally, all Greek specimens exhibited a non-synonymous transversion (T to A) at nucleotide position 293 of ND2, resulting in a codon change. Specifically, all Greek ND2 sequences had asparagine (a polar amino acid) at position 98 of the translated sequences, whereas isoleucine (a nonpolar amino acid) was present at the same position in the other samples. This substitution is not subject to selection pressure, but results in a change to the physiochemical properties of the amino acid at position 98. Isoleucine (Ile, I) and asparagine (Asn, N) are both small amino acids, but there are some important differences between them. Isoleucine is a non-polar, hydrophobic amino acid whose aliphatic side chain can play a crucial role in the recognition of substrates, particularly hydrophobic ligands. It is also flexible enough to be packed into the interior of a protein. Conversely, asparagine (Asn, N) is a neutral polar amino acid that can form hydrogen bonds with other protein chains or the main chain of the same protein. As it is polar, it is mostly located in a position that exposes it to an aqueous environment and it is frequently found in protein-active or binding sites, mostly because the polar side of the molecule can interact with other charged atoms [37].
Examining the sites under positive selection, the MEME and FUBAR models revealed evidence of diversifying positive selection in only one codon of the ND2 gene of X. gladius: codon 324, which is in the C-terminal tail within the intermembrane space. Signs of diversifying positive selection were also detected using FUBAR in codon 19 of the ND2 gene of I. platypterus, located at the end of the first α-helix near intermembrane space (IMS). Comparing the amino acid sequences at position 324 in the two species, reveals that proline (apolar) is replaced by arginine (polar) in some X. gladius individuals, whereas proline is consistently present in I. platypterus. Conversely, whereas isoleucine is consistently present at position 19 in X. gladius, valine is almost always present in I. platypterus, although in some individuals of this species, valine is replaced by isoleucine and leucine.
Arginine is an amino acid that is both hydrophobic and polar, which is why it is described as amphipathic. This condition could allow arginine to play a dual role: buried with the aliphatic side chain in the protein and exposed to water with the other part. Proline is a cyclic nonpolar amino acid often found at the end of α-helix or in turns or loops. Valine, leucine, and isoleucine are aliphatic amino acids, with a non-reactive side chain, which are more likely to be found in protein hydrophobic cores and may be involved in the binding and recognition of hydrophobic ligands (i.e., lipids) [37].
However, the results of the analysis conducted using SIFT and ChimeraX confirmed that these amino acid substitutions are not deleterious for the stability and functionality of the protein and do not lead to a change in its tridimensional structure. In this context it is essential to acknowledge that many sites of the three OXPHOS genes examined were under purifying selection. Indeed, out of a total of 828 codons analyzed (227 for COI, 267 for Cytb, and 334 for ND2), 21 were under purifying selection (approximately 2.54% of the total). This confirms that any negative changes would have been immediately eliminated by selection events, and that the amino acid change does not result in a change in the functionality of the protein or result in its improvement. Among the 13 mitochondrial protein coding genes, the ones encoding for the subunits of the NADH dehydrogenase complex (complex I) often show the highest number of sites under positive selection [38]. One of the reasons why the ND genes are more susceptible to mutations may be linked to their position in the mitochondrial genome: being located upstream to the origin of light strand replication (OL) and/or downstream to the origin of heavy strand replication (OH), they are more vulnerable to mutations because they remain single stranded for more time in comparison with other genes during DNA replication [38]. Similar studies aimed at analyzing protein-coding genes of the OXPHOS system in European sardines (Sardina pilchardus) highlighted that the number of codons under purifying selection was between 12.9% and 21.1% of the 3826 total codons analyzed [39]. Numerous sites under purifying selection in OXPHOS genes such as COI, Cytb and ND1, were also found in Aphanius fasciatus and Mullus barbatus, two teleosts widely distributed in the Mediterranean Sea [8,40].
However, when interpreting the above data, the functional importance of the NADH complex and its subunits of the OXPHOS pathway must be considered because it is the first complex involved in mitochondrial electron transport system (ETS) that transfers electrons from NADH to O2. Evidence of positive selection in ND1, ND3, and ND4 genes was found in the Atlantic salmon (Salmo salar) because of the need for increased metabolic efficiency at low temperatures [41]. ND2, ND4, and ND5 genes were found under significant positive selection in the herring (Clupea arengus) [42]. The authors highlight that the NADH complex may be useful for examining population adaptation and population structuring on a broad geographic scale.
In migratory species, like X. gladius and I. platypterus, due to their lifestyle characterized by above-average energy expenditure, selective pressures on mitochondrial OXPHOS genes can be fundamental to fulfill metabolic demands and adapt to novel environments. A study conducted on tunas and billfishes aimed to discover any selective pressures on the COX-II gene that could have increased their aerobic capacity and ability to adapt to different thermal niches, considering that they swim through waters with significant differences in their temperature. The study showed the presence of many positively selected sites that may influence how different COX subunits interact with each other, potentially affecting the yield of this enzyme complex [43].
Our study revealed some similarities in the results of selection analyses in X. gladius and I. platypterus, in that negatively selected codons were detected in all three genes, whereas positively selected codons were only detected in ND2. These results indicate that complex I of the OXPHOS system is often involved in the adaptive processes of migratory species, as also observed in S. salar [41] and C. harengus [42], and suggest that it is involved in improving metabolic efficiency under variable environmental conditions. Nevertheless, as the codons for which selection was detected in swordfish and sailfish were mostly different, this suggests that selection acted differently in the two species, presumably due to the different adaptive pressures acting on them. Furthermore, we found that all the amino acid changes in the ND2 protein are not deleterious for the stability and the functionality of the protein as shown by SIFT and ChimeraX analyses. Therefore, we hypothesize that these changes may result in an improvement in the functionality of the protein.
These results are preliminary and require confirmation through analysis of a larger number of specimens and of the functional aspects involving the amino acid changes found in ND2. Furthermore, any correlation between oceanic environmental variables and the selection pressure on the ND2 gene will need to be explored.
5. Conclusions
The results obtained from the analysis of the COI, Cytb, and ND2 genes in X. gladius and I. platypterus add significant data in gaining a better understanding of genetic differences between specimens living in widely separated regions. We confirmed the presence of (i) sites under purifying selection in all the three genes and positive selection only in the ND2 gene and (ii) a non-synonymous amino acid change in the X. gladius Greek population, for which the functional implications remain to be explored. Moreover, it would be interesting and necessary to conduct more in-depth analyses, using a larger sampling and correlating genetic analyses and the migratory behavior of the species, in order to assess whether these genetic peculiarities depend on an adaptation to local environmental conditions or on the fact that the X. gladius Greek population could be partially isolated in this region. Additionally, the relationships of the difference in sites under selection between X. gladius and I. platypterus and their migratory behavior and distribution will be the focus of future studies.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/d17110747/s1; Table S1: Xiphias gladius COI haplotypes used in this study; Table S2: Xiphias gladius Cytb haplotypes detected in this study. Table S3: Xiphias gladius ND2 haplotypes detected in this study. Figure S1: Amino acid alignment of ND2 gene sequences in X. gladius and I. platypterus.
Author Contributions
Conceptualization, A.M.P. and V.F.; methodology, G.S.C. and G.M.; software, M.M.; formal analysis, M.M.; writing—original draft preparation, A.M.P. and V.F.; writing—review and editing, A.M.P., V.F. and G.S.C.; supervision, V.F.; funding acquisition, V.F. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the University of Catania (PIACERI Linea 2, 2022, No. 22722132148).
Institutional Review Board Statement
Ethical approval was not required for the study involving animals in accordance with the local legislation and institutional requirements because ethical review and approval were waived due to the fact that the research did not involve any kind of experimentation on fish.
Data Availability Statement
The data presented in this study are available on request from the corresponding authors.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Gross, M.R. Evolution of diadromy in fishes. Am. Fish. Soc. Symp. 1987, 1, 14–25. [Google Scholar]
- Sun, Y.B.; Shen, Y.Y.; Irwin, D.M.; Zhang, Y.P. Evaluating the roles of energetic functional constraints on teleost mitochondrial-encoded protein evolution. Mol. Biol. Evol. 2011, 28, 39–44. [Google Scholar] [CrossRef] [PubMed]
- Zhang, F.; Broughton, R.E. Heterogeneous natural selection on oxidative phosphorylation genes among fishes with extreme high and low aerobic performance. BMC Evol. Biol. 2015, 15, 173. [Google Scholar] [CrossRef] [PubMed]
- Ballard, J.W.O.; Whitlock, M.C. The incomplete natural history of mitochondria. Mol. Ecol. 2004, 13, 729–744. [Google Scholar] [CrossRef] [PubMed]
- Kummer, E.; Ban, N. Mechanisms and regulation of protein synthesis in mitochondria. Nat. Rev. Mol. Cell Biol. 2021, 22, 307–325. [Google Scholar] [CrossRef] [PubMed]
- Signes, A.; Fernandez-Vizarra, E. Assembly of mammalian oxidative phosphorylation complexes I–V and supercomplexes. Essays Biochem. 2018, 62, 255–270. [Google Scholar] [CrossRef]
- Elbassiouny, A.A.; Lovejoy, N.R.; Chang, B.S. Convergent patterns of evolution of mitochondrial oxidative phosphorylation (OXPHOS) genes in electric fishes. Philos. Trans. R. Soc. B 2020, 375, 20190179. [Google Scholar] [CrossRef]
- Calogero, G.S.; Mancuso, M.; Segvic-Bubic, T.; Ferrito, V.; Pappalardo, A.M. OXPHOS genes analysis in the red mullet (Mullus barbatus Linnaeus, 1758). Front. Mar. Sci. 2025, 12, 1577491. [Google Scholar] [CrossRef]
- Garvin, M.R.; Bielawski, J.P.; Sazanov, L.A.; Gharrett, A.J. Review and meta-analysis of natural selection in mitochondrial complex I in metazoans. J. Zool. Syst. Evol. Res. 2015, 53, 1–17. [Google Scholar] [CrossRef]
- Mukundan, L.; Sukumaran, S.; Raj, N.; Jose, A.; Gopalakrishnan, A. Positive selection in the mitochondrial protein coding genes of teleost regional endotherms: Evidence for adaptive evolution. J. Mar. Biol. Assoc. India 2022, 64, 10–18. [Google Scholar] [CrossRef]
- Wegner, N.C.; Snodgrass, O.E.; Dewar, H.; Hyde, J.R. Whole-body endothermy in a mesopelagic fish, the opah, Lampris guttatus. Science 2015, 348, 786–789. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.F.; Yu, H.Y.; Ma, S.B.; Lin, Q.; Wang, D.Z.; Wang, X. Phylogenetic and evolutionary comparison of mitogenomes reveal adaptive radiation of lampriform fishes. Int. J. Mol. Sci. 2023, 24, 8756. [Google Scholar] [CrossRef]
- Palko, B.J.; Beardsley, G.L.; Richards, W.J. Synopsis of the Biology of the Swordfish, Xiphias gladius (L.); U.S. Department of Commerce NOAA Technical Report; National Marine Fisheries Service: Silver Spring, MD, USA, 1981; Volume 441, p. 21. [Google Scholar]
- Collette, B.; Graves, J.; Kells, V.A. Tunas Billfishes World; Johns Hopkins University Press: Baltimore, MD, USA, 2019. [Google Scholar]
- Beardsley, G.L., Jr.; Merrett, N.R.; Richards, W.J. Synopsis of the Biology of the Sailfish, Istiophorus platypterus (Shaw and Nodder, 1791); NOAA Technical Report NMFS SSRF: Washington, DC, USA, 1975; Volume 675, p. 95. [Google Scholar]
- Block, B.A. Endothermy in Tunas, Billfishes, and Sharks. In Encyclopedia of Fish Physiology: From Genome to Environment; Farrell, A.P., Ed.; Academic Press: San Diego, CA, USA, 2011; Volume 3, pp. 1914–1920. [Google Scholar]
- Righi, T.; Splendiani, A.; Fioravanti, T.; Casoni, E.; Gioacchini, G.; Carnevali, O.; Caputo Barucchi, V. Loss of mitochondrial genetic diversity in overexploited mediterranean swordfish (Xiphias gladius, 1759) population. Diversity 2020, 12, 170. [Google Scholar] [CrossRef]
- Neilson, J.; Arocha, F.; Cass-Calay, S.; Mejuto, J.; Ortiz, M.; Scott, G.; Smith, C.; Travassos, G.T.; Andrushchenko, I. The recovery of Atlantic swordfish: The comparative roles of the regional fisheries management organization and species biology. Rev. Fish. Sci. 2013, 21, 59–97. [Google Scholar] [CrossRef]
- Abascal, F.J.; Mejuto, J.; Quintans, M.; García-Cortés, B.; Ramos-Cartelle, A. Tracking of the broadbill swordfish, Xiphias gladius, in the central and eastern North Atlantic. Fish. Res. 2015, 162, 20–28. [Google Scholar] [CrossRef]
- Ward, R.D.; Zemlak, T.S.; Innes, B.H.; Last, P.R.; Hebert, P.D. DNA barcoding Australia’s fish species. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2005, 360, 1847–1857. [Google Scholar] [CrossRef]
- Messing, J. New M13 vectors for cloning. Meth. Enzymol. 1983, 101, 20–78. [Google Scholar] [CrossRef]
- Sevilla, R.G.; Diez, A.; Norén, M.; Mouchel, O.; Jérome, M.; Verrez-Bagnis, V.; Van Pelt, H.; Favre-Krey, L.; Krey, G.; The Fishtrace Consortium; et al. Primers and polymerase chain reaction conditions for DNA barcoding teleost fish based on the mitochondrial cytochrome b and nuclear rhodopsin genes. Mol. Ecol. Notes 2007, 7, 730–734. [Google Scholar] [CrossRef]
- Bradman, H.; Grewe, P.; Appleton, B. Direct comparison of mitochondrial markers for the analysis of swordfish population structure. Fish. Res. 2011, 109, 95–99. [Google Scholar] [CrossRef]
- Katoh, K.; Rozewicki, J.; Yamada, K.D. MAFFT online service: Multiple sequence alignment, interactive sequence choice and visualization. Brief. Bioinform. 2019, 20, 1160–1166. [Google Scholar] [CrossRef] [PubMed]
- Kozlov, A.M.; Darriba, D.; Flouri, T.; Morel, B.; Stamatakis, A. RAxML-NG: A fast, scalable and user-friendly tool for maximum likelihood phylogenetic inference. Bioinformatics 2019, 35, 4453–4455. [Google Scholar] [CrossRef]
- Darriba, D.; Taboada, G.L.; Doallo, R.; Posada, D. jModelTest 2: More models, new heuristics and parallel computing. Nat. Methods 2012, 9, 772. [Google Scholar] [CrossRef]
- Nei, M.; Gojobori, T. Simple methods for estimating the numbers of synonymous and non-synonymous nucleotide substitutions. Mol. Biol. Evol. 1986, 3, 418–426. [Google Scholar] [CrossRef] [PubMed]
- Stecher, G.; Tamura, K.; Kumar, S. Molecular evolutionary genetics analysis (MEGA) for macOS. Mol. Biol. Evol. 2020, 37, 1237–1239. [Google Scholar] [CrossRef]
- Kosakovsky Pond, S.L.; Frost, S.D.W. Datamonkey: Rapid Detection of Selective Pressure on Individual Sites of Codon Alignments. Bioinformatics 2005, 21, 2531–2533. [Google Scholar] [CrossRef]
- Murrell, B.; Moola, S.; Mabona, A.; Weighill, T.; Sheward, D.; Kosakovsky Pond, S.L.; Scheffler, K. FUBAR: A Fast, Unconstrained Bayesian AppRoximation for Inferring Selection. Mol. Biol. Evol. 2013, 30, 1196–1205. [Google Scholar] [CrossRef]
- Murrell, B.; Wertheim, J.O.; Moola, S.; Weighill, T.; Scheffler, K.; Sergei, L.; Pond, K.S.L. Detecting individual sites subject to episodic diversifying selection. PLoS Genet. 2012, 8, e1002764. [Google Scholar] [CrossRef]
- Silva, G.; Lima, F.P.; Martel, P.; Castilho, R. Thermal Adaptation and Clinal Mitochondrial DNA Variation of European Anchovy. Proc. R. Soc. B 2014, 281, 20141093. [Google Scholar] [CrossRef] [PubMed]
- Waterhouse, A.; Bertoni, M.; Bienert, S.; Studer, G.; Tauriello, G.; Gumienny, R.; Heer, F.T.; de Beer, T.A.P.; Rempfer, C.; Bordoli, L.; et al. SWISS-MODEL: Homology modelling of protein structures and complexes. Nucleic Acids Res. 2018, 46, W296–W303. [Google Scholar] [CrossRef] [PubMed]
- Lomize, M.A.; Lomize, A.L.; Pogozheva, I.D.; Mosberg, H.I. OPM: Orientations of proteins in membranes database. Bioinformatics 2006, 22, 623–625. [Google Scholar] [CrossRef]
- Ng, P.C.; Henikoff, S. SIFT: Predicting amino acid changes that affect protein function. Nucleic Acids Res. 2003, 31, 3812–3814. [Google Scholar] [CrossRef] [PubMed]
- DeLano, W.L. Pymol: An Open-Source Molecular Graphics Tool; CCP4 Newsletter on Protein Crystallography; Charles Ballard and Maeri Howard-Eales Daresbury Laboratory: Warrington, UK, 2002; Volume 40, pp. 82–92. [Google Scholar]
- Betts, M.J.; Russel, R.B. Amino acid properties and consequences of substitutions. In Bioinformatics for Geneticists; Barnes, M.R., Gray, I.C., Eds.; John Wiley & Sons, Ltd.: Hoboken, NJ, USA, 2003; pp. 289–314. [Google Scholar]
- Marshall, H.D.; Coulson, M.W.; Carr, S.M. Near neutrality, rate heterogeneity, and linkage govern mitochondrial genome evolution in Atlantic cod (Gadus morhua) and other gadine fish. Mol. Biol. Evol. 2008, 26, 579–589. [Google Scholar] [CrossRef][Green Version]
- Vieira, A.R.; de Sousa, F.; Bilro, J.; Viegas, M.B.; Svanbäck, R.; Gordo, L.S.; Paulo, O.S. Mitochondrial genomes of the European sardine (Sardina pilchardus) reveal Pliocene diversification, extensive gene flow and pervasive purifying selection. Sci. Rep. 2024, 14, 30977. [Google Scholar] [CrossRef]
- Pappalardo, A.M.; Santa Calogero, G.; Šanda, R.; Giuga, M.; Ferrito, V. Evidence for Selection on Mitochondrial OXPHOS Genes in the Mediterranean Killifish Aphanius fasciatus Valenciennes, 1821. Biology 2024, 13, 212. [Google Scholar] [CrossRef] [PubMed]
- Consuegra, S.; John, E.; Verspoor, E.; de Leaniz, C.G. Patterns of natural selection acting on the mitochondrial genome of a locally adapted fish species. Genet. Select. Evol. 2015, 47, 58. [Google Scholar] [CrossRef]
- Teacher, A.G.; André, C.; Merilä, J.; Wheat, C.W. Whole mitochondrial genome scan for population structure and selection in the Atlantic herring. BMC Evol. Biol. 2012, 12, 248. [Google Scholar] [CrossRef] [PubMed]
- Dalziel, A.C.; Moyes, C.D.; Fredriksson, E.; Lougheed, S.C. Molecular evolution of cytochrome c oxidase in high-performance fish (Teleostei: Scombroidei). J. Mol. Evol. 2006, 62, 319–331. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).