The Complete Chloroplast Genome of the Green Algae Desmodesmus spinosus (Chodat) E.Hegewald: Genome Structure, Phylogeny, and Comparative Analysis
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Materials, DNA Extraction and Genome Sequencing
2.2. De novo Chloroplast Genome Assemblies
2.3. Genome Annotation and Visualization
2.4. Codon Usage Bias, Simple Sequence Repeat, Long Tandem Repeat and Dispersed Repeat Analysis
2.5. Predicting RNA-Editing Sites
2.6. Comparative Analysis of cp Genomes
2.7. Phylogenetic Analysis
3. Results
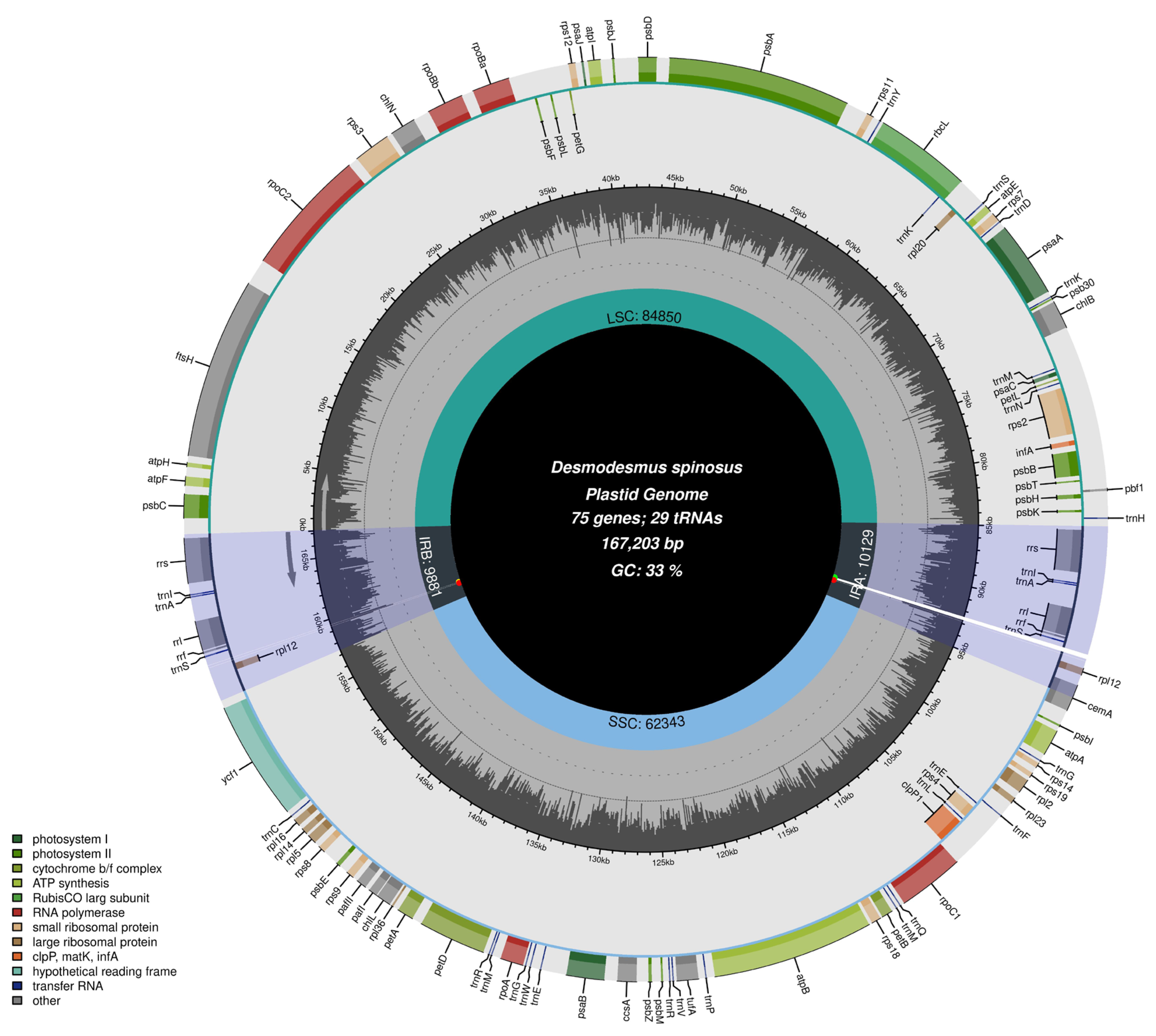
3.1. Features of the cp Genome and Its Base Composition
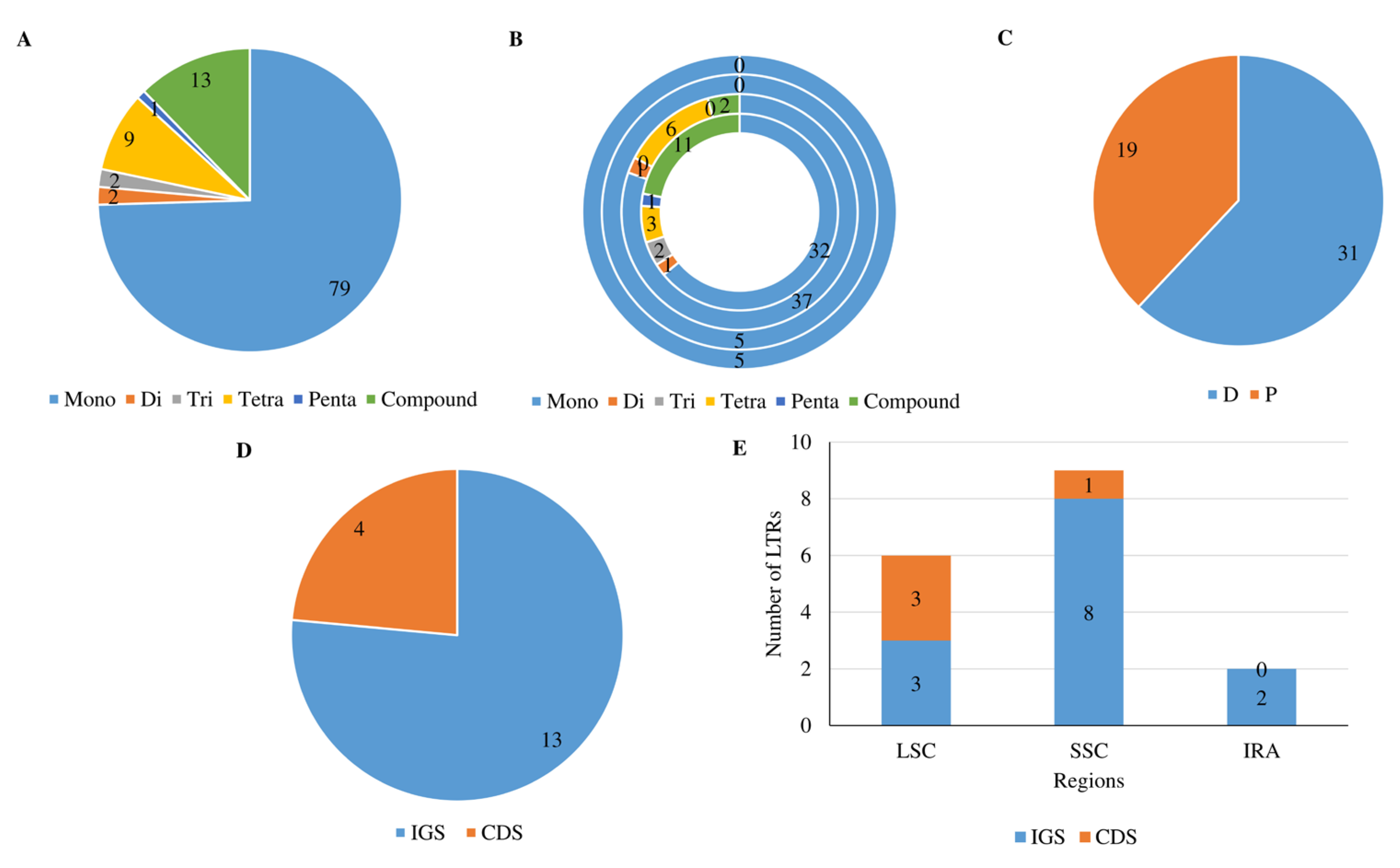
3.2. Diversity and Composition of SSR, LTR and DR
3.3. Prediction of RNA-Editing Sites
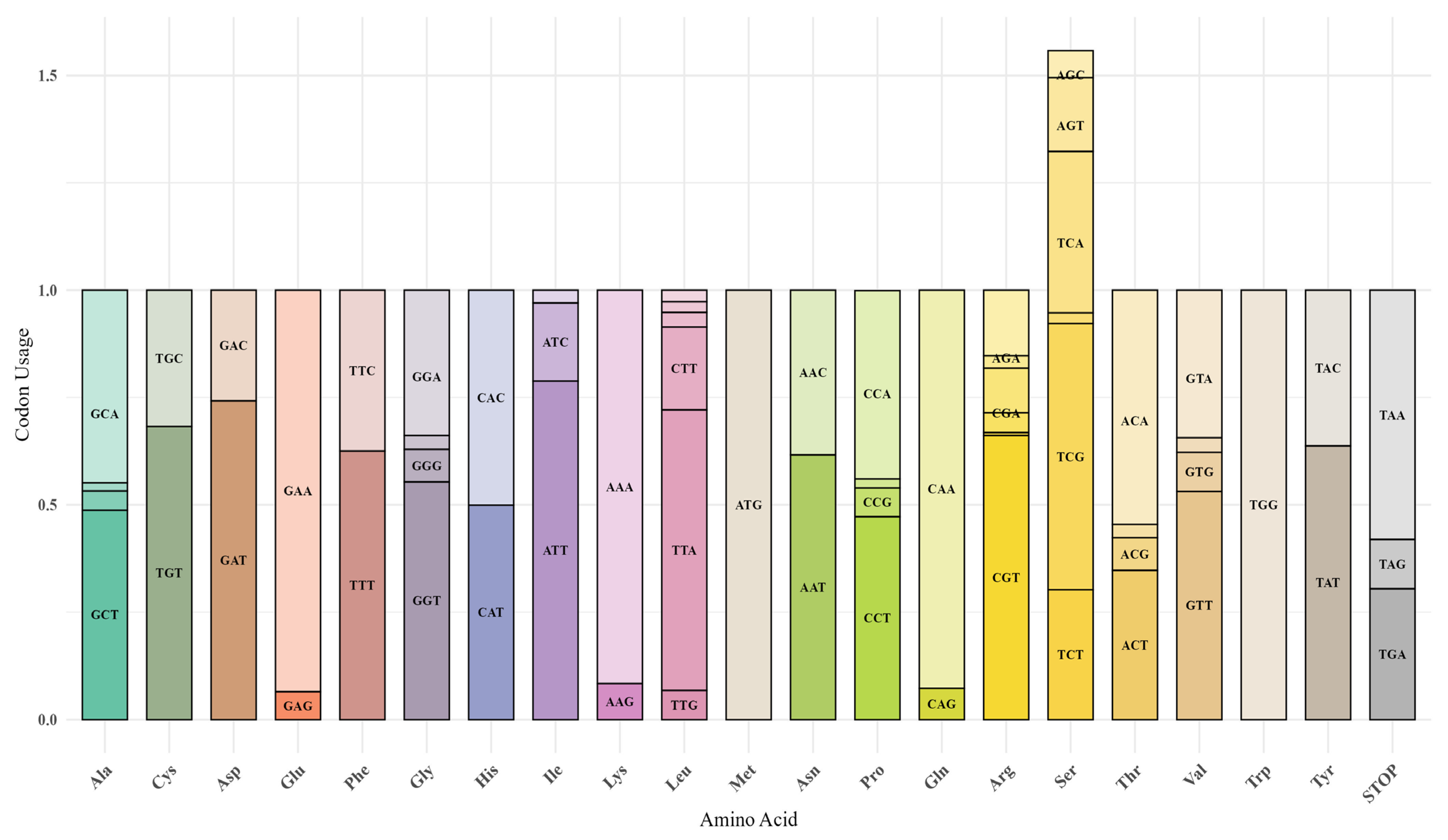
3.4. Relative Synonymous Codon Usage (RSCU)
3.5. IR-Boundaries and Adjacent Genomic Structures
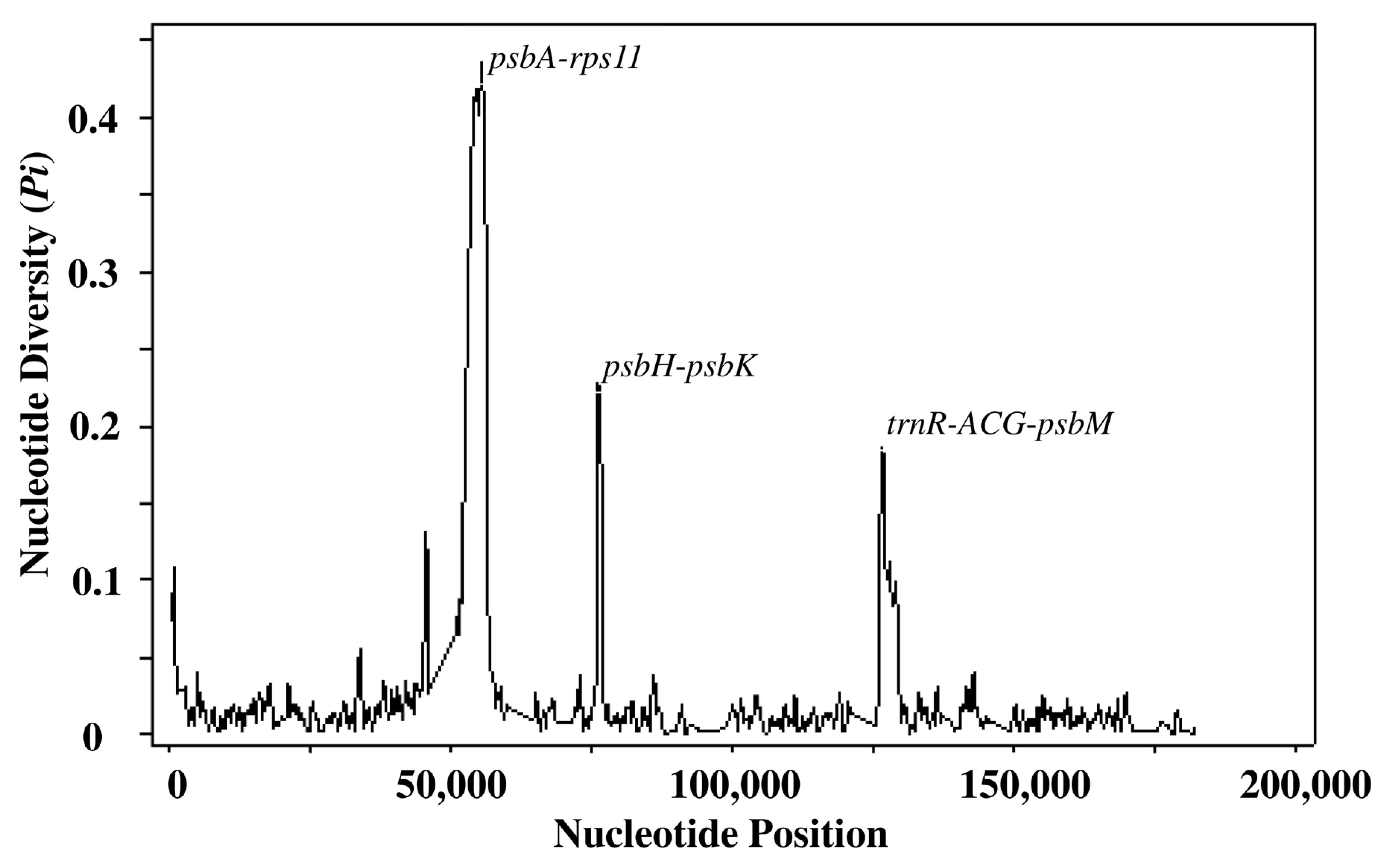
3.6. Nucleotide Diversity
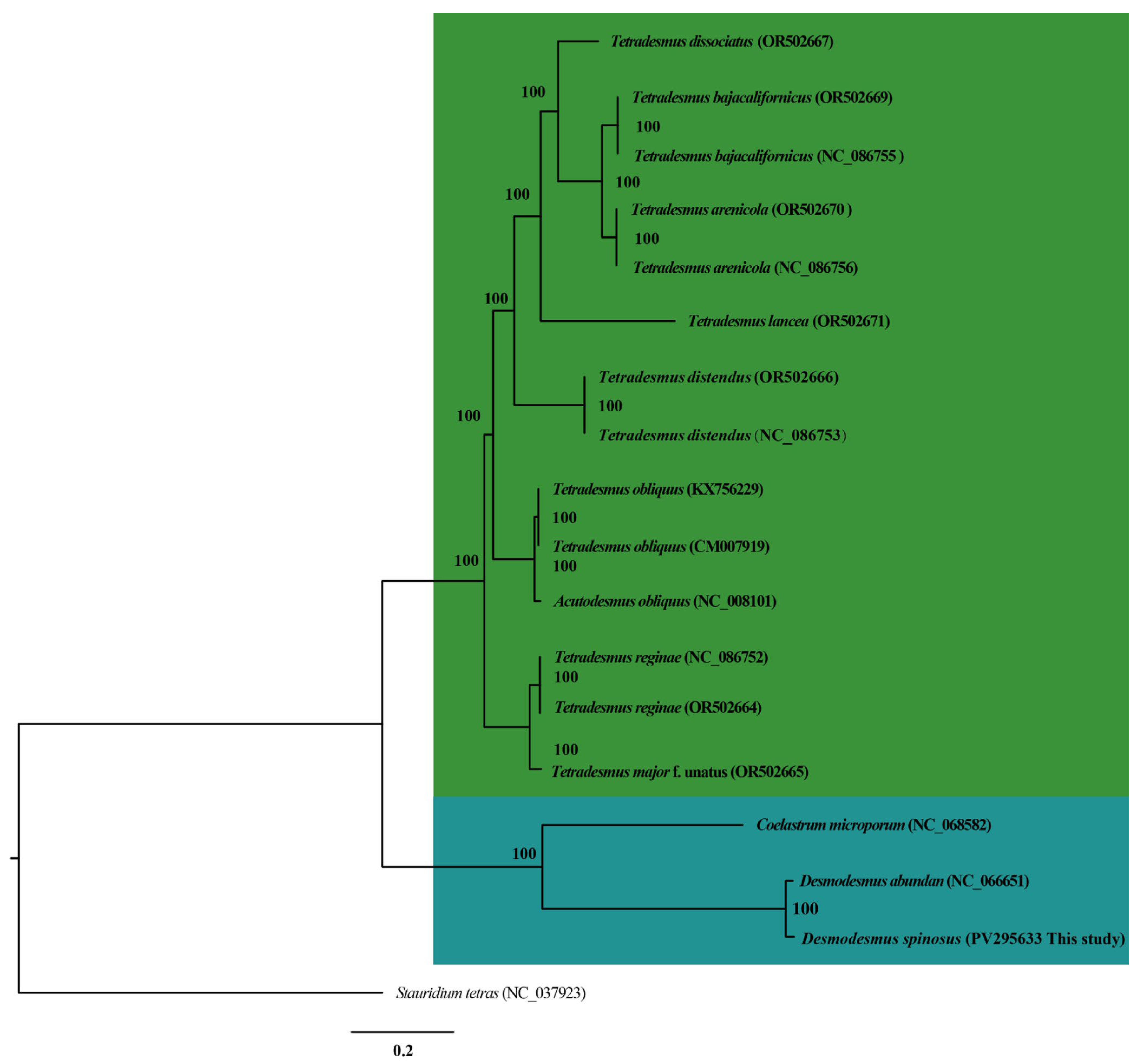
3.7. Phylogenetic Analyses
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Schoch, C.L.; Ciufo, S.; Domrachev, M.; Hotton, C.L.; Kannan, S.; Khovanskaya, R.; Leipe, D.; Mcveigh, R.; O’nEill, K.; Robbertse, B.; et al. NCBI Taxonomy: A Comprehensive Update on Curation, Resources And Tools. Database 2020, 2020, baaa062. [Google Scholar] [CrossRef]
- Soós, V.; Shetty, P.; Maróti, G.; Incze, N.; Badics, E.; Bálint, P.; Ördög, V.; Balázs, E. Biomolecule Composition and Draft Genome of a Novel, High-Lipid Producing Scenedesmaceae Microalga. Algal Res. 2021, 54, 102181. [Google Scholar] [CrossRef]
- Leite, G.B.; Paranjape, K.; Hallenbeck, P.C. Breakfast of Champions: Fast Lipid Accumulation by Cultures of Chlorella and Scenedesmus Induced By Xylose. Algal Res. 2016, 16, 338–348. [Google Scholar] [CrossRef]
- Tsarenko, P.M.; Borysova, O.V.; Korkhovyi, V.I.; Blume, Y.B. High-Efficiency Ukrainian Strains of Microalgae for Biodiesel Fuel Production (Overview). Open Agric. J. 2020, 14, 209–218. [Google Scholar] [CrossRef]
- Pezzolesi, L.; Samorì, C.; Zoffoli, G.; Xamin, G.; Simonazzi, M.; Pistocchi, R. Semi-Continuous Production of Polyhydroxybutyrate (PHB) in the Chlorophyta Desmodesmus Communis. Algal Res. 2023, 74, 103196. [Google Scholar] [CrossRef]
- Hess, S.K.; Lepetit, B.; Kroth, P.G.; Mecking, S. Production of chemicals from microalgae lipids—Status and perspectives. Eur. J. Lipid Sci. Technol. 2018, 120, 1700152. [Google Scholar] [CrossRef]
- Akgül, R.; Akgül, F.; Kızılkaya, I.T. Effects of different phosphorus concentrations on growth and biochemical composition of Desmodesmus communis (E.Hegewald) E.Hegewald. Prep. Biochem. Biotechnol. 2021, 51, 705–713. [Google Scholar] [CrossRef]
- Burgel, G.; Ribas, P.G.; Ferreira, P.C.; Passos, M.F.; Santos, B.; Savi, D.C.; Ludwig, T.A.V.; Vargas, J.V.C.; Galli-Terasawa, L.V.; Kava, V.M. Morphology, molecular phylogeny and biomass evaluation of Desmodesmus abundans (Scenedesmaceae-Chlorophyceae) from Brazil. Braz. J. Biol. 2022, 82, e265235. [Google Scholar] [CrossRef]
- Najeeb, M.I.; Ahmad, M.-D.; Anjum, A.A.; Maqbool, A.; Ali, M.A.; Nawaz, M.; Ali, T.; Manzoor, R. Distribution, screening and biochemical characterization of indigenous microalgae for bio-mass and bio-energy production potential from three districts of Pakistan. Braz. J. Biol. 2024, 84, e261698. [Google Scholar] [CrossRef]
- Shin, Y.-S.; Do, J.-M.; Noh, H.-S.; Yoon, H.-S. Exploring the Potential of Desmodesmus sp. KNUA231 for Bioenergy and Biofertilizer Applications and Its Adaptability to Environmental Stress. Appl. Sci. 2025, 15, 5097. [Google Scholar] [CrossRef]
- Yang, B.; Li, Y.; Wang, Z.; Yue, Z.; Wen, J.; Zhao, X.; Zhang, H.; Wang, X.; Wang, X.; Zhang, M. Strong Inhibitory Effects of Desmodesmus sp. on Microcystis blooms: Potential as a Biological Control Agent in Aquaculture. Aquac. Rep. 2025, 40, 102579. [Google Scholar] [CrossRef]
- Krienitz, L. The Contribution of Eberhard Hegewald (1942–2024) to the Systematics of Coccoid Green Algae—An Obituary. Nova Hedwig. 2024, 119, 261–268. [Google Scholar] [CrossRef]
- Nguyen, M.L.; Mai, X.C.; Chu, N.H.; Trinh, D.M.; Liu, C.-L.; Shen, C.-R. DNA signaturing derived from the internal transcribed spacer 2 (ITS2): A novel tool for identifying Desmodesmus species (Scenedesmaceae, Chlorophyta). Fottea 2023, 23, 1–7. [Google Scholar] [CrossRef]
- Porebski, S.; Bailey, L.G.; Baum, B.R. Modification of a CTAB DNA extraction protocol for plants containing high polysaccharide and polyphenol components. Plant Mol. Biol. Rep. 1997, 15, 8–15. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A Flexible Trimmer for Illumina Sequence Data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef]
- Dierckxsens, N.; Mardulyn, P.; Smits, G. NOVOPlasty: De Novo Assembly of Organelle Genomes from Whole Genome Data. Nucleic Acids Res. 2017, 45, e18. [Google Scholar] [CrossRef]
- Tillich, M.; Lehwark, P.; Pellizzer, T.; Ulbricht-Jones, E.S.; Fischer, A.; Bock, R.; Greiner, S. GeSeq—Versatile and Accurate Annotation of Organelle Genomes. Nucleic Acids Res. 2017, 45, W6–W11. [Google Scholar] [CrossRef]
- Greiner, S.; Lehwark, P.; Bock, R. OrganellarGenomeDRAW (OGDRAW) Version 1.3.1: Expanded Toolkit for the Graphical Visualization of Organellar Genomes. Nucleic Acids Res. 2019, 47, W59–W64. [Google Scholar] [CrossRef]
- Shi, L.; Chen, H.; Jiang, M.; Wang, L.; Wu, X.; Huang, L.; Liu, C. CPGAVAS2, an integrated plastome sequence annotator and analyzer. Nucleic Acids Res. 2019, 47, W65–W73. [Google Scholar] [CrossRef]
- Beier, S.; Thiel, T.; Münch, T.; Scholz, U.; Mascher, M. MISA-web: A web server for microsatellite prediction. Bioinformatics 2017, 33, 2583–2585. [Google Scholar] [CrossRef]
- Benson, G. Tandem repeats finder: A program to analyze DNA sequences. Nucleic Acids Res. 1999, 27, 573–580. [Google Scholar] [CrossRef]
- Kurtz, S. The Vmatch large scale sequence analysis software. Ref Type Comput. Program 2003, 412, 297. [Google Scholar]
- Liu, S.; Ni, Y.; Li, J.; Zhang, X.; Yang, H.; Chen, H.; Liu, C. CPGView: A package for visualizing detailed chloroplast genome structures. Mol. Ecol. Resour. 2023, 23, 694–704. [Google Scholar] [CrossRef] [PubMed]
- Edera, A.A.; Small, I.; Milone, D.H.; Sanchez-Puerta, M.V. Deepred-Mt: Deep representation learning for predicting C-to-U RNA editing in plant mitochondria. Comput. Biol. Med. 2021, 136, 104682. [Google Scholar] [CrossRef] [PubMed]
- Zheng, S.; Poczai, P.; Hyvönen, J.; Tang, J.; Amiryousefi, A. Chloroplot: An online program for the versatile plotting of organelle genomes. Front. Genet. 2020, 11, 576124. [Google Scholar] [CrossRef] [PubMed]
- Katoh, K.; Rozewicki, J.; Yamada, K.D. MAFFT online service: Multiple sequence alignment, interactive sequence choice and visualization. Briefings Bioinform. 2017, 20, 1160–1166. [Google Scholar] [CrossRef]
- Rozas, J.; Ferrer-Mata, A.; Sánchez-DelBarrio, J.C.; Guirao-Rico, S.; Librado, P.; Ramos-Onsins, S.E.; Sánchez-Gracia, A. DnaSP 6: DNA sequence polymorphism analysis of large data sets. Mol. Biol. Evol. 2017, 34, 3299–3302. [Google Scholar] [CrossRef]
- Menéndez, C.D.; Poczai, P.; Williams, B.; Myllys, L.; Amiryousefi, A. IRplus: An augmented tool to detect inverted repeats in plastid genomes. Genome Biol. Evol. 2023, 15, evad177. [Google Scholar] [CrossRef]
- Huerta-Cepas, J.; Serra, F.; Bork, P. ETE 3: Reconstruction, Analysis, and Visualization of Phylogenomic Data. Mol. Biol. Evol. 2016, 33, 1635–1638. [Google Scholar] [CrossRef]
- Katoh, K.; Kuma, K.I.; Toh, H.; Miyata, T. MAFFT Version 5: Improvement in Accuracy of Multiple Sequence Alignment. Nucleic Acids Res. 2005, 33, 511–518. [Google Scholar] [CrossRef]
- Nguyen, L.-T.; Schmidt, H.A.; Von Haeseler, A.; Minh, B.Q. IQ-TREE: A Fast and Effective Stochastic Algorithm for Estimating Maximum-Likelihood Phylogenies. Mol. Biol. Evol. 2015, 32, 268–274. [Google Scholar] [CrossRef] [PubMed]
- Wang, R.-J.; Cheng, C.-L.; Chang, C.-C.; Wu, C.-L.; Su, T.-M.; Chaw, S.-M. Dynamics and evolution of the inverted repeat-large single copy junctions in the chloroplast genomes of monocots. BMC Evol. Biol. 2008, 8, 36. [Google Scholar] [CrossRef] [PubMed]
- Park, S.; Jun, M.; Park, S.; Park, S. Lineage-specific variation in IR boundary shift events, inversions, and substitution rates among Caprifoliaceae s.l. (Dipsacales) Plastomes. Int. J. Mol. Sci. 2021, 22, 10485. [Google Scholar] [CrossRef] [PubMed]
- Liu, M.-L.; Fan, W.-B.; Wang, N.; Dong, P.-B.; Zhang, T.-T.; Yue, M.; Li, Z.-H. Evolutionary analysis of plastid genomes of seven Lonicera L. species: Implications for sequence divergence and phylogenetic relationships. Int. J. Mol. Sci. 2018, 19, 4039. [Google Scholar] [CrossRef]
- Chen, S.; Azam, F.M.S.; Akter, M.L.; Ao, L.; Zou, Y.; Qian, Y. The first complete chloroplast genome of Thalictrum fargesii: Insights into phylogeny and species identification. Front. Plant Sci. 2024, 15, 1356912. [Google Scholar] [CrossRef]
- Xu, Y.; Chen, X.; Melkonian, M.; Wang, S.; Sahu, S.K. Comparative chloroplast genome analysis of two Desmodesmus species reveals genome diversity within Scenedesmaceae (Sphaeropleales, Chlorophyceae). Protist 2024, 175, 126073. [Google Scholar] [CrossRef]
- Cho, H.S.; Lee, J. Taxonomic Reinvestigation of the Genus Tetradesmus (Scenedesmaceae; Sphaeropleales) Based on Morphological Characteristics and Chloroplast Genomes. Front. Plant Sci. 2024, 15, 1303175. [Google Scholar] [CrossRef]
- Lee, C.; Cooper, J.T.; Moroni, F.; Salim, A.M.; Lee, C.; Spanbauer, T.; Theriot, E.C. Complete Plastome of Coelastrum microporum Nägeli (Scenedesmaceae, Sphaeropleales). Mitochondrial DNA Part B 2024, 8, 948–951. [Google Scholar] [CrossRef]
- Guo, Y.-Y.; Yang, J.-X.; Bai, M.-Z.; Zhang, G.-Q.; Liu, Z.-J. The chloroplast genome evolution of Venus slipper (Paphiopedilum): IR expansion, SSC contraction, and highly rearranged SSC regions. BMC Plant Biol. 2021, 21, 248. [Google Scholar] [CrossRef]
- Qin, Q.; Dong, Y.; Chen, J.; Wang, B.; Peng, Y.; Zhang, X.; Wang, X.; Zeng, J.; Zhong, G.; Zhang, S.; et al. Comparative analysis of chloroplast genomes reveals molecular evolution and phylogenetic relationships within the Papilionoideae of Fabaceae. BMC Plant Biol. 2025, 25, 157. [Google Scholar] [CrossRef]
- Park, H.; Park, J.H.; Kang, Y.J. Characterization of the complete chloroplast genome of Wolffia arrhiza and comparative genomic analysis with relative Wolffia species. Sci. Rep. 2024, 14, 5873. [Google Scholar] [CrossRef] [PubMed]
- Wang, T.; Feng, H.; Zhu, H.; Zhong, B. Molecular phylogeny and comparative chloroplast genome analysis of the type species Crucigenia quadrata. BMC Plant Biol. 2025, 25, 64. [Google Scholar] [CrossRef] [PubMed]
- Song, H.; Peng, H.; Fang, Z.; Zhang, B.; Zhu, Z.; Xiao, Z.; Liu, G.; Hu, Y. Koliella bifissiva sp. nov (Chlorellaceae, Chlorophyta) and analysis of its organelle genomes. Plants 2024, 13, 2604. [Google Scholar] [CrossRef] [PubMed]
- Odom, O.W.; Holloway, S.P.; Deshpande, N.N.; Lee, J.; Herrin, D.L. Mobile self-splicing group I introns from the psbA gene of Chlamydomonas reinhardtii: Highly efficient homing of an exogenous intron containing its own promoter. Mol. Cell. Biol. 2001, 21, 3472–3481. [Google Scholar] [CrossRef]
- Chu, D.; Wei, L. The chloroplast and mitochondrial C-to-U RNA editing in Arabidopsis thaliana shows signals of adaptation. Plant Direct 2019, 3, e00169. [Google Scholar] [CrossRef]
- Saleh, M.M. Effect of Electromagnetic Field on Growth and Morphology of Some Algal Species. Ph.D. Thesis, Suez Canal University, Ismailia, Egypt, 2006. [Google Scholar]
- Hegewald, E. New Combinations in the Genus Desmodesmus (Chlorophyceae, Scenedesmaceae). Arch. Für Hydrobiol. 2000, 96, 1–18. [Google Scholar] [CrossRef]
- Wang, F.; Qi, Y.; Yu, F. The biological functions of FtsH in plant organelle protein homeostasis. J. Exp. Bot. 2025, 76, 4220–4231. [Google Scholar] [CrossRef]
- Krynická, V.; Komenda, J. The role of FtsH complexes in the response to abiotic stress in cyanobacteria. Plant Cell Physiol. 2024, 65, 1103–1114. [Google Scholar] [CrossRef]

| Category | Gene Group | Gene Name * |
|---|---|---|
| Photosynthesis | Subunits of photosystem I | psaA i, psaB, psaC, psaJ |
| Subunits of photosystem II | psbA q, psbB, psbC, psbD, psbE, psbF, psbH, psbI, psbJ, psbK, psbL, psbM, psbT, psbZ | |
| Subunits of cytochrome b/f complex | petA, petB, petD i, petG, petL | |
| Subunits of ATP synthase | atpA, atpB e, atpE, atpF, atpH, atpI | |
| Large subunit of rubisco | rbcL i | |
| light-independent protochlorophyllide reductase (DPOR) enzyme complex | chlB, chlL, chlN | |
| Photosystem I assembly protein | pafI, pafII | |
| Photosystem II reaction center protein | psb30 | |
| Self-replication | Large ribosomal subunit | rpl2, rpl5, rpl12 d, rpl14, rpl16, rpl20, rpl23, rpl36 |
| Small ribosomal subunit | rps11, rps12, rps14, rps18, rps19, rps2, rps3, rps4, rps7, rps8, rps9 | |
| Subunits of RNA polymerase | rpoA, rpoBa, rpoBb, rpoC1, rpoC2 | |
| Ribosomal RNAs | Rrs d, rrl d, rrf d | |
| Transfer RNAs | trnY-GUA, trnK-UUU d, trnS-UGA, trnD-GUC, trnM-CAU t, trnN-GUU, trnH-GUG, trnI-GAU d, trnA-UGC d, trnS-GCU d, trnG-UCC, trnF-GAA, trnE-UUC d, trnL-UAG, trnQ-UUG, trnP-UGG, trnV-UAC, trnR-ACG, trnW-CCA, trnG-GCC, trnR-UCU, trnC-GCA | |
| Other genes | ATP-dependent CLP protease | clpP1 |
| ATP-dependent zinc metalloprotease | ftsH | |
| N-terminal nucleophile amino hydrolase superfamily | pbf1 | |
| Envelope membrane protein | cemA | |
| c-type cytochrome synthesis gene | ccsA | |
| Translational initiation factor | infA | |
| Elongation factor | tufA | |
| Genes of unknown function | Conserved hypothetical chloroplast ORF | ycf1 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, S.; Tan, J.; Safiul Azam, F.M.; Li, A.; Zhang, R.; Li, B. The Complete Chloroplast Genome of the Green Algae Desmodesmus spinosus (Chodat) E.Hegewald: Genome Structure, Phylogeny, and Comparative Analysis. Diversity 2025, 17, 721. https://doi.org/10.3390/d17100721
Chen S, Tan J, Safiul Azam FM, Li A, Zhang R, Li B. The Complete Chloroplast Genome of the Green Algae Desmodesmus spinosus (Chodat) E.Hegewald: Genome Structure, Phylogeny, and Comparative Analysis. Diversity. 2025; 17(10):721. https://doi.org/10.3390/d17100721
Chicago/Turabian StyleChen, Shixi, Jiang Tan, Fardous Mohammad Safiul Azam, Ao Li, Renqing Zhang, and Bin Li. 2025. "The Complete Chloroplast Genome of the Green Algae Desmodesmus spinosus (Chodat) E.Hegewald: Genome Structure, Phylogeny, and Comparative Analysis" Diversity 17, no. 10: 721. https://doi.org/10.3390/d17100721
APA StyleChen, S., Tan, J., Safiul Azam, F. M., Li, A., Zhang, R., & Li, B. (2025). The Complete Chloroplast Genome of the Green Algae Desmodesmus spinosus (Chodat) E.Hegewald: Genome Structure, Phylogeny, and Comparative Analysis. Diversity, 17(10), 721. https://doi.org/10.3390/d17100721







