Genetic Diversity, Kinship, and Polychromatism in the Spotted Eagle Ray Aetobatus ocellatus of Fiji
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Collection
2.2. DNA COI Barcoding for Species Identification
2.3. DArT-SeqTM: Extraction and SNP Sequencing
2.4. SNP Quality Control Filtering
2.5. Allelic Diversity, Relatedness, and Structure
3. Results
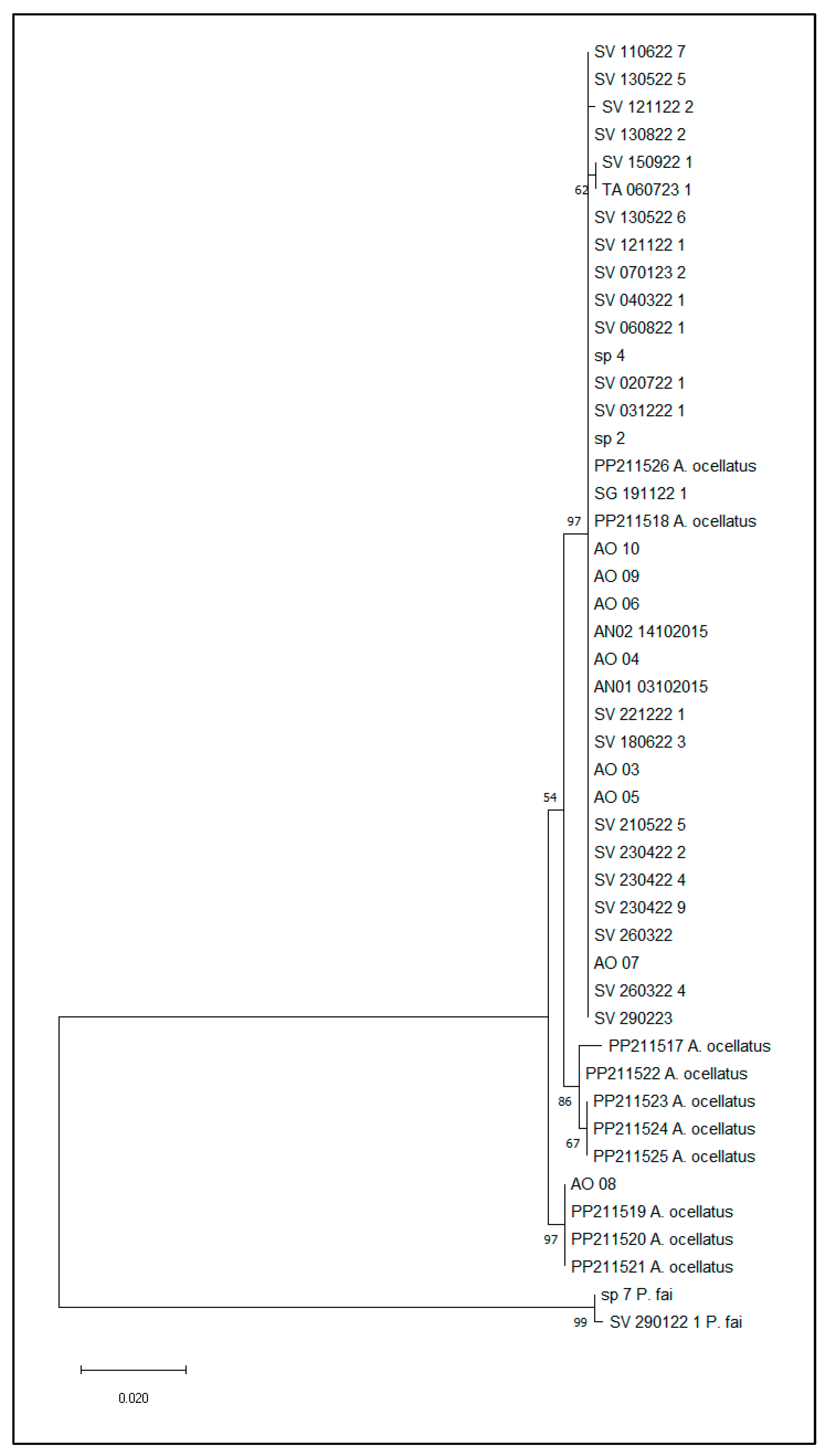
3.1. DNA COI Barcoding for Species Identification
3.2. SNP Filtering
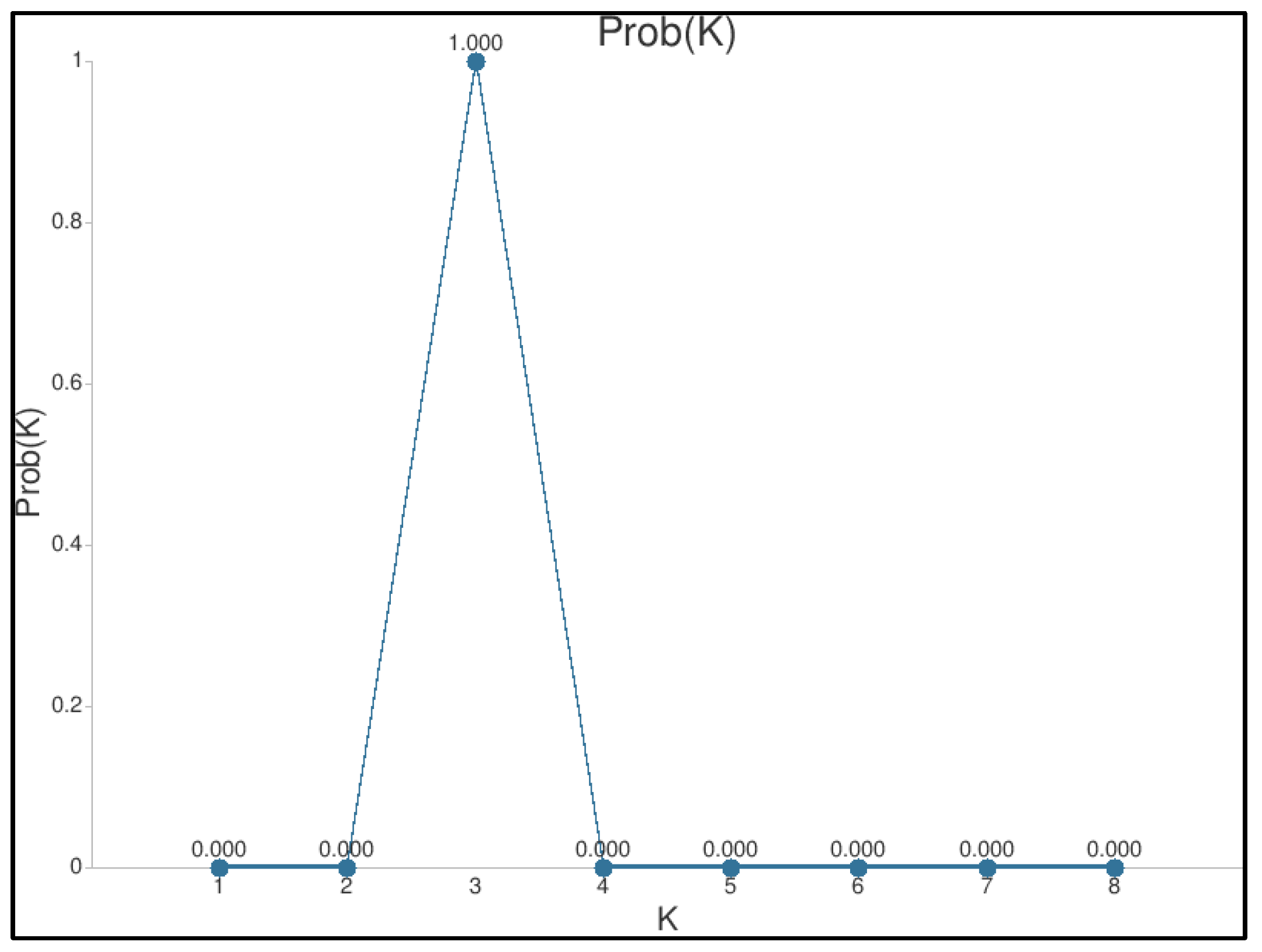
3.3. Genomic Diversity, Relatedness, and Structure
3.4. Intraspecific Polychromatism
4. Discussion
4.1. COI Barcoding and Genetic Diversity Estimates
4.2. Kinship
4.3. Intraspecific Polychromatism
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Dulvy, N.K.; Pacoureau, N.; Rigby, C.L.; Pollom, R.A.; Jabado, R.W.; Ebert, D.A.; Finucci, B.; Pollock, C.M.; Cheok, J.; Derrick, D.H. Overfishing drives over one-third of all sharks and rays toward a global extinction crisis. Curr. Biol. 2021, 31, 4773–4787.e8. [Google Scholar] [PubMed]
- Sherman, C.S.; Simpfendorfer, C.A.; Pacoureau, N.; Matsushiba, J.H.; Yan, H.F.; Walls, R.H.; Rigby, C.L.; VanderWright, W.J.; Jabado, R.W.; Pollom, R.A. Half a century of rising extinction risk of coral reef sharks and rays. Nat. Commun. 2023, 14, 15. [Google Scholar]
- Worm, B.; Orofino, S.; Burns, E.S.; D’Costa, N.G.; Manir Feitosa, L.; Palomares, M.L.; Schiller, L.; Bradley, D. Global shark fishing mortality still rising despite widespread regulatory change. Science 2024, 383, 225–230. [Google Scholar] [PubMed]
- Sherman, C.S.; Heupel, M.R.; Moore, S.K.; Chin, A.; Simpfendorfer, C.A. When sharks are away, rays will play: Effects of top predator removal in coral reef ecosystems. Mar. Ecol. Prog. Ser. 2020, 641, 145–157. [Google Scholar]
- Simpfendorfer, C.A.; Heithaus, M.R.; Heupel, M.R.; MacNeil, M.A.; Meekan, M.; Harvey, E.; Sherman, C.S.; Currey-Randall, L.M.; Goetze, J.S.; Kiszka, J.J. Widespread diversity deficits of coral reef sharks and rays. Science 2023, 380, 1155–1160. [Google Scholar]
- Allendorf, F.W.; Luikart, G.H.; Aitken, S.N. Conservation and the Genetics of Populations; John Wiley & Sons: Hoboken, NJ, USA, 2012. [Google Scholar]
- Dudgeon, C.L.; Blower, D.; Broderick, D.; Giles, J.; Holmes, B.J.; Kashiwagi, T.; Krück, N.; Morgan, J.; Tillett, B.; Ovenden, J. A review of the application of molecular genetics for fisheries management and conservation of sharks and rays. J. Fish Biol. 2012, 80, 1789–1843. [Google Scholar] [PubMed]
- Luikart, G.; Kardos, M.; Hand, B.K.; Rajora, O.P.; Aitken, S.N.; Hohenlohe, P.A. Population genomics: Advancing understanding of nature. In Population Genomics: Concepts, Approaches and Applications; Springer: Cham, Switzerland, 2019; pp. 3–79. [Google Scholar]
- Frankham, R. Genetics and conservation biology. Comptes Rendus Biol. 2003, 326, 22–29. [Google Scholar]
- DiBattista, J.D. Patterns of genetic variation in anthropogenically impacted populations. Conserv. Genet. 2008, 9, 141–156. [Google Scholar]
- Hernández, S.; Daley, R.; Walker, T.; Braccini, M.; Varela, A.; Francis, M.P.; Ritchie, P.A. Demographic history and the South Pacific dispersal barrier for school shark (Galeorhinus galeus) inferred by mitochondrial DNA and microsatellite DNA mark. Fish. Res. 2015, 167, 132–142. [Google Scholar]
- Vrijenhoek, R. Genetic diversity and fitness in small populations. In Conservation Genetics; Springer: Berlin/Heidelberg, Germany, 1994; pp. 37–53. [Google Scholar]
- Bräutigam, A.; Callow, M.; Campbell, I.R. Global Priorities for Conserving Sharks and Rays: A 2015–2025 Strategy. IUCN: International Union for Conservation of Nature. Global Sharks and Rays Initiative, IUCN Species Survival Commission (SSC), Shark Specialist Group, The Shark Trust, UK, TRAFFIC International, Wildlife Conservation Society (WCS), US, WWF. 2016. Available online: https://coilink.org/20.500.12592/s532n0 (accessed on 18 August 2024).
- Hirschfeld, M.; Dudgeon, C.; Sheaves, M.; Barnett, A. Barriers in a sea of elasmobranchs: From fishing for populations to testing hypotheses in population genetics. Glob. Ecol. Biogeogr. 2021, 30, 2147–2163. [Google Scholar]
- Last, P.; Naylor, G.; Séret, B.; White, W.; de Carvalho, M.; Stehmann, M. Rays of the World; CSIRO Publishing: Clayton, Australia, 2016. [Google Scholar]
- McEachran, J.D.M.R.C. Myliobatidae. The Living Marine Resources of the Western Central Atlantic. Volume 1: Introduction, Mollusks, Crustaceans, Hagfishes, Sharks, Batoid Fishes, and Chimaeras; Carpenter, K.E., Ed.; California Academy of Sciences: San Francisco, CA, USA; FAO: Rome, Italy, 2002; pp. 578–582. [Google Scholar]
- Marie, A.D.; Justine, J.-L. Monocotylids (Monogenea: Monopisthocotylea) from Aetobatus cf. narinari off New Caledonia, with a description of Decacotyle elpora n. sp. Syst. Parasitol. 2005, 60, 175–185. [Google Scholar] [PubMed]
- Richards, V.P.; Henning, M.; Witzell, W.; Shivji, M.S. Species delineation and evolutionary history of the globally distributed spotted eagle ray (Aetobatus narinari). J. Hered. 2009, 100, 273–283. [Google Scholar] [PubMed]
- Schluessel, V.; Broderick, D.; Collin, S.; Ovenden, J. Evidence for extensive population structure in the white-spotted eagle ray within the Indo-Pacific inferred from mitochondrial gene sequences. J. Zool. 2010, 281, 46–55. [Google Scholar]
- White, W.; Last, P.; Naylor, G.; Jensen, K.; Caira, J. Clarification of Aetobatus ocellatus (Kuhl, 1823) as a valid species, and a comparison with Aetobatus narinari (Euphrasen, 1790)(Rajiformes: Myliobatidae). CSIRO Mar. Atmos. Res. Pap. 2010, 32, 141. [Google Scholar]
- Berthe, C. First ecological, biological and behavioral insights of the ocellated eagle ray Aetobatus ocellatus in French Polynesia. Biodivers. Ecol. 2017. Available online: https://ephe.hal.science/hal-01690359/ (accessed on 18 August 2024).
- Berthe, C.; Waqalevu, V.P.; Latry, L.; Besson, M.; Lerouvreur, F.; Siu, G.; Lecellier, G.; Rummer, J.L.; Bertucci, F.; Iglésias, S. Distribution patterns of ocellated eagle rays, Aetobatus ocellatus, along two sites in Moorea Island, French Polynesia. Cybium 2018, 42, 313–320. [Google Scholar]
- White, W.T.; Last, P.R.; Stevens, J.D.; Yearsly, G. Economically Important Sharks and Rays of Indonesia; ACIAR Publishing: Canberra, Australia, 2006. [Google Scholar]
- Theivasigamani, M.; Subbiah, S. Elasmobranch fishery resources of Gulf of Mannar, southeast coast of India. World J. Fish Mar. Sci. 2014, 6, 24–29. [Google Scholar]
- Spaet, J.L.; Berumen, M.L. Fish market surveys indicate unsustainable elasmobranch fisheries in the Saudi Arabian Red Sea. Fish. Res. 2015, 161, 356–364. [Google Scholar]
- Kyne, P.M.; Dudgeon, C.L.; Ishihara, H.; Dudley, S.F.J.; White, W.T. Spotted Eagle Ray. 2016. Available online: https://www.iucnredlist.org/species/42566169/42566212 (accessed on 10 June 2024).
- Sales, J.B.L.; de Oliveira, C.N.; dos Santos, W.C.R.; Rotundo, M.M.; Ferreira, Y.; Ready, J.; Sampaio, I.; Oliveira, C.; Cruz, V.P.; Lara-Mendoza, R.E. Phylogeography of eagle rays of the genus Aetobatus: Aetobatus narinari is restricted to the continental western Atlantic Ocean. Hydrobiologia 2019, 836, 169–183. [Google Scholar]
- Glaus, K.; Gordon, L.; Vierus, T.; Marosi, N.D.; Sykes, H. Rays in the Shadows: Batoid Diversity, Occurrence, and Conservation Status in Fiji. Biology 2024, 13, 73. [Google Scholar] [CrossRef]
- Glaus, K.; Savou, R.; Brunnschweiler, J.M. Characteristics of Fiji’s small-scale ray fishery and its relevance to food security. Mar. Policy 2024, 163, 106082. [Google Scholar]
- Morin, P.A.; Luikart, G.; Wayne, R.K.; Group, S.W. SNPs in ecology, evolution and conservation. Trends Ecol. Evol. 2004, 19, 208–216. [Google Scholar]
- Pazmiño, D.A.; Maes, G.E.; Simpfendorfer, C.A.; Salinas-de-León, P.; van Herwerden, L. Genome-wide SNPs reveal low effective population size within confined management units of the highly vagile Galapagos shark (Carcharhinus galapagensis). Conserv. Genet. 2017, 18, 1151–1163. [Google Scholar]
- Devloo-Delva, F.; Burridge, C.; Kyne, P.; Brunnschweiler, J.; Chapman, D.; Charvet, P.; Chen, X.; Cliff, G.; Daly, R.; Drymon, M. From rivers to ocean basins: The role of ocean barriers and philopatry in the genetic structuring of a cosmopolitan coastal predator. Authorea Prepr. 2022. Available online: https://www.researchgate.net/publication/364575245_From_rivers_to_ocean_basins_the_role_of_ocean_barriers_and_philopatry_in_the_genetic_structuring_of_a_cosmopolitan_coastal_predator (accessed on 18 August 2024).
- Postaire, B.D.; Devloo-Delva, F.; Brunnschweiler, J.M.; Charvet, P.; Chen, X.; Cliff, G.; Daly, R.; Drymon, J.M.; Espinoza, M.; Fernando, D. Global genetic diversity and historical demography of the Bull Shark. J. Biogeogr. 2023, 51, 632–648. [Google Scholar]
- Jones, O.R.; Wang, J. COLONY: A program for parentage and sibship inference from multilocus genotype data. Mol. Ecol. Resour. 2010, 10, 551–555. [Google Scholar] [PubMed]
- Dale, J.; Holland, K. Age, growth and maturity of the brown stingray (Dasyatis lata) around Oahu, Hawai’i. Mar. Freshw. Res. 2012, 63, 475–484. [Google Scholar]
- Ward, R.D.; Holmes, B.H.; White, W.T.; Last, P.R. DNA barcoding Australasian chondrichthyans: Results and potential uses in conservation. Mar. Freshw. Res. 2008, 59, 57–71. [Google Scholar]
- Ward, R.D.; Zemlak, T.S.; Innes, B.H.; Last, P.R.; Hebert, P.D. DNA barcoding Australia's fish species. Philos. Trans. R. Soc. B Biol. Sci. 2005, 360, 1847–1857. [Google Scholar]
- Wang, Y.; Zhao, Y.; Bollas, A.; Wang, Y.; Au, K.F. Nanopore sequencing technology, bioinformatics and applications. Nat. Biotechnol. 2021, 39, 1348–1365. [Google Scholar]
- Ratnasingham, S.; Hebert, P.D. BOLD: The Barcode of Life Data System (http://www.barcodinglife.org). Mol. Ecol. Notes 2007, 7, 355–364. [Google Scholar] [PubMed]
- Madden, T. The BLAST sequence analysis tool. In The NCBl Handbook; National Center for Biotechnology Information (US): Bethesda, MD, USA, 2003. [Google Scholar]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547. [Google Scholar]
- Kimura, M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 1980, 16, 111–120. [Google Scholar]
- Marie, A.D.; Herbinger, C.; Fullsack, P.; Rico, C. First reconstruction of kinship in a scalloped hammerhead shark aggregation reveals the mating patterns and breeding sex ratio. Front. Mar. Sci. 2019, 6, 676. [Google Scholar]
- Foll, M.; Gaggiotti, O. A genome-scan method to identify selected loci appropriate for both dominant and codominant markers: A Bayesian perspective. Genetics 2008, 180, 977–993. [Google Scholar]
- Belkhir, K. GENETIX, Logiciel Sous WindowsTM Pour la Génétique des Populations. 1999. Available online: https://kimura.univ-montp2.fr/genetix/ (accessed on 18 August 2024).
- Pritchard, J.K.; Stephens, M.; Donnelly, P. Inference of population structure using multilocus genotype data. Genetics 2000, 155, 945–959. [Google Scholar] [PubMed]
- Falush, D.; Stephens, M.; Pritchard, J.K. Inference of population structure using multilocus genotype data: Linked loci and correlated allele frequencies. Genetics 2003, 164, 1567–1587. [Google Scholar]
- Hubisz, M.J.; Falush, D.; Stephens, M.; Pritchard, J.K. Inferring weak population structure with the assistance of sample group information. Mol. Ecol. Resour. 2009, 9, 1322–1332. [Google Scholar]
- Kopelman, N.M.; Mayzel, J.; Jakobsson, M.; Rosenberg, N.A.; Mayrose, I. Clumpak: A program for identifying clustering modes and packaging population structure inferences across K. Mol. Ecol. Resour. 2015, 15, 1179–1191. [Google Scholar]
- Evanno, G.; Regnaut, S.; Goudet, J. Detecting the number of clusters of individuals using the software STRUCTURE: A simulation study. Mol. Ecol. 2005, 14, 2611–2620. [Google Scholar]
- Carson, A.R.; Smith, E.N.; Matsui, H.; Brækkan, S.K.; Jepsen, K.; Hansen, J.-B.; Frazer, K.A. Effective filtering strategies to improve data quality from population-based whole exome sequencing studies. BMC Bioinform. 2014, 15, 125. [Google Scholar]
- Glaus, K.; Loganimoce, E.; Mescam, G.; Appleyard, S.A. Genetic diversity of an undescribed cryptic maskray (Neotrygon sp.) species from Fiji. Pac. Conserv. Biol. 2024, in press. [Google Scholar]
- Sellas, A.B.; Bassos-Hull, K.; Pérez-Jiménez, J.C.; Angulo-Valdés, J.A.; Bernal, M.A.; Hueter, R.E. Population structure and seasonal migration of the spotted eagle ray, Aetobatus narinari. J. Hered. 2015, 106, 266–275. [Google Scholar] [PubMed]
- Glaus, K.B.; Appleyard, S.A.; Stockwell, B.; Brunnschweiler, J.M.; Shivji, M.; Clua, E.; Marie, A.D.; Rico, C. Insights Into Insular Isolation of the Bull Shark, Carcharhinus leucas (Müller and Henle, 1839), in Fijian Waters. Front. Mar. Sci. 2020, 7, 586015. [Google Scholar]
- Canfield, S.J.; Galván-Magaña, F.; Bowen, B.W. Little sharks in a big world: Mitochondrial DNA reveals small-scale population structure in the California horn shark (Heterodontus francisci). J. Hered. 2022, 113, 298–310. [Google Scholar]
- Swift, D.G.; O’Leary, S.J.; Grubbs, R.D.; Frazier, B.S.; Fields, A.T.; Gardiner, J.M.; Drymon, J.M.; Bethea, D.M.; Wiley, T.R.; Portnoy, D.S. Philopatry influences the genetic population structure of the blacktip shark (Carcharhinus limbatus) at multiple spatial scales. Mol. Ecol. 2023, 32, 4953–4970. [Google Scholar] [PubMed]
- Wang, I.J. Recognizing the Temporal Distinctions between Landscape Genetics and Phylogeography; Wiley Online Library: Hoboken, NJ, USA, 2010. [Google Scholar]
- Benestan, L. Population genomics applied to fishery management and conservation. In Population Genomics: Marine Organisms; Springer: Cham, Switzerland, 2020; pp. 399–421. [Google Scholar]
- Brown, K.T.; Seeto, J.; Lal, M.M.; Miller, C.E. Discovery of an important aggregation area for endangered scalloped hammerhead sharks, Sphyrna lewini, in the Rewa River estuary, Fiji Islands. Pac. Conserv. Biol. 2016, 22, 242–248. [Google Scholar]
- Glaus, K.B.; Brunnschweiler, J.M.; Piovano, S.; Mescam, G.; Genter, F.; Fluekiger, P.; Rico, C. Essential waters: Young bull sharks in Fiji's largest riverine system. Ecol. Evol. 2019, 9, 7574–7585. [Google Scholar]
- Gordon, L.; Vierus, T. First photographic evidence of oceanic manta rays (Mobula birostris) at two locations in the Fiji islands. PeerJ 2022, 10, e13883. [Google Scholar]
- Martins, A.; Heupel, M.; Chin, A.; Simpfendorfer, C. Batoid nurseries: Definition, use and importance. Mar. Ecol. Prog. Ser. 2018, 595, 253–267. [Google Scholar]
- Utrera-López, N. Estimación de la Edad y Crecimiento de la raya águila Aetobatus Narinari (Euphrasen, 1790) en el sur del Golfo de México; Universidad Autónoma de Baja California Sur: La Paz, Mexico, 2015. [Google Scholar]
- Boggio-Pasqua, A.; Bassos-Hull, K.; Aeberhard, W.H.; Hoopes, L.A.; Swider, D.A.; Wilkinson, K.A.; Dureuil, M. Whitespotted eagle ray (Aetobatus narinari) age and growth in wild (in situ) versus aquarium-housed (ex situ) individuals: Implications for conservation and management. Front. Mar. Sci. 2022, 9, 960822. [Google Scholar]
- Schluessel, V.; Bennett, M.; Collin, S. Diet and reproduction in the white-spotted eagle ray Aetobatus narinari from Queensland, Australia and the Penghu Islands, Taiwan. Mar. Freshw. Res. 2010, 61, 1278–1289. [Google Scholar]
- Torres, Y.; Charvet, M.; Faria, V.; Charvet, P. Dots in the dark: Dorsal polychromatism in the endemic Xingu Freshwater Stingray. J. Zool. 2023, 321, 165–174. [Google Scholar]
- Conover, D.; Clarke, L.; Munch, S.; Wagner, G. Spatial and temporal scales of adaptive divergence in marine fishes and the implications for conservation. J. Fish Biol. 2006, 69, 21–47. [Google Scholar]
- Erickson, D.L.; Fenster, C.B.; Stenøien, H.K.; Price, D. Quantitative trait locus analyses and the study of evolutionary process. Mol. Ecol. 2004, 13, 2505–2522. [Google Scholar] [PubMed]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Glaus, K.; Appleyard, S.A. Genetic Diversity, Kinship, and Polychromatism in the Spotted Eagle Ray Aetobatus ocellatus of Fiji. Diversity 2024, 16, 588. https://doi.org/10.3390/d16090588
Glaus K, Appleyard SA. Genetic Diversity, Kinship, and Polychromatism in the Spotted Eagle Ray Aetobatus ocellatus of Fiji. Diversity. 2024; 16(9):588. https://doi.org/10.3390/d16090588
Chicago/Turabian StyleGlaus, Kerstin, and Sharon A. Appleyard. 2024. "Genetic Diversity, Kinship, and Polychromatism in the Spotted Eagle Ray Aetobatus ocellatus of Fiji" Diversity 16, no. 9: 588. https://doi.org/10.3390/d16090588
APA StyleGlaus, K., & Appleyard, S. A. (2024). Genetic Diversity, Kinship, and Polychromatism in the Spotted Eagle Ray Aetobatus ocellatus of Fiji. Diversity, 16(9), 588. https://doi.org/10.3390/d16090588





