Abstract
Studies performed in North America, Africa, and South America have led to the isolation of four new species of Kastovskya, a filamentous cyanobacterial genus that before this manuscript had only one species, Kastovskya adunca from Chile. Kastovskya nitens and K. viridissima were isolated from soils on San Nicolas Island, K. sahariensis was isolated from hypolithic habitats from the Sahara Desert in Algeria, and K. circularithylacoides was isolated from hypolithic habitats in Chile. The molecular analyses are corroborated by morphological data, morphometric analysis, and ecological and biogeographical considerations for robust polyphasic descriptions of all taxa. The peculiar transatlantic distribution of this genus bears similarity to other taxa in recently published studies and is in agreement with a hypothesis suggesting that cyanobacteria in Africa may disperse to the Americas on dust particles during windstorms. This work is unusual in that species in a single rare cyanobacterial genus with a disjunct distribution are described simultaneously from three continents. The 16S rRNA gene analyses performed for this study also revealed that another recent genus, Arizonema, is clearly a later synonym of Symplocastrum. This issue is resolved here with the collapsing of the type species Arizonema commune into Symplocastrum flechtnerae.
1. Introduction
In dryland ecosystems, cyanobacteria are ecologically significant primary producers that serve a number of other critical functions. In these ecosystems, cyanobacteria, along with other microbes and bryophytes, form biological soil crusts (BSCs), which are diverse communities of microorganisms that form a crusted layer on the uppermost surface of the soil [1,2]. These BSCs perform an array of ecologically crucial functions, particularly in preventing erosion by stabilizing soil and increasing nutrient availability via biogeochemical cycling [2,3,4,5,6]. Although the ecological importance of BSCs has been known for decades [7,8], the biodiversity contained in them remains understudied. Disproportionate investigative effort has been given to aquatic algal groups, in part due to the historical assumption that soils lacked significant diversity [9]. Environments with extreme conditions in particular, such as hyperarid deserts, were assumed to be more or less absent of life due to their array of challenging characteristics such as extreme temperatures, significant diurnal temperature variation, unrelenting UV radiation without protection from higher plants, and periods of drought [10]. In recent years, however, the cyanobacteria present in such soils have seen a rapid taxonomic expansion due to increased sampling efforts, the progressively increasing attainability of sequencing, and the subsequent increase in the number of high-quality DNA sequences available for robust phylogenetic analyses [11,12]. Consequently, taxa previously thought to be geographically restricted, or endemic have begun to show up in biodiversity surveys and metagenomic studies [13,14,15].
Kastovskya, a genus of filamentous, terrestrial cyanobacteria in the family Coleofasciculaceae, was one taxon previously determined to be geographically restricted and endemic to a relatively small area in the Atacama Desert in Chile. Up until this point, the genus has had only one described species, K. adunca (basionym: Schizothrix adunca Schwabe [16]), which was isolated from biocrusts in the region [16]. Until now, strains and sequences representing the genus have remained completely absent from biodiversity surveys and metagenomic studies in similar arid environments. Now, however, Kastovskya strains have been isolated from San Nicolas Island, California, USA, and from the Sahara Desert in Algeria. Upon further phylogenetic and morphological analyses, it was determined that these recently isolated strains represent two novel species from San Nicolas Island, and one new species from the Sahara Desert. We also determined during our analyses that the two strains originally assigned to K. adunca [17] actually represent two semi-cryptic species. This study significantly expands the known distribution of the genus from one species in Chile to five species on three continents.
The two Californian species, K. nitens and K. viridissima, were isolated as part of a larger effort to characterize the algal diversity on San Nicolas Island (SNI), CA, USA. A previous study on SNI algal diversity was performed without the use of molecular data yet still showed the island’s algal flora to be highly diverse [18]. This current SNI study serves as a continuation of the former, this time with the addition of genetic data for molecular phylogenetic analysis. As a result of the current effort, two papers have been recently published, including a description of six species in three genera (Pycnacronema, Konicacronema, and Atlanticothrix) previously restricted to Brazil [14,19,20], as well as the description of a novel species of Pseudoacaryochloris [21], which is a recently described coccoid genus with one prior existing species, P. sahariensis, isolated from the Sahara Desert in Algeria [22,23]. Due to the disjunct distribution and vast geographical space between observations of these taxa, it was hypothesized in the previous publications that the progenitive common ancestors of cyanobacterial species sharing this distribution most likely arose in Africa and were carried across the Atlantic Ocean on dust particles during rare but large-scale Saharan dust storms where subsequent lineage divergence occurred. This peculiar distribution has also been observed in works unrelated to the SNI study [24,25,26].
In this paper, four novel species of Kastovskya are described using a total-evidence, polyphasic approach to taxonomy [27] that considers genetic, morphological, ecological, and biogeographical data. Sequences of both the 16S rRNA gene and the associated 16S–23S internal transcribed spacer (ITS) gene were used to perform robust phylogenetic analyses, and these were supported by morphological analyses, including a morphometric principal component analysis (PCA) to determine if strains could be separated into species based on average measurements of various features. The data from these analyses, along with ecological and biogeographical data, are presented herein to construct robust, well-supported arguments for lineage separation and taxonomic description of these novel taxa. With the description of two species from California, one from the Atacama, and one from the Sahara, we are now for the first time as a result of this study describing congeneric species isolated from three continents simultaneously. Furthermore, the distributional pattern, niche differentiation, and other characteristics of the genus are discussed herein.
While performing the 16S rRNA gene phylogenetic analyses, it was also determined that the genus Arizonema [28] is clearly a later synonym of Symplocastrum (Gomont) Kirchner [29] and, moreover, that the only described species, A. commune, is a later synonym of S. flechtnerae [30]. This taxonomic issue is resolved in this publication with the collapsing of the genus and the relocation of all Arizonema strains to Symplocastrum.
2. Materials and Methods
2.1. Field Collection
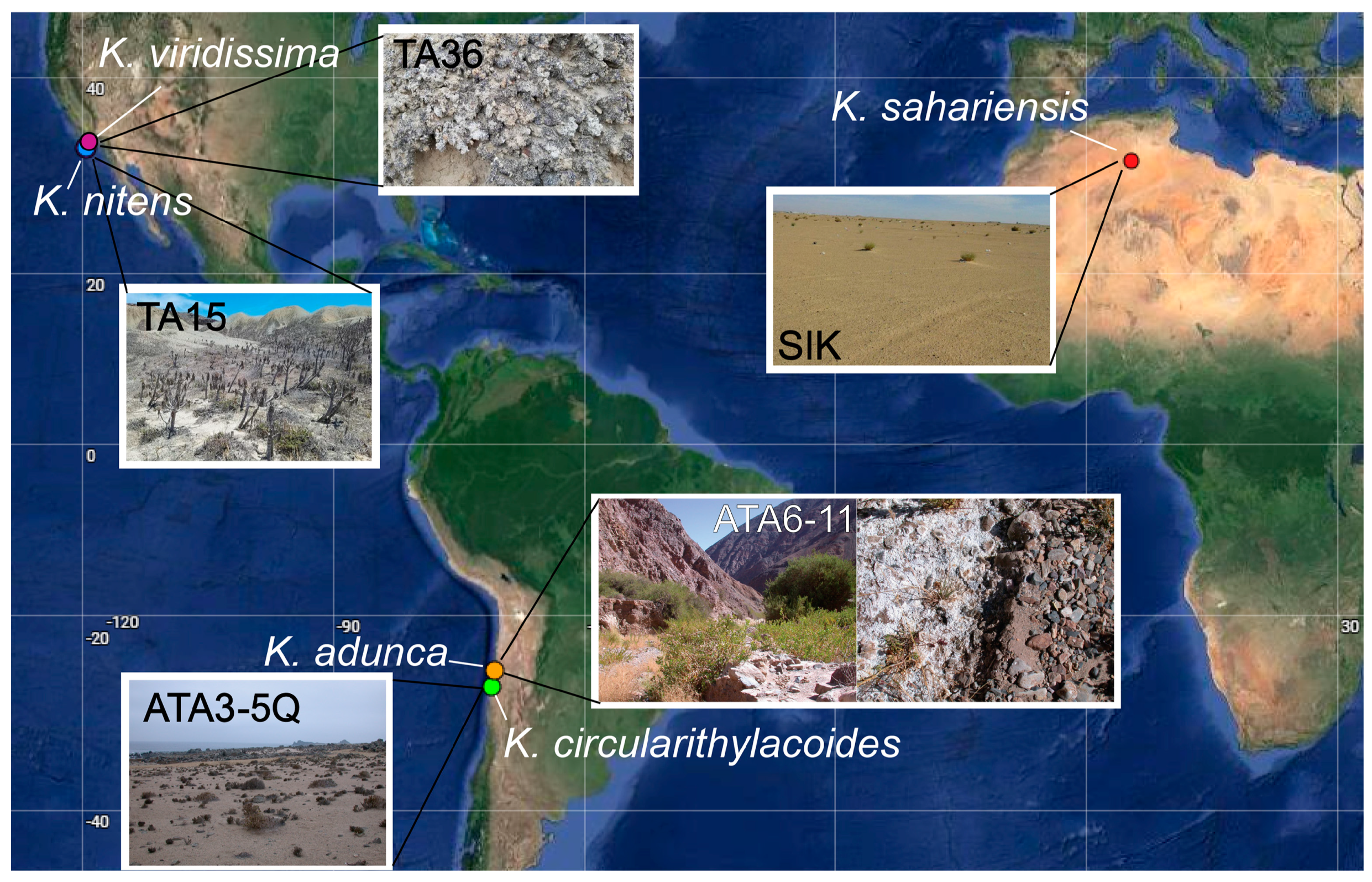
The samples originating from San Nicolas Island were collected on 25 May 2021 as part of a larger biodiversity study, and the details of the sampling effort have been previously reported elsewhere in detail [14]. The sample containing K. nitens specifically (TA15) consisted of biological soil crust collected from just above a natural spring near sea level and with sparse vascular plant cover (33.230701, −119.518852, Figure 1). The sample containing K. viridissima was collected from a highly developed, blackened, rugose, and pedicled BSC (Figure 1) sample from the bottom of a north-facing canyon (33.26033, −119.48643). The sample containing K. circularithylacoides was collected in the Atacama Desert on 13 May 2009 from a sandy soil sample found underneath quartz rocks near lichen-covered cacti (−27.783889, −71.034722, 9 m above sea level). Further details on this sampling effort have been previously reported [17,31]. The sample containing K. sahariensis was collected in December 2018 from the Sidi Khouiled (SIK) sampling site in the Sahara Desert in northeastern Algeria (31.994389, 5.415139) on a quaternary alluvial terrace with a slight slope. Additional strains representing the species were also collected in December 2018, but from a dried saline lake bed (a “chott”) in the Oued Souf area (33.717861 N, 7.441167 E) of the Sahara Desert in Algeria. These last samples were collected as part of studies on hypolithic and biocrust cyanobacterial communities in the Sahara Desert, and additional details on the methods and sampling sites have been previously reported elsewhere [22,32].

Figure 1.
Coordinates of sampling sites from which each species of Kastovskya was isolated superimposed on a map of the Americas and Africa. Representative images of sites TA15, TA36, ATA3−5, ATA6−11, and SIK are shown.
2.2. Isolation and Culture
From each biocrust or soil sample from San Nicolas Island, 1.0 g subsamples were assembled by breaking and combining fragments of larger pieces of material. Subsamples were homogenized by crushing, diluted in Erlenmeyer flasks with 100 mL sterile Z8 media [33], and agitated for 30 min on a rotary shaker to liberate cyanobacteria from the soil matrix. Enrichment plates of agar-solidified Z8 media were inoculated with 0.1 mL of the soil solution diluted to 10-4 via serial dilution in fresh, sterile Z8. Plates were sealed with Parafilm (Bemis Company, Inc., Neeneh, WI, USA) and placed under a 12 h:12 h light–dark cycle at 20 °C until macroscopic algal colonies were visible (about 5 weeks). Well-isolated uniagal colonies were picked from plates under an Olympus SZ40 stereoscope (Olympus Corporation, Tokyo, Japan) with pulled pipets and placed into capped test tubes containing 5 mL sterile liquid Z8 media. When significant growth was observed, strains were assigned strain identifiers based on the sampling site (e.g., SNI-TA36) and initials of the person who isolated the culture (e.g., SNI-TA36-BJ6, representing the sixth sample Brian Jusko isolated from site TA36 on San Nicolas Island). Strains were then assigned a putative taxonomic designation based on morphology and were again transferred to capped test tubes containing slants of agar-solidified Z8. Cultures are maintained in the Algal Culture Collection at John Carroll University in University Heights, Ohio. Selected strains of K. circularithylacoides were submitted to the Culture Collection of Autotrophic Organisms (CCALA) in Třeboň, Czech Republic. Herbarium vouchers of immobilized, air-dried material were prepared. Vouchers for California strains were submitted to the Clifton Smith Herbarium at the Santa Barbara Botanical Garden. A holotype voucher for K. circularithylacoides was deposited in the herbarium at the University of South Bohemia.
Samples from the Saharan sampling sites were ground with a mortar, and 0.2 g of the resulting composite was mixed with 0.5 mL of BG11 culture medium [34], incubated for 30 min at room temperature, and then centrifuged at 3000× g for 30 s. Two aliquots of 0.2 mL of the supernatant were cultured on separate 1% agar-solidified BG11 plates prepared with cycloheximide (0.1 mg/mL) to prevent fungal contamination. The plates were incubated in a growth chamber at 28 °C and 20–50 µmol photon m−2·s−1 and allowed to grow for about four weeks. Unialgal cultures were isolated from the plates by picking single trichomes or groups of cells using pulled capillary pipets under a Leica dissecting microscope (Leica Microsystems, Wetzler, Germany). The isolates were transferred to multiwall plates with liquid BG11 and maintained under the same conditions after sufficient growth was observed, at which point the strains were transferred to flasks and maintained under the same conditions. Cultures were named for the sampling site from which they were isolated (e.g., SIK77, for Sidi Khouiled isolate 77). K. sahariensis was immobilized via cryopreservation and placed into the Algal Culture Collection of Universidad Autónoma de Madrid with code UAM 900.
2.3. Morphological Characterization
Strains from the Atacama Desert and from San Nicolas Island were observed over time, photographed, and characterized morphologically using an Olympus BX-60 microscope equipped with Nomarski DIC optics and CellSense Dimension 1.12 software (Olympus Corporation, Tokyo). Each strain investigated for this publication was photographed along microscope slide transects over two months to capture the full extent of morphological variation and development over time. Morphological features of each strain were noted, and measurements of vegetative cells, apical cells, and sheaths were taken of each strain. The morphological characterization of K. sahariensis was performed as previously reported [22,32].
A morphometric analysis based on our complete set of photomicrographs was performed to determine if strains could be separated into species based on cell, trichome, and sheath dimensions. Four strains of K. adunca, including the reference strain (ATA6-11-RM4), and the strains belonging to the novel taxa presented in this publication were measured for vegetative cell length and width (n = 130), apical cell length and width (n = 100), and sheath width (n = 100). Additionally, length and width ratios were calculated for vegetative and apical cells, and the trichome tapering percentage was quantified from the midpoint of the apical cell to both the fourth and eighth vegetative cells (n = 45). Vegetative cell measurements were taken for every fifth cell along each trichome at the midpoint of the cell on each axis. Apical cell length measurements were taken in the same manner, while width measurements were taken at the widest point of the cell, usually near the crosswall. Sheath width was measured at multiple points along each trichome on both sides and measured from the outer cell wall to the outer extent of the sheath. Trichomes without sheaths were not measured to reduce the skewing effect of trichomes with zero sheath width. Measurements were taken from microscope images taken at 1000× magnification that were subsequently scaled up in size using Adobe Photoshop (v. 23.4.2, Adobe Systems, San Jose, CA, USA) and standardized such that 10 µm on each image was represented by 5 cm in Photoshop. Measurements were taken on the same computer by the same researcher to standardize measurements and reduce the effect of bias, and strains were photographed at the same stage of development to account for life-stage variation and phenotypic plasticity. Average values were calculated for the above characteristics and were used to conduct a principal component analysis (PCA) in R (v. 4.2.1).
2.4. Molecular Characterization
For the San Nicolas Island and Atacama Desert strains, genomic DNA was extracted from strains using Qiagen DNeasy Powersoil Pro Kits (Qiagen, Hilden, Germany), following the manufacturer’s protocol, and was eluted in 50 µL of the included elution buffer. K. sahariensis genomic DNA was extracted in the same manner described above but was additionally subjected to previously described modifications to this process to disrupt the exopolysaccharides surrounding the cells [32,35,36]. Polymerase chain reaction (PCR) was used to amplify a partial sequence of about 2000 base pairs of the 16S rRNA gene and the complete associated 16S–23S rRNA internal transcribed spacer (ITS) region using primers VRF1R and VRF2F [37,38]. The PCR reaction mixture containing 1 µL of each primer at 0.01 mM concentration was combined with 12.5 µL of LongAmpTM Taq 2x Mastermix (NEB, Ipswich, MA), 1 µL template DNA (50 ng·mL−1), and 9.5 µL nuclease-free water. The PCR mix was subjected to 35 cycles of denaturing (94 °C for 45 s), annealing (at 57 °C for 45 s), extension (at 72 °C for 135 s), and a final extension (at 72 °C for 5 min). The PCR products were cloned into the lacZ gene of plasmid pSC-A-amp.kan, and the resulting recombinant plasmid was used to transform StrataClone (Agilent, Santa Clara, CA, USA) -competent Escherichia coli cells via heat shock following the manufacturer’s protocol. The transformed E. coli cells were plated on agar-solidified LB-ampicillin plates prepared with 40 µL X-Gal. For each strain, a total of five colonies were picked via blue–white screening. Overnight cultures were prepared, and plasmid DNA containing the gene of interest was extracted and purified with Qiagen QIAprep Miniprep kits. Insertions were confirmed by EcoRI restriction enzyme digestion followed by visualization on 1% TBE agarose gels. Five clones of each strain were sent to Functional Biosciences (Madison, WI, USA) for Sanger sequencing. Primers M13 forward and M13 reverse, along with internal primers VRF5 (5′–TGT ACA CAC CGG CCC GTC–3′), VRF7 (5′–AATGGG ATT AGA TAC CCC AGT AGT C–3′), and VFR8 (5′–AAG GAG GTG ATC CAG CCA CA–3′) [37,39], were used to obtain partial overlapping sequences. Overlapping sequences were error-proofed using Chromas software (v. 2.6.6) and assembled into contigs by alignment with ClustalW [40]. When possible, multiple clones were used to construct consensus sequences.
The molecular analysis of K. sahariensis strains following DNA extraction was performed in a comparable manner; however, the forward primer 27F (5′–AGA GTT TGA TCC TGG-CTC AG–3′) [41] and reverse primer B23SR (5′–CTT CGC CTC CTG TGT GCC TAG GT–3′) [40] were used to amplify the 16S rRNA gene and the 16S–23S rRNA ITS gene under conditions previously described [39]. Additional details on this process have been published [32].
2.5. Phylogenetic Analyses
Sequences of the 16S rRNA gene (a partial sequence of 1157 nucleotides) were subjected to both Bayesian inference (BI) and maximum likelihood (ML) analyses using the CIPRES Science Gateway [42] to obtain posterior probability and bootstrap support values, respectively, for each node in the tree. Bayesian inference analysis was performed using MrBayes on XSEDE 3.2.6 [43], and ML analysis was performed using RAxML-HPC2 on XEDE 8.2.10 [44]. In both cases, the GTR + I + G evolutionary model was used. The BI analysis was run for 95 million generations with the first 25% of samples being discarded as burn-in. The ML analysis was performed using the same alignment as the BI analysis and was run with 1000 bootstrapping iterations. For both analyses, an alignment of 226 taxa was prepared from sequences obtained as a result of this effort along with sequences available in GenBank. Taxa were chosen to be included in the alignment if they either belonged to genera in the same (or a closely related) family, or if they had high sequence identity, as determined by the BLAST search tool in GenBank. The tree resulting from the BI analysis was used as a backbone, and both posterior probability values and bootstrap support values were mapped onto the appropriate nodes. The BI analysis had a mean estimated sample size (ESS) exceeding 4080 for all parameters (range 4083–16,240), far exceeding the 100 accepted as sufficient [45]. The average standard deviation of split frequencies was 0.033, and the potential scale reduction factor (PSRF) was an average of 1.004 for all parameters, indicating nearly complete convergence of the MCMC chains was achieved [46]. The 16S–23S rRNA internal transcribed spacer region sequences were subjected to both BI and maximum parsimony (MP) analyses using an alignment of 498 nucleotides with sequences from all Kastovskya ITS sequences available in GenBank as of 15 June 2024. Maximum parsimony analyses were run in PAUP using the parameters GAP = NEWSTATE, NREPS = 10,000, MULTREES = YES, SWAP = TBR, and STEEPEST = NO. The BI analysis used the GTR + I + G evolutionary model with two data partitions: one for the DNA sequences and the other for coding indels as standard data (1 = nucleotides, 0 = indel, and ? = missing). Bootstrap support values from the MP analysis were mapped onto the respective nodes on the tree produced from the BI analysis. All phylogenetic trees were visualized with Fig Tree [47] and post-edited in Adobe Illustrator (v. 26.4.1; Adobe Systems, San Jose, CA, USA).
Percent identity values among 16S rRNA gene sequences and percent dissimilarity values among 16S–23S rRNA ITS gene sequences were determined using the SHOWDIST command in PAUP [48]. Additionally, hypothetical 16S–23S ITS rRNA region secondary structures for the D1-D1′, Box-B, and V3 helices were identified by alignment of the conserved basal clamps. The basal clamp sequence was determined from previous literature and aligned with the newly acquired sequences in ClustalW [40]. Secondary structures were then visualized with MFold [49] on the UNAFold Web Server (http://www.unafold.org). All settings were set to default except drawing mode, which was set to untangle with loop fix. Structures determined with MFold were then post-edited in Adobe Illustrator. Lines connecting bases were used to represent canonical base pairings, and dots were used to represent noncanonical U-G pairings.
3. Results
3.1. Descriptions of Species in the Genus Kastovskya
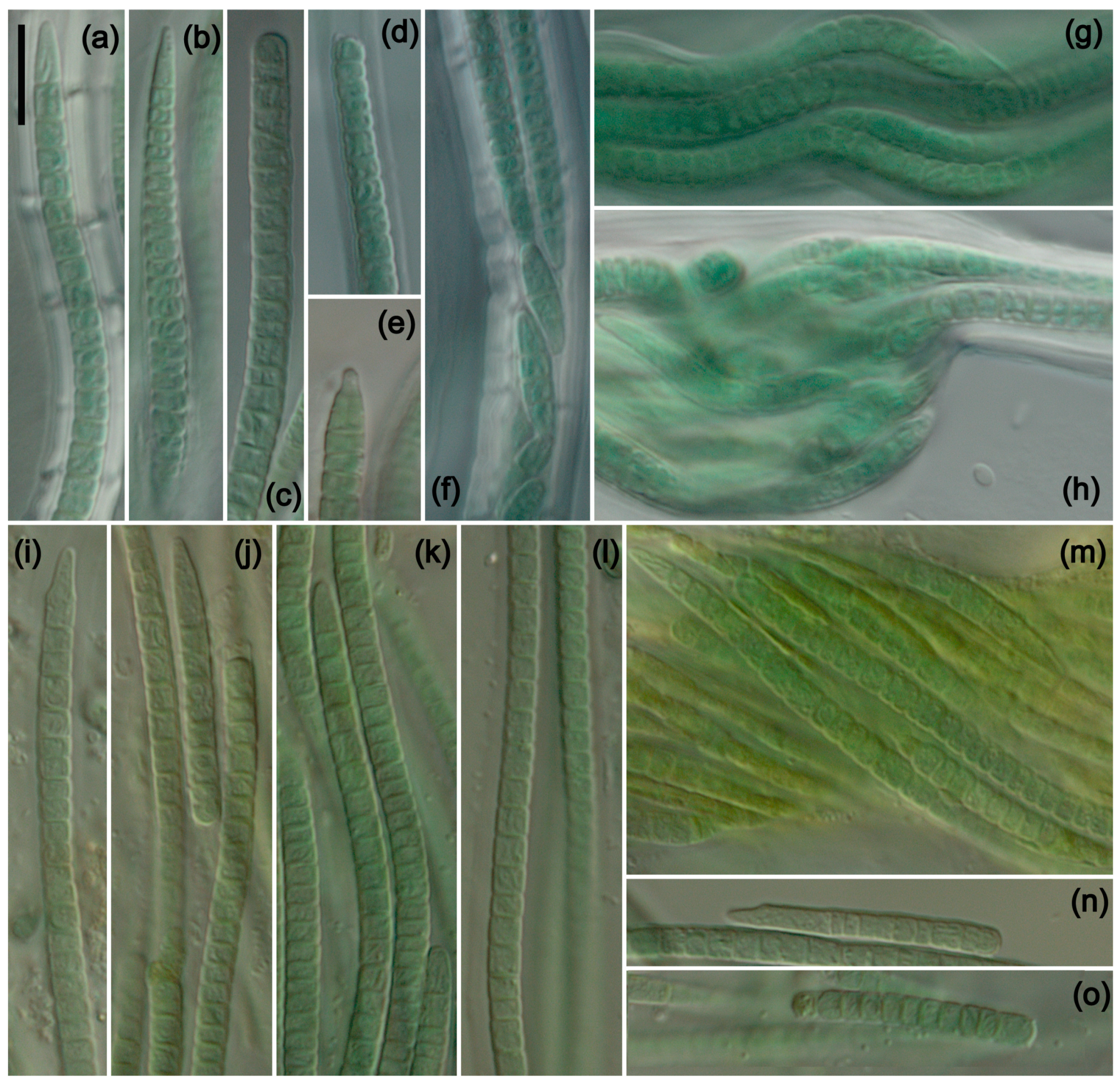
Kastovskya nitens Jusko et J.R. Johans. sp. nov. Figure 2a–h.
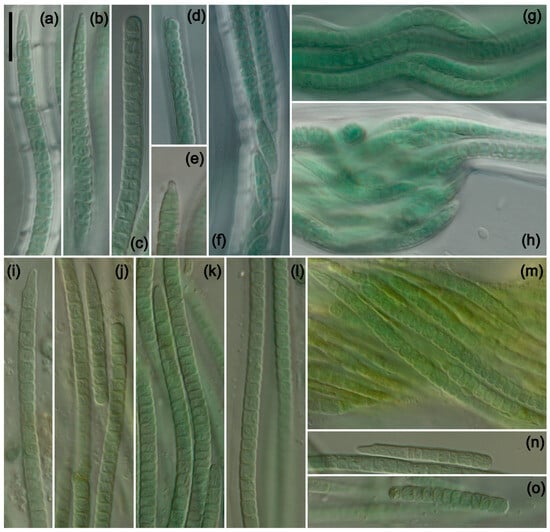
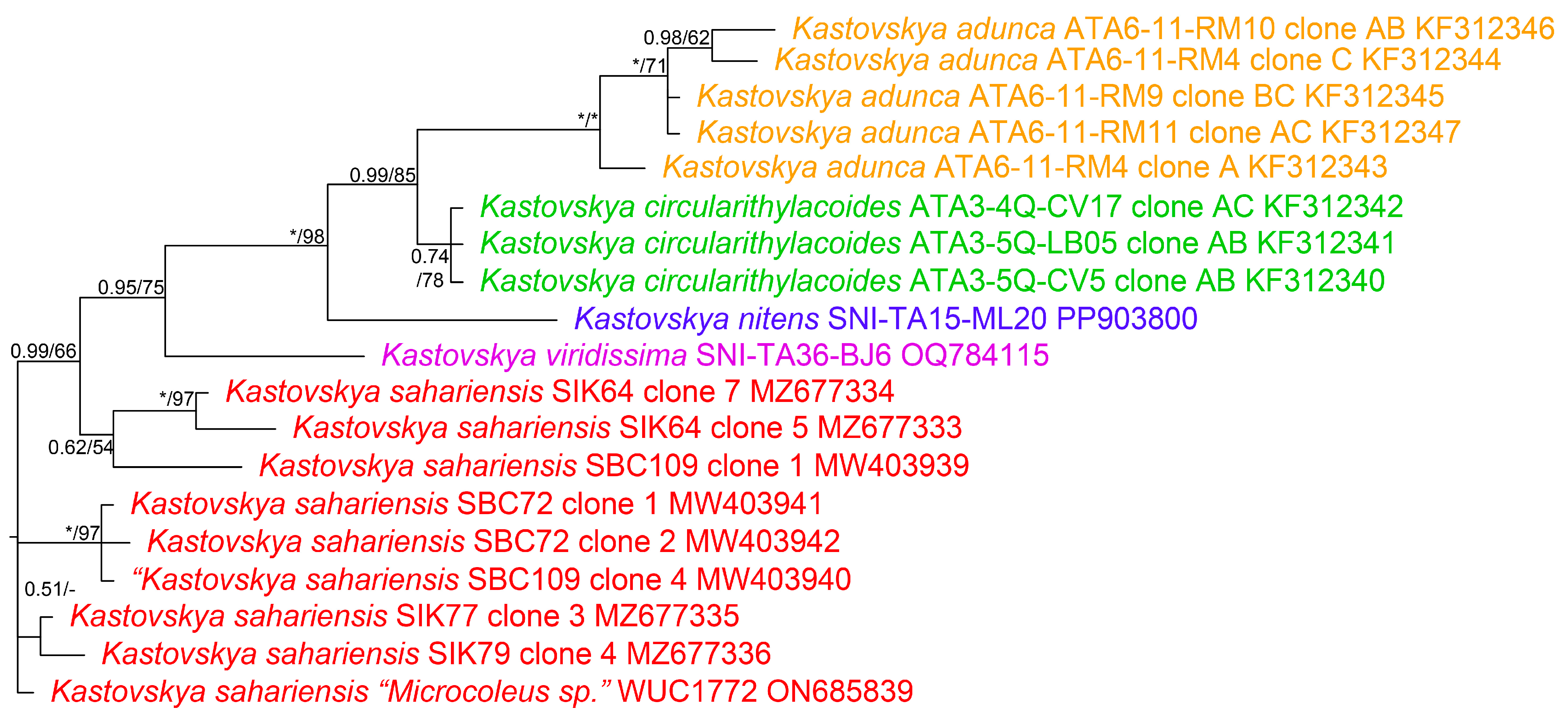
Figure 2.
Photomicrographs of species from San Nicolas Island. (a–h) K. nitens and (i–o) K. viridissima. Scale bar = 10 μm, applies to all panels. (a–e) Single-trichome filaments of K. nitens showing variation in apical cell and sheath morphology. (f) Multiple-trichome filament with hormogonia formation. (g) Characteristic wavy filament with multiple trichomes. (h) Nodule formation along a multiple-trichome filament. (i–k) Filaments of K. viridissima showing the characteristic green color and variation in apical cell morphology. (l) Characteristic sheath of K. viridissima. (m) Ropelike twisted bundle of multiple trichomes. (n,o) Hormogonia.
DIAGNOSIS: Most similar in dimensions and collection sites with K. viridissima, but differing from that taxon in its phylogenetic placement, blue–green color, nodule production, and firm lamellated sheath.
DESCRIPTION: Colonies on agar a green to blue green mat, comprised of parallel-growing thin and medium-length fascicles averaging 4 mm in length, completely and uniformly covering surface of agar, covered in a thick homogenous mucilage layer, appearing distinctly shiny, growth only observed in culture. Filaments containing one to multiple trichomes, from straight to wavy or loosely coiled, with parallel or tightly twisted trichomes, occasionally with a single-trichome filament wrapped tightly around a multiple-trichome filament, regularly forming elongated Nodosilinea-like nodules in which trichomes become knotted, 3.0–33 µm wide. Sheath facultative, attached, colorless, from thin to thick, usually firm, with thick sheaths appearing longitudinally lamellated as well as crosswise lamellated, with edges smooth in straight filaments but becoming buckled when filament bends, 0.3–4 (mean = 1.6) µm wide. Trichomes exhibiting slight gliding motility, especially when young, green or pale to deep blue–green, straight to curled to wavy, with constriction at crosswalls slight to distinct, with crosswalls becoming yellowish with maturity, tapering an average 21% from the midpoint of the apical cell to the midpoint of the eighth vegetative cell, unbranched, 2.4–4.6 (mean = 3.3) µm wide. Cells on average 30% shorter than wide but can be isodiametric to longer than wide, thylakoids often appearing fasciculated, successive phormidiacean cell division occasionally present, 1.2–3.6 (mean = 2.3) µm long. Apical cells on average 50% longer than wide, bluntly rounded to elongated and conical, occasionally with a straight and bulbous or hooked protuberance at apex, 1.6–3.8 (mean = 2.8) µm wide, 2.0–8.6 (mean = 4.0) µm long. Necridia absent. Reproduction by hormogonia that form in series in a common sheath, typically 3–9 cells long.
HOLOTYPE HERE DESIGNATED: SBBG247941! Dried immobilized material prepared from the reference strain, SNI-TA15-ML20.
TYPE LOCALITY: BSC from sample SNI-TA15, San Nicolas Island, Ventura County, CA, USA, 33.230701, −119.518852.
ETYMOLOGY: L. nitens = shiny, referring to the shiny mucilaginous appearance of populations cultured on agar.
Kastovskya viridissima Jusko et J.R. Johans. sp. nov. Figure 2i–o.
DIAGNOSIS: Differing from all other species in the production of a diffluent sheath and yellowish-green color.
DESCRIPTION: Colonies on agar a dark green to dark blue–green mat, creeping on agar but not completely covering surface, composed of both medium-length fascicles and spherical formations of tightly entangled fascicles, appearing matte to slightly glossy, growth only observed in culture. Filaments usually straight, most often solitary but can contain few to many trichomes, trichomes growing parallel or forming tightly twisted ropelike fascicles, with nodule formation not observed, 2.5–36 µm wide. Sheath facultative, attached, colorless, difficult to detect under microscope, thin to medium thickness, diffluent, most often with smooth edges and without lamellation but can rarely exhibit rough edges, sometimes extending beyond trichome, 0.3–4.0 (mean = 1.6) µm wide. Trichomes exhibiting gliding motility especially when young, pale yellow–green to green, slightly constricted at crosswalls, tapering an average of 17% from midpoint of apical cell to midpoint of eighth cell, unbranched, 2.3–3.5 (mean = 3.0) µm wide. Cells on average 27% shorter than wide, cylindrical to barrel shaped, typically shorter than wide but can be isodiametric or longer than wide, thylakoids appearing somewhat fasciculated or concentrated in parietal region of cell, cell division isolated or rarely in meristematic regions, 1.2–3.2 (mean = 2.2) µm long. Apical cells on average 48% longer than wide, bluntly rounded to sharply conical, occasionally with asymmetrical but only minutely hooked protuberance at terminus, 1.7–3.6 (mean = 2.6) µm wide, 1.8–10.2 (mean = 3.7) µm long. Necridia absent. Reproduction by hormogonia, typically with rounded apices, most often 3–8 cells long.
HOLOTYPE HERE DESIGNATED: SBBG238567! Dried immobilized material prepared from the reference strain, SNI-TA36-BJ6.
TYPE LOCALITY: BSC from sample SNI-TA36, San Nicolas Island, Ventura County, California, USA, 33.26033, −119.48643.
ETYMOLOGY: L. viridis = green, −issima = superlative suffix, referring to color of the species being the greenest of all species in the genus.
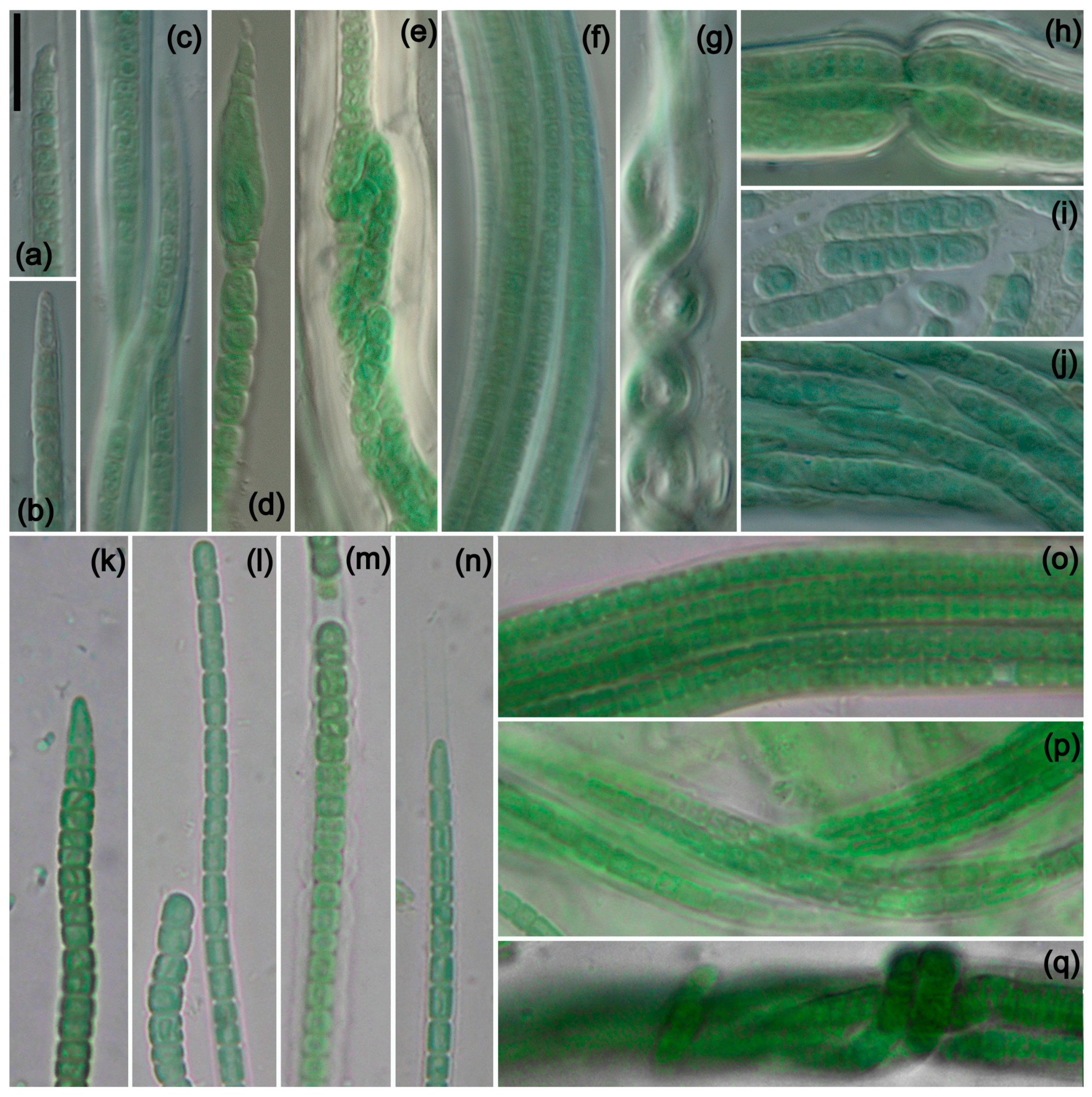
Kastovskya circularithylacoides Jusko et J.R. Johans. sp. nov. Figure 3a–j.
Figure 3.
Photomicrographs of species from South America and Africa. Scale bar = 10 μm, applies to all panels. (a–i) K. circularithylacoides. (k–q) K. sahariensis. (a) Filament with sheath extending beyond trichome, with hook-shaped apical cell characteristic of the genus. (b) Conical apical cell. (a–c,j) Various filaments with characteristic circular formations formed by thylakoids. (d) Rare and unusual enlarged cell near apex of trichome. (e,h) Nodule formation with wide and lamellated sheath. (f) Multiple-trichome filament. (g) Helix formed by twisting of a filament. (i) Hormogonia. (k–n) Trichomes of K. sahariensis showing morphological variation in vegetative and apical cells. (m) Thick sheath with undulated edges. (n) Thin sheath extending beyond trichome. (o–q) Variation in filaments with self-knotting visible in (q). (a–h) Strain ATA3-5Q-CV5. (i,j) Strain ATA3-5Q-LB5. (k,m,q) Strain SIK64. (l,n) Strain SBC72. (o,p) Strain SIK77.
DIAGNOSIS: Differs from all other taxa in the production of circular thylakoids, and is the only hypolithic species that produces nodules.
DESCRIPTION: Colonies on agar a pale green to green mat comprised of short fascicles growing in multiple directions, creeping on agar and eventually covering surface, covered with a thin and uniform mucilaginous layer, appearing slightly glossy, growth only observed in culture. Filaments containing one to multiple trichomes, typically straight and rigid, with trichomes loosely to tightly twisted into ropelike bundles within a common sheath, regularly forming ovular to thin and elongated nodules with thickened sheaths, 2.4–24 µm wide. Sheath facultative, usually attached but sometimes becoming unattached and widely opening near apices, colorless, usually thick and firm, with smooth edges, lamellation rare and typically only present when multiple trichomes are in a common sheath, sometimes extending beyond trichome, 0.2–5.4 (mean = 1.3) µm wide. Trichomes exhibiting slight gliding motility especially when young, green to blue–green, with minute to distinct constrictions at crosswalls, tapering an average of 13% from midpoint of apical cell to midpoint of eighth cell, unbranched, 2.0–3.8 (mean = 2.9) µm wide. Cells cylindrical to barrel shaped, 7% longer than wide, approximately isodiametric but can be shorter or longer than wide, sometimes becoming slightly constricted at cell midpoint with maturity, occasionally minutely granular, thylakoids appearing fasciculated and often forming one to multiple distinct circular formations, successive cell division rare and typically restricted to apical region of trichome, 1.3–5.4 (mean = 3.0) µm long. Apical cells 97% longer than wide on average, bluntly rounded to conical, often becoming elongated, commonly with straight or irregularly hooked protuberance at terminus, 1.6–3.8 (mean = 2.5) µm wide, 2.2–15.2 (mean = 4.8) µm long. Necridia present. Reproduction by hormogonia, typically 3–8 cells in length.
HOLOTYPE HERE DESIGNATED: CBFS A239-1! Dried immobilized material prepared from the reference strain, ATA3-5Q-CV5 (=CCALA 1026).
TYPE LOCALITY: BSC from sample ATA3, Atacama Desert, Chile, −27.783889, −71.034722.
ETYMOLOGY: L. circularis = circular, G. thylakoides = thylakoids, relating to the circular arrangement often formed by the thylakoids.
ADDITIONAL STRAINS: ATA3-5Q-LB5, ATA3-4Q-CV17 (=CCALA 1027).
Kastovskya sahariensis Mehda and Muñoz-Martín sp. nov. Figure 3k–q.
DIAGNOSIS: Most similar in habitat to K. circularithylakoides (both hypolithic beneath quart stones), back lacking nodule formation.
DESCRIPTION: Colonies forming underneath quartz stones, growing in a community with other filamentous and coccoid cyanobacterial taxa, adhering to irregular surface features of stones, light green in color but heterogeneous, appearing granular, large dark green to black bundles up to hundreds of microns visible under medium magnification, growth on agar not observed. Filaments containing one to multiple trichomes, from straight to wavy, with trichomes often becoming twisted into ropelike bundles, occasionally with trichomes becoming tightly coiled around filaments, nodule formation not observed, 2.5–23 µm wide. Sheath facultative, colorless, from thin to thick, typically firm, with straight to undulated edges, longitudinal lamellation rare but present, can extend beyond trichome, 0.4–2.5 (mean = 1.1) µm wide. Trichomes exhibiting gliding motility, green to blue–green, typically distinctly constricted at crosswalls, tapering an average of 17% from midpoint of apical cell to midpoint of eighth cell, unbranched, 2.3–4.8 (mean = 3.0) µm wide. Cells barrel shaped to cylindrical, slightly shorter than wide on average but can be isodiametric or longer than wide, occasionally with slight constriction at midpoint, minutely granular, with thylakoids appearing fasciculated, cell division solitary, rarely in short meristematic regions, 1.4–5.9 (mean = 3.1) µm long. Apical cells 61% longer than wide on average, bluntly rounded to conical, often becoming elongated with maturity, occasionally with asymmetrical growth at terminus growing to one direction, 1.7–4.0 (mean = 2.8) µm wide, 2.4–7.6 (mean = 4.50) µm long. Necridia absent. Reproduction via hormogonia, typically 3–8 cells long.
HOLOTYPE HERE DESIGNATED: UAM 900! Cryopreserved material in a metabolically inactive state prepared from the reference strain, SIK64, deposited into the Algal Culture Collection of Universidad Autónoma de Madrid.
TYPE LOCALITY: Hypolithic biofilm community under quartz rocks on quaternary alluvial terrace with a light (<10%) gradient from sample SIK, Sidi Khouiled, Sahara Desert, Algeria, 31.994389 N, 5.415139 E.
ADDITIONAL LOCALITY: Saline biocrusts from dried salt lake “Chott Kralla”.
ADDITIONAL STRAINS: SIK 77, SIK79, SBC 72, SBC 109.
ETYMOLOGY: L. saharaensis, referring to this species growing in the Sahara Desert.
3.2. Phylogenetic Analyses
3.2.1. Analysis of the 16S rRNA Gene Sequences
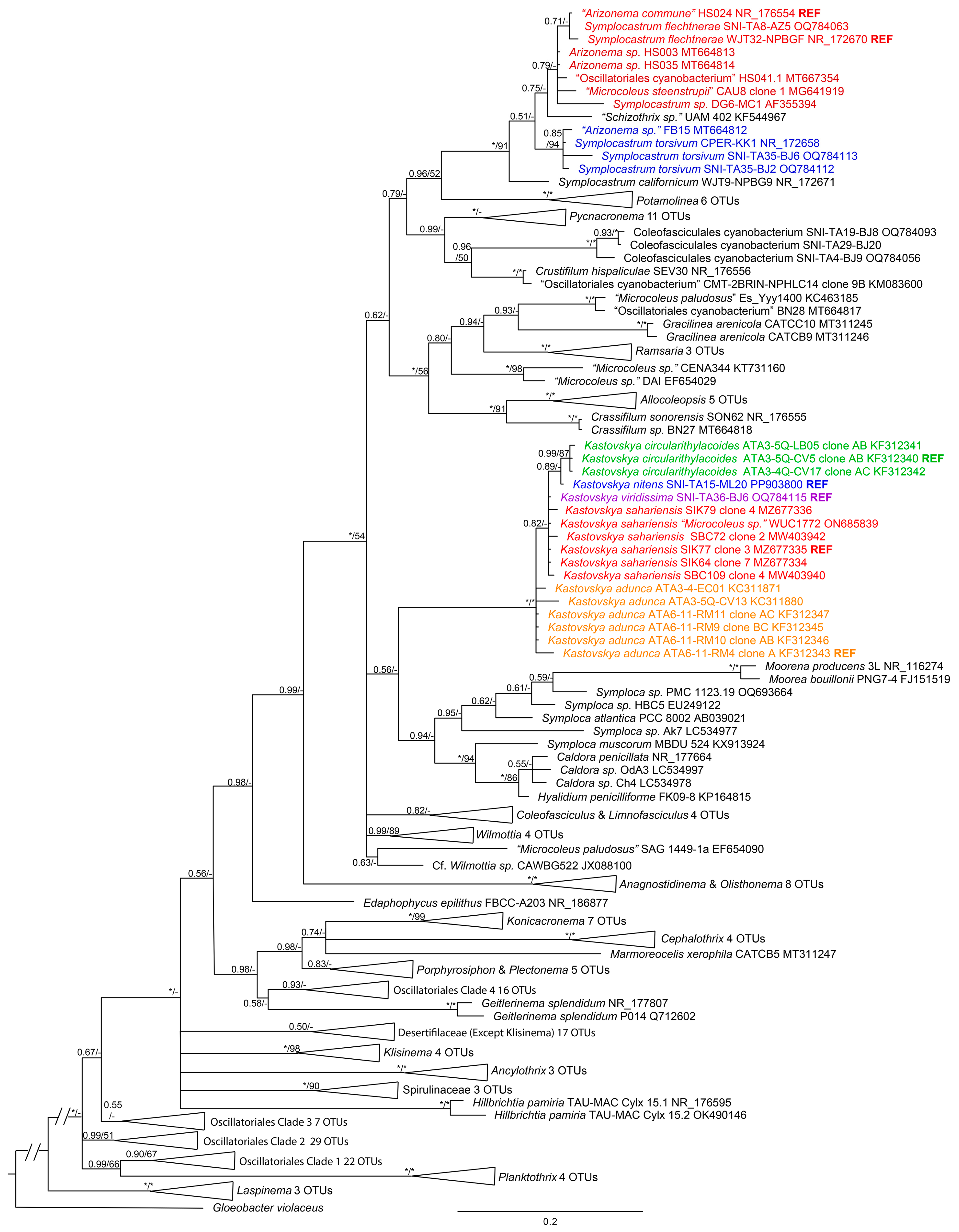
The 16S rRNA gene analysis (Figure 4) performed with all new and existing Kastovskya strains produced a fully supported (1.0 BI posterior probability, 100 ML bootstrap support) genus-level clade sister to the clade containing the genera Symploca, Moorena, and Caldora, which are also in Coleofasciculaceae [12]. However, the species-level relationships were only partially resolved by 16S rRNA gene analysis. K. circularithylacoides strains formed a highly supported (0.99 BI posterior probability, 87 ML bootstrap support) and monophyletic species-level clade within the genus, sister to K. nitens, which also fell onto a single-strain branch. This group fell sister to all SIK and SBC strains (K. sarahiensis), a strain from India identified as Microcoleus vaginatus WUC1772 (which we identify as K. sahariensis), and K. viridissima, with all of these strains forming an unresolved paraphyletic polytomy. The remaining strains, representative of K. adunca, fell sister to the rest of the strains but formed a basal paraphyletic cline. All strains in all species had >98.7% (min.
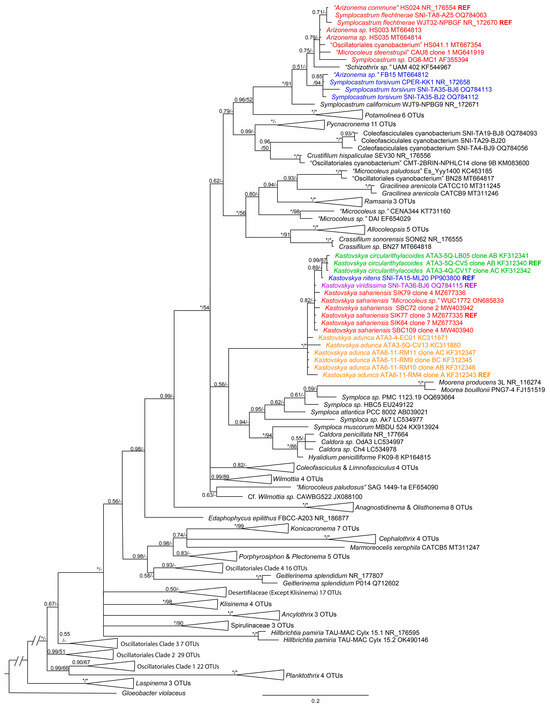
Figure 4.
Bayesian inference (BI) analysis based on 16S rRNA gene sequence alignment for 226 cyanobacterial taxa, with posterior probabilities and maximum likelihood (ML) bootstrap support values superimposed on nodes. Full nodal support (1.0 BI, 100 ML) is indicated by (*). Support below 0.5 BI and 50 ML is indicated by (-). Strains in same color font are the same species. The uncollapsed tree for this analysis is presented in the Supplementary Materials (Figure S1). 98.8% identity in the 16S rRNA gene; however, percent identity was higher within a species than among species in all cases (overall average 99.8% identity within species, 99.4% among species, see Table 1), highlighting the necessity for further molecular analysis.
3.2.2. Analysis of the 16S–23S ITS rRNA Region
The phylogenetic analysis of the 16S–23S ITS region (Figure 5) was more informative in resolving relationships at the species level. The strains belonging to K. adunca formed a fully supported (1.0 BI posterior probability, 100 MP bootstrap support) clade sister to the set of strains belonging to K. circularithylacoides. Kastovskya circularithylacoides strains formed a well-supported (0.74 BI posterior probability, 78 MP bootstrap support) clade sister to K. nitens and K. adunca. Both K. nitens and K. viridissima fell on lone branches with high node support (1.0 BI posterior probability, 98 MP bootstrap support and 0.95 BI posterior probability, 75 MP bootstrap support, respectively). All four of the Chilean and Californian species were in a well-supported clade (0.95 BI posterior probability, 75 MP bootstrap support), separating these species from all K. sahariensis strains, which had internal structure but represented a monophyletic species (Figure 5).
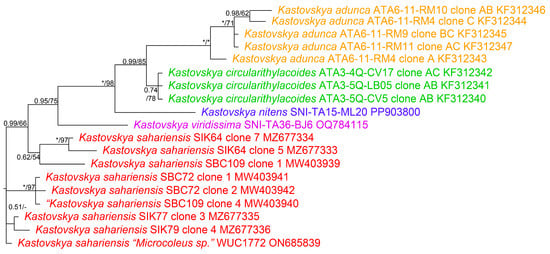
Figure 5.
Bayesian inference (BI) analysis based on 16S–23S rRNA internal transcribed spacer gene sequence alignment of 19 taxa in genus Kastovskya, with posterior probability and maximum parsimony (MP) bootstrap support values superimposed on nodes. Full nodal support (1.0 BI, 100 MP) is indicated by (*). Support below 0.5 BI and 50 MP is indicated by (-). Strains in same color font are the same species.
Percent dissimilarity among the Saharan and Punjab strains was 1.56%, further supporting their placement in a single species. All K. circularithylacoides strains were exactly identical in the ITS region and an average of 4.2% dissimilar to other strains (Table 1). Kastovskya nitens and K. viridissima were only represented by one strain each but were an average of 4.3% and 4.4% dissimilar to other strains, respectively. All species groups averaged between 4% and 5% dissimilarity from other groups (Table 1), which is good evidence of lineage separation. There was no evidence of multiple operons being included in the analysis, and thus these dissimilarity values showed these taxa are distinct from each other.

Table 1.
Percent similarity of 16S rRNA gene sequences and percent dissimilarity of 16S–23S ITS regions among Kastovskya strains. Taxa belonging to the same species share the same color of highlighting. The boldface font indicates comparisons among reference strains for the species shown.
3.2.3. Analysis of the ITS Secondary Structures
In general, the hypothetical ITS-region secondary structures (Figure 6) were highly conserved among taxa. In the D1-D1′ helix, K. nitens (Figure 6a) was unique from other taxa in having only four nucleotides in the terminal loop, while the remainder of the taxa had five. Additionally, this was the only taxon with unpaired guanines followed by a noncanonical U-G pairing along the stem. It also is the only strain with only three A-U pairings prior to the large subapical bilateral bulge along the stem. Both K. nitens and K. viridissima (Figure 6a,b) had four base pairs directly beneath the terminal loop, whereas all K. sahariensis, K. adunca, and K. circularithylacoides had three (Figure 6c–e). Kastovskya viridissima (Figure 6b) was the only strain with a U-G pairing directly at the base of the subapical bilateral bulge, and was also the only strain with only four bases on the 5′ side of the bilateral bulge and five on the 3′ side, while the rest had five and six, respectively. Kastovskya circularithylacoides (Figure 6d) was separate from K. adunca (Figure 6e) in that it had differing sequences on both the 5′ side of the subapical bilateral bulge and on the basal 3′ unilateral bulge. Both of these taxa differed from K. sahariensis (Figure 6c) in the presence of an unpaired G-A along the stem, as well as in the sequence of the subapical bilateral bulge. There was some variation in secondary structure among K. sahariensis strains; however, all shared the same and unique 5′–A-C-A-A–3′ sequence for the four bases on the 5′ side of the subapical bilateral bulge.
The Box-B helices (Figure 6f–j) were relatively uninformative for separating taxa. Kastovskya nitens shared an exactly identical helix with all of the K. circularithylacoides strains (Figure 6f,i). Kastovskya adunca (Figure 6j) was the only species with a G in place of a C in the second mismatch across from an unpaired 3′ A (Figure 6g). Kastovskya viridissima was unique in the sequence of its terminal loop and was the most divergent helix. The V3 helices (Figure 6k–o) were only slightly variable among taxa. In this case, K. circularithylacoides strains were exactly identical to K. nitens (Figure 6k,n), and K. viridissima was unique from all other strains with four adenines on the 5′ side of the largest bilateral bulge (Figure 6l). Kastovskya circularithylacoides and K. viridissima (Figure 6l,n) were separated from K. adunca by stem sequence. K. adunca had four base pairings after the large bilateral bulge followed by an unpaired A-A (Figure 6o), while K. circularithylacoides and K. viridissima had only three base pairings followed by an unpaired C-A (Figure 6l,n). All but one strain of K. sahariensis (SIK64) shared an unpaired uracil just upstream of the terminal loop, which is absent in all strains of other species (Figure 6m). This species was also the only to exhibit an A-U pairing rather than a G-C pairing beneath the terminal loop. While there was significant overlap among the secondary structures, each species had a unique combination of structures and a number of features only seen within each taxon.
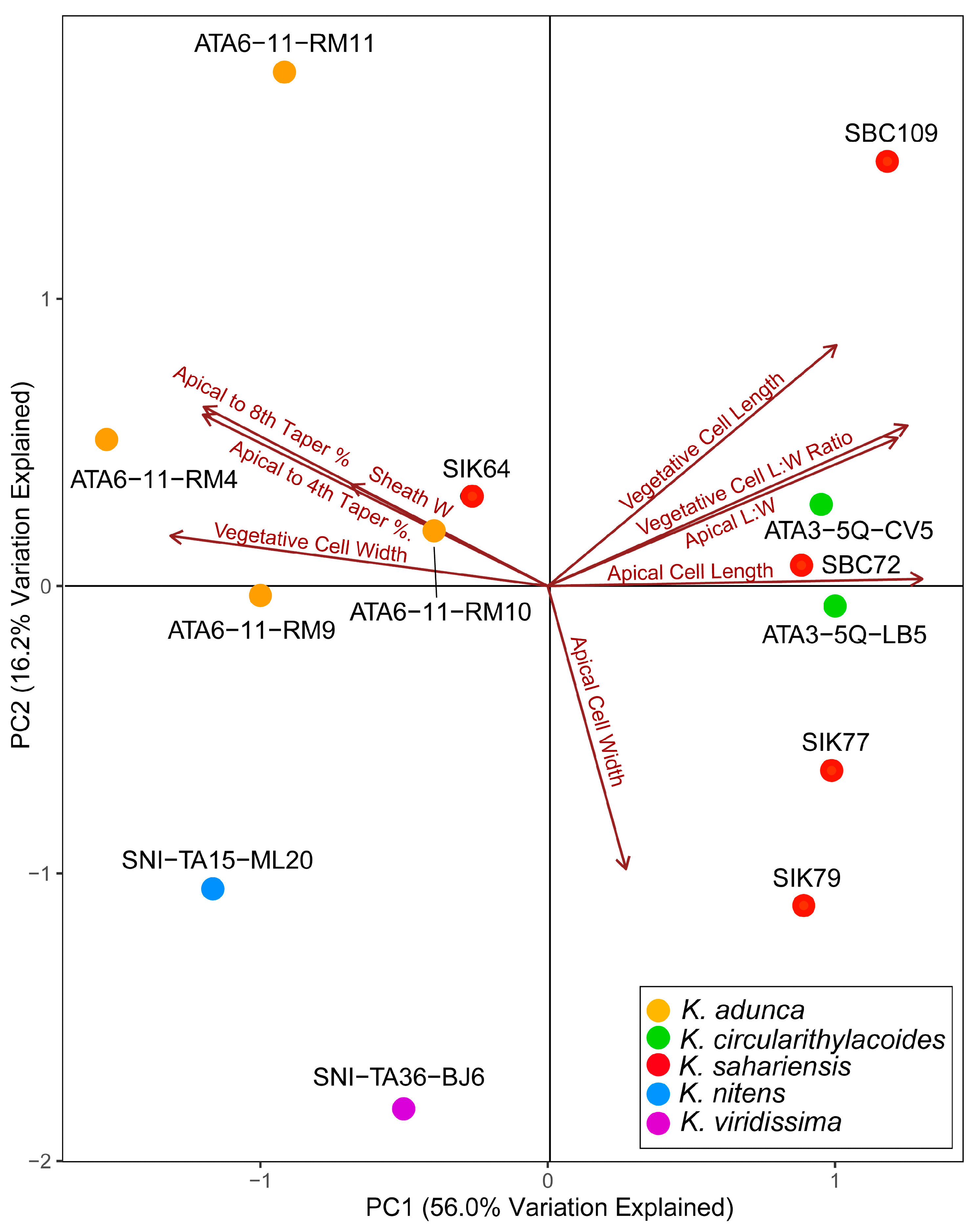
3.3. Morphological Analysis
Morphology was compared among strains to determine if they could be separated into species based on size, size ratios, and degree of tapering. The principal component analysis conducted with average values showed relatively strong separation of strains into their respective species (Figure 7). Axes PC1 and PC2 explained 56.0% and 16.2% of the variation in the data, respectively, for a total of 72.2% variation explained. Apical cell length and vegetative cell width were the traits most closely associated with PC1. Apical cell width was the trait most strongly associated with PC2. The remaining traits contributed to both axes but were more strongly associated with PC1. In general, strains were fairly strongly separated into species based on the morphometric PCA. Kastovskya adunca strains fell exclusively into the upper left quadrant, with only strain SIK64 present as an outlier also in that quadrant. Both K. circularithylacoides strains fell on the far right of PC1, near the axis of PC2. All of the K. sahariensis strains except SIK64 fell in nearly identical positions along PC1 axis on the right side. Both K. nitens and K. viridissima fell into the lower left quadrant. These strains were separated along both PC1 and PC2 and were the lone strains in the quadrant. Although the strains did not form perfect species groupings, it is clear that each lineage has identifiable trends in morphology and size. This, along with other qualitative differences in morphology, provides additional support for distinct lineages worthy of taxonomic recognition.
3.4. Ecological and Biogeographical Considerations
The genetic and morphological analyses alone were able to separate strains into species, but ecology and biogeography were also considered. Kastovskya adunca and K. circularithylacoides were both isolated from the Atacama Desert in Chile in relative proximity; however, there were significant differences in life habits. Kastovskya adunca was isolated from a stream-moistened and consolidated soil sample in an open intershrub space, while K. circularithylacoides was found growing underneath quartz rocks with moisture supplied primarily by the coastal fog. Kastovskya nitens and K. viridissima were both isolated from San Nicolas Island; however, K. nitens was isolated from a poorly developed algal crust sample just above a natural spring, while K. viridissima was isolated from a lichenized BSC near the bottom of a canyon. These species are separated from the Atacama by vast geographical distance, and the same is true for K. sahariensis, which was isolated in Algeria.
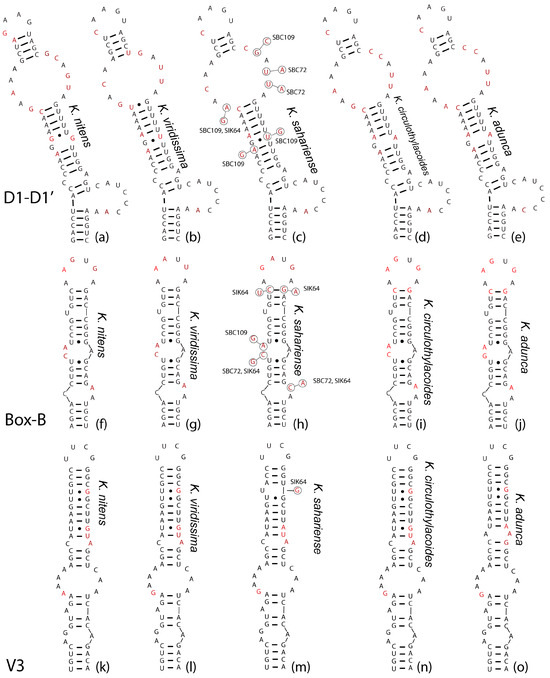
Figure 6.
Secondary structures of conserved helices in the 16S–23S internal transcribed spacer (ITS) regions of all Kastovskya species, including the D1-D1′, Box-B, and V3. (a–e): D1-D1′ helices. (f–j): Box-B helices. (k–o): V3 helices. Bubbles on K. sahariensis strains represent variation among strains within the species. Nucleotides that were variable among species are in red font to improve ability to see where sequence differences exist.
Figure 6.
Secondary structures of conserved helices in the 16S–23S internal transcribed spacer (ITS) regions of all Kastovskya species, including the D1-D1′, Box-B, and V3. (a–e): D1-D1′ helices. (f–j): Box-B helices. (k–o): V3 helices. Bubbles on K. sahariensis strains represent variation among strains within the species. Nucleotides that were variable among species are in red font to improve ability to see where sequence differences exist.
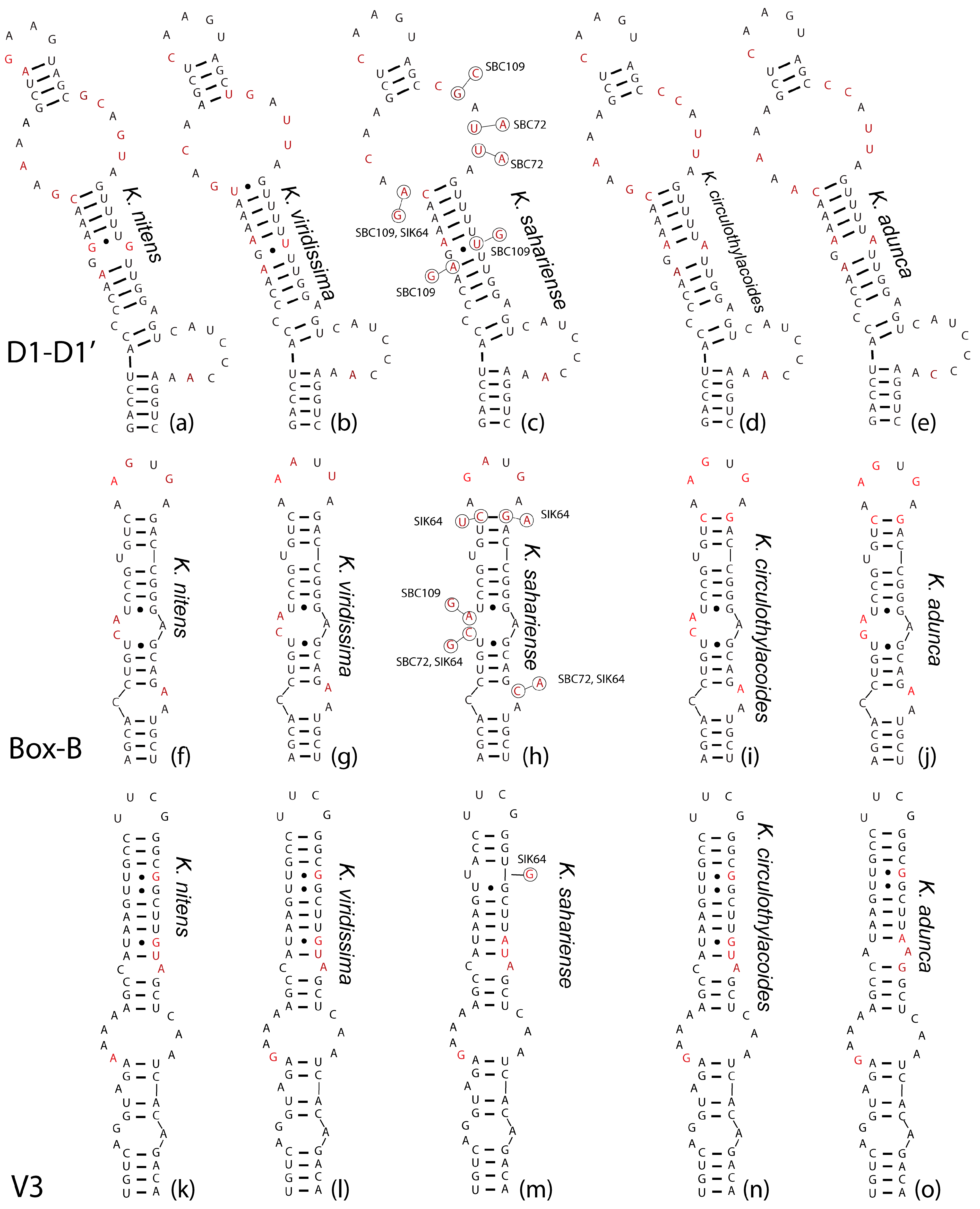
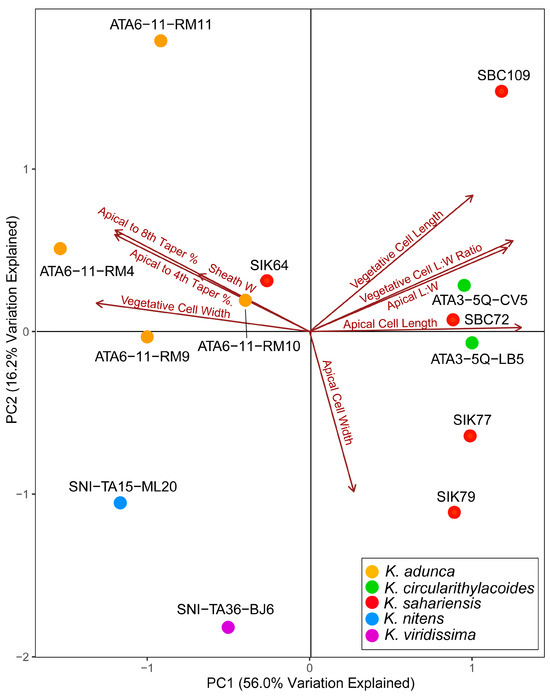
Figure 7.
Morphometric principal component analysis (PCA) of Kastovskya strains based on average measurements of strain apical cell length and width, vegetative cell length and width, sheath width, taper from the midpoint of the apical cell to the midpoint of the fourth and eighth vegetative cells, and length–width ratios for vegetative and apical cells. Points represent individual strains and are colorized to their respective species.
Figure 7.
Morphometric principal component analysis (PCA) of Kastovskya strains based on average measurements of strain apical cell length and width, vegetative cell length and width, sheath width, taper from the midpoint of the apical cell to the midpoint of the fourth and eighth vegetative cells, and length–width ratios for vegetative and apical cells. Points represent individual strains and are colorized to their respective species.
3.5. Genus Symplocastrum
It was determined by 16S rRNA gene phylogenetic analysis that the genus Arizonema is clearly a later synonym for Symplocastrum (Gomont) Kirchner [29]. In the 16S rRNA phylogenetic analysis (Figure 2), Arizonema commune, the type species for the genus [28], together with Arizonema spp. HS003 and HS035, fell into a well-supported clade (0.71 BI posterior probability) with the reference strain (WJT32-NPBGF) for Symplocastrum flechtnerae Pietrasiak et Mühlsteinová [30], with which it also shares 99.66% nucleotide identity. Arizonema sp. FB15 can be phylogenetically placed in Symplocastrum torsivum Pietrasiak et Johansen [30]. Given the phylogenetic placement, high 16S rRNA gene percent identity, and lack of ITS data for the all Arizonema strains, there is insufficient evidence for Arizonema to be recognized as taxonomically separate from Symplocastrum. Given that Arizonema commune is a later synonym of both the genus and the species S. flechtneriae, a novel combination is not warranted.
4. Discussion
4.1. Polyphasic Approach
It remains important to consider multiple sets of evidence for taxonomic determinations, and the polyphasic approach [27,50] remains a key tool to decide when species are unique lineages worthy of taxonomic recognition. In the case of Kastovskya, each part of the evidence in isolation provided underwhelming support for describing the new species; however, taken collectively, the full set of evidence provides a clear and robust argument for taxonomic description. It has been shown that the 16S rRNA gene historically used for prokaryotic species delimitation is insufficiently variable for this purpose [51], and the number of cryptic species that have been described recently shows the shortcomings of a morphology-alone taxonomic framework [52,53]. Given that using each of these metrics alone would lead to considerably different sets of species being described, it is clear that the tandem use of these data sets is the most preferable means by which to make taxonomic decisions.
4.2. Molecular Analyses
The 16S rRNA gene percent similarity values among Kastovksya species are higher than the current <98.7% threshold for the separation of bacterial species [54], but this is unsurprising given that similar trends have been observed in other groups, such as members of the Nostocales [55,56,57]. Although the arbitrary percent identity threshold for species separation was not met, it was nevertheless clear that 16S rRNA gene percent identity was higher among conspecific strains than among different species (Table 1). Although the 16S rRNA gene was insufficient for resolving species-level identities in most cases, it was able to group species into a clear genus-level clade and was able to resolve species in the cases of K. circularithylacoides and K. nitens (Figure 2). It should be noted that while the 16S rRNA gene remains a useful tool for taxonomy, evidence of its limitations is mounting. It has been shown that the gene evolves more slowly than other genes and that changes in this gene may not reflect significant overall genomic divergence among taxa [58,59]. The average within-species identity value (99.8%) is artificially deflated both by a somewhat divergent clone of the K. adunca reference strain and by the somewhat variable 16S rRNA gene sequence among K. sahariensis strains that were isolated from nearby but different sampling sites. These strains were grouped together as a single species in the absence of sufficient evidence to separate them (affecting overall identity values); however, it is likely that strains from the SIK and SBC sampling sites are different lineages on their way to accumulating diagnosable differences. If the scope is narrowed, the reference strain of K. sahariensis is 99.14% identical to the reference strain of K. adunca, while on average, K. sahariensis strains share 99.4% identity to K. adunca.
The 16S–23S rRNA ITS region remains a better tool to resolve finer, species-level relationships, as it is conserved at the species level but variable among congeneric species [55,60,61,62]. Strains belonging to different species averaged 4.5% dissimilarity from each other if all comparisons were considered, whereas strains belonging to the same species averaged about 0.8% dissimilarity (Table 1). Dissimilarity values also aligned to an extent with geographical distribution and ecological niche. K. adunca was most similar to K. circularithylacoides (also from the Atacama Desert) at 2.94%, but K. sahariensis was more similar to K. circularithylacoides than it was to K. adunca, an interesting observation given that the two were both isolated from underneath translucent quartz rocks. Similarly, the SIK strains of K. sahariensis that were isolated from a hypolithic community were more similar to K. circularithylacoides on average than the SBC strains that were isolated from biocrusts, at 4.2% and 4.6%, respectively. Although a dissimilarity of >7% has been considered strong evidence of lineage separation that justifies separating species [24,63,64,65,66], a dissimilarity of <7% but >3.0% has been used to separate taxa if other data also indicate lineage divergence [14]. The ITS phylogenetic analyses (Figure 3) clearly placed strains into highly supported and monophyletic clades, and the tree aligned with the 16S rRNA tree. Given that 16S rRNA phylogeny, 16S–23S ITS phylogeny, ITS dissimilarity, ITS secondary structure, morphology, ecology, and biogeography all aligned well, the evidence is strong for taxonomic-description-worthy lineage separation. It has become clear that 16S rRNA gene identity and 16S–23S ITS gene identity do not necessarily change proportionally to each other, nor do they behave the same across orders and families. Consequently, it remains important to consider both genes when performing phylogenetic analyses for the purpose of taxonomic descriptions.
4.3. ITS Structures
Analysis of the hypothetical ITS secondary structures (Figure 4) provided qualitative evidence of lineage separation among the species, and each species has a unique combination of features in their helices distinct from other species. Overall, the structures appeared highly conserved with only relatively minor differences among them. Although we do not yet understand the function of the ITS secondary structures, they are consistently present in all cyanobacterial ITS regions and consistently not present in the ITS regions in other bacteria. This, in conjunction with highly similar conserved structures among congeneric ITS operon orthologs, suggests that they do serve some critical function in ribosome assembly or otherwise. This suggests the region is subjected to stabilizing evolutionary pressure and that mutations are rare and probably deleterious in most cases. As such, accumulated differences in these regions suggest lineage separation. It was observed that dissimilarity values in the helices tended to be less than the overall ITS dissimilarity, suggesting that while the structures provide visual evidence of divergence, it is important to assess overall ITS dissimilarity when comparing orthologs, rather than just the structures. Analyses of the 16S–23S ITS region are highly subject to the effects of multiple ribosomal operons in a genome, and evidence is mounting that widespread issues may have arisen due to the lack of consideration of this phenomenon [66]. Kastovskya strains were checked for the presence of multiple operons via alignment, and although there was no evidence of multiple operons among the sequences, it is crucial that this be considered in every publication using ITS data for systematics.
4.4. Morphology, Ecology, and Biogeography
Although the Kastovskya species look quite similar under the microscope, they each have unique morphological features that can be used to identify them to the species level based on morphology alone. K. nitens has a regularly lamellated and wide sheath that is quite different from the other taxa (Figure 2a,b,f). This species also commonly forms highly wavy filaments (Figure 2g), a feature that was not observed in the other species with regularity. Additionally, the macroscopic growth of the species is the only one that exhibited a thick, highly glossy mucilaginous layer on the surface. K. viridissima has the softest, most diffluent, and thinnest sheath on average (Figure 2k,l), which was immediately discernable from all other species via microscopy. Perhaps even more obvious is the color of this species (Figure 2i–o), which is by far the lightest and greenest (as opposed to blue–green in the other species), and this trait was therefore chosen as the species epitaph. K. circularithylacoides consists of strains originally assigned to K. adunca; however, upon further analysis, it was determined that they are discernable from each other by morphology. K. circularithylacoides vegetative cells are over 30% longer on average than those of K. adunca, and vegetative cells are around 15% longer. As another obvious difference between these two species, K. adunca tapers an average of 22% from the midpoint of the apical cell to the midpoint of the eighth cell, while K. circularithylacoides tapers an average of only 13%. Some interesting patterns were observed among the taxa, such as the lack of necridia in both K. viridissima and K. nitens (Figure 2) but the presence thereof in the Atacama Desert taxa. This may be related to their preferred means of dispersal, as necridic cells have been linked to both hormogonia release and subsequent adhesion to a substrate [67,68]. Furthermore, nodule formation was observed in K. nitens (Figure 2h), K. circularithylacoides (Figure 3e), and K. adunca but never observed in K. viridissima, even in identical growth conditions and high photographic sampling of the strain. Another interesting pattern observed among the Kastovskya species was the varying degrees to which each formed the characteristic curved, hook-like protuberance on the apical cells. The type species, K. adunca, was named for this feature (L. adunca = “curved”) and tends to form them regularly and with a severe curve. Although all species of Kastovskya formed asymmetrical apical cells to some extent, some, such as K. viridissima, formed them only very rarely and with only a minor asymmetry (Figure 2n). Generally, they were more bluntly rounded and curved to a lesser degree. The function of these apical cells is unclear, but their form and frequency seem to vary significantly among taxa in the genus.
Ecological and biogeographical differences were also an important consideration in taxonomic decision making. While obvious, several of the species are separated by vast, nearly impossible-to-traverse geographical distances, and there is no evidence of the presence of species outside of their narrow sampling sites, let alone on different continents. There is also significant ecological variability, even among species living in close proximity. K. circularithylacoides strains are quite obviously separated genetically and morphologically from K. adunca; however, they may be even further separated by their niche. K. circularithylacoides was found growing under quartz in the Atacama, while K. adunca was not. Interestingly, the reference strain of K. sahariensis was also found living under quartz in the Sahara, suggesting this trait may be an evolutionarily relevant characteristic shared between these species, but not by every member of the genus. As a note, both K. circularithylacoides and K. sahariensis had similar measurement values for their apical cells and vegetative cells and were also the two species with the lowest degree of tapering. Strains representing these two species clustered closely in the principal component analysis, suggesting that the overall morphometric similarity among them is quite high. While it is unclear why this is the case, this overlap may suggest that these taxa are receiving some advantage by sharing these features when it comes to living under quartz rocks, either as a result of being a symplesiomorphy or as a case of convergent evolution among species.
The utility of morphometric analyses has been potentially overlooked and underestimated as a tool in taxonomic studies. Several of the species presented in this paper appeared to be morphologically cryptic relative to each other with similar measurements (e.g., K. adunca and K. circularithylacoides), yet they were able to be well separated based on high sample size measurement averages of multiple morphological features subjected to a principal component analysis (PCA) (Figure 5). Without such an analysis, it would be difficult to determine visually that the cell sizes were both different and relatively consistent across strains belonging to the same species. The PCA axis 1 explained 56% of the variation in the data. Interestingly, K. adunca and K. circularithylacoides were strongly separated along PC1, even though they were isolated near to each other (183 km), while K. circularithylacoides and K. sahariensis were not well separated. This is notable given that both species were found growing underneath quartz but are separated from each other geographically by nearly 10,500 km. K. viridissima and K. nitens fell into the same quadrant on the PCA but were separated along both PC1 and PC2, providing evidence of lineage separation even between taxa living in the closest proximity. While the PCA did not perfectly group taxa into species, such as with K. sahariensis outlier strain SIK64, the clustering is certainly nonrandom and conforms well to the genetic data. It should be noted that this type of analysis is highly subject to sampling bias and the effects of phenotypic variation, so it is of the utmost importance to standardize both growth conditions and the sampling process itself to ensure the observed values closely represent true population means. Morphometry of terrestrial cyanobacteria can also be potentially problematic given that strains may appear differently in nature than they do in culture, and generally only cultured growth is observed [11,69,70]. The analysis performed in this publication also suggests that typically overlooked yet quantifiable features (e.g., the extent to which a trichome tapers) have taxonomic utility. Reporting that species are morphologically cryptic only suggests they are indiscernible by the extent to which their morphologically has been investigated; however, it may be the case that finer, more intentional morphological analyses using data ordination techniques may reveal diagnosable differences where they have been presumed to be lacking.
The distribution of Kastovskya bears close resemblance to that of other genera that have been isolated in both the Americas and in Africa. While the exact means of dispersal are unclear, the most likely scenario seems to be that large-scale dust storms in the Sahara can carry cyanobacteria on dust particles across the Atlantic, as previously proposed as a part of the San Nicolas Island study [14,21]. The observed distribution of Kastovskya further supports this hypothesis. As an interesting note, all of the Kastovskya species were isolated between 25° and 35° latitude, north or south. This approximately lines up with the descending portion of Hadley cells, in which warm equatorial air is lifted, cooled, and descends [71,72]. This same phenomenon is partially responsible for the creation of deserts [73], suggesting an instance of correlation over causation. It is possible that the progenitors of the current species may have been dropped at these latitudes before subsequently becoming established.
In this publication, we have now provided a direct link between multiple congeneric species on three continents, and these species share a distinctly similar distribution to other taxa observed as a part of this study. While we do not have direct evidence (e.g., aerial collections) of cyanobacteria during a transatlantic dispersal event, the evidence continues to support this hypothesis. There is a chance that this distributional pattern is a consequence of patchy and limited sampling efforts. The number of cyanobacterial taxa being described from locations known to have high overall biodiversity is relatively low, largely due to the lack of sampling [74]. Africa in particular is greatly understudied due to the lack of active research and risks associated with sampling in politically unstable areas. However, a number of biodiversity surveys have been performed in the regions between the now-multiple Kastovskya isolation sites, and representative strains have not yet been found to exist. This suggests that dispersal and colonization events are rare and that establishment is not due to gradual, small-scale dispersal.
4.5. Further Afield
It was determined during the course of the Kastovskya 16S rRNA gene phylogenetic analysis (Figure 2) that Arizonema [28] is clearly a later synonym for Symplocastrum, which was described long before and thus has taxonomic priority [30]. Furthermore, the only existing species in the genus, A. commune, was determined to be a later synonym of the species Symplocastrum flechtnerae [30]. The 16S rRNA gene percent identity between the type strains of A. commune and S. flechtnerae was 99.66%, which is higher than the similarity among Arizonema strains, and the percent identity between A. commune and species S. torsivum and S. californicum is 98.54% and 98.11%, respectively, indicating that all three are clearly members of the same genus. As such, the genus Arizonema should not be recognized.
5. Conclusions
This study is in agreement with other studies that desert soils are repositories of undiscovered microbial diversity. Four new species of the rare, until-now monospecific genus Kastovskya were discovered and described using a polyphasic approach. Although molecular data were the primary means of recognizing the diversity, morphological data confirmed conclusions based on the molecular analysis. The observation that none of the species in this rare genus occurred in geographically distant locations supports the hypothesis that either dispersal or establishment (or both) is limiting, and upon establishment, colonists in this genus are isolated long enough to become locally adapted and molecularly separated lineages worthy of taxonomic recognition. The two species with hypolithic niches (growing beneath quartz rocks) were morphologically similar, suggesting that this particular niche may have selected for convergent morphologies. Intraspecific variation in the ITS sequence was evident in several populations of K. sahariensis, although the secondary structure of the conserved domains was retained.
The high similarity and phylogenetic placement of the recently described genus Arizonema with several species in the long established genus Symplocastrum indicates that this more recent taxon should be subsumed into Symplocastrum.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/d16080474/s1, Figure S1: Uncollapsed 16S rRNA BI analysis with ML bootstrap values mapped to shared nodes.
Author Contributions
Conceptualization, methodology, validation, and formal analysis, B.M.J. and J.R.J.; investigation, B.M.J., S.M., E.P. and M.Á.M.-M.; resources, J.R.J.; data curation, B.M.J.; writing—original draft preparation, B.M.J.; writing—review and editing, J.R.J., B.M.J. and M.Á.M.-M.; supervision, J.R.J.; project administration, J.R.J.; funding acquisition, J.R.J. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by a subcontract from the Santa Barbara Botanical Garden with funding from U.S. Navy Cooperative Agreement Number N62473-21-2-0002 and additional support from Grantová Agentura České Republiky, Grant/Award Number: GAČR 22-06374S. The research on cyanobacteria of Sahara Desert strains was supported by project CGL2017-86258-R of the Spanish Government and European Regional Funds and by the Directorate General of Scientific Research and Technological Development, Algeria (DGRSDT), through the Val-Ped-Oasis project (Valorisation des données pédologiques des oasis du Bas Sahara) (grant number FNRSDT-2017).
Institutional Review Board Statement
Not applicable.
Data Availability Statement
The uncollapsed 16S rRNA phylogeny can found in the Supplementary Materials, Figure S1, which also contains information about NCBI GenBank accession numbers.
Acknowledgments
William Hoyer and Kristen Lehman assisted in the collection of the soil samples from San Nicolas Island; Karina Osorio-Santos and Lauren Baldarelli assisted in the collection of soil samples and the isolation of strains from the Atacama Desert, Chile. Faculty and students at John Carroll University provided advice, training, and support, and we thank in particular Chris Sheil, Rebecca Drenovsky, Amanda Szinte, Mathew Luknis, and Salvadore Peron.
Conflicts of Interest
The authors declare no conflicts of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
References
- Johansen, J.R. Cryptogamic crusts of semiarid and arid lands of North America. J. Phycol. 1993, 29, 140–147. [Google Scholar] [CrossRef]
- Weber, B.; Belnap, J.; Büdel, B.; Antoninka, A.J.; Barger, N.N.; Chaudhary, V.B.; Darrouzet-Nardi, A.; Eldridge, D.J.; Faist, A.M.; Ferrenberg, S.; et al. What is a biocrust? A refined, contemporary definition for a broadening research community. Biol. Rev. 2022, 97, 1768–1785. [Google Scholar] [CrossRef] [PubMed]
- Evans, R.D.; Johansen, J.R. Microbiotic crusts and ecosystem processes. Crit. Rev. Plant Sci. 1999, 18, 183–225. [Google Scholar] [CrossRef]
- Harper, K.T.; Marble, J.R. Effect of timing of grazing on soil surface cryptogamic communities in a Great Basin low-shrub desert: A preliminary report. Great Basin Nat. 1989, 49, 104–107. [Google Scholar]
- Kleiner, E.F.; Harper, K.T. Environment and community organization in the grasslands of Canyonlands National Park. Ecology 1972, 53, 299–309. [Google Scholar] [CrossRef]
- West, N.E. Structure and function of microphytic soil crusts in wildland ecosystems of arid to semiarid regions. Adv. Ecol. Res. 1990, 20, 179–223. [Google Scholar] [CrossRef]
- Booth, W.E. Algae as pioneers in plant succession and their importance in erosion control. Ecology 1941, 22, 38–46. [Google Scholar] [CrossRef]
- Fletcher, J.E.; Martin, W.P. Some effects of algae and molds in the rain-crust of desert soils. Ecology 1948, 29, 95–100. [Google Scholar] [CrossRef]
- Fritsch, F.E. The terrestrial alga. J. Ecol. 1922, 10, 220–236. [Google Scholar] [CrossRef]
- Navarro-González, R.; Rainey, F.A.; Molina, P.; Bagaley, D.R.; Hollen, B.J.; De La Rosa, J.; Small, A.M.; Quinn, R.C.; Grunthaner, F.J.; Cáceres, L.; et al. Mars-like soils in the Atacama Desert, Chile, and the dry limit of microbial life. Science 2003, 302, 1018–1021. [Google Scholar] [CrossRef]
- Kaštovský, J. Welcome to the jungle!: An overview of modern taxonomy of cyanobacteria. Hydrobiologia 2024, 851, 1063–1077. [Google Scholar] [CrossRef]
- Strunecký, O.; Ivanova, A.P. An updated classification of cyanobacterial orders and families based on phylogenomic and polyphasic analysis. J. Phycol. 2023, 59, 12–51. [Google Scholar] [CrossRef] [PubMed]
- Brown, A.O.; Romanis, C.S.; Dvořák, P.; Foss, A.J.; Gibson, Q.A.; Villanueva, C.D.; Durden, W.N.; Garvey, A.D.; Jenkins, P.; Hašler, P.; et al. A new species of cryptic cyanobacteria isolated from the epidermis of a bottlenose dolphin and as a bioaerosol. Phycologia 2021, 60, 603–618. [Google Scholar] [CrossRef]
- Jusko, B.M.; Johansen, J.R. Description of six new cyanobacterial species from soil biocrusts on San Nicolas Island, California, in three genera previously restricted to Brazil. J. Phycol. 2024, 60, 133–151. [Google Scholar] [CrossRef] [PubMed]
- Shalygin, S.; Shalygina, R.; Redkina, V.V.; Gargas, C.B.; Johansen, J.R. Description of Stenomitos kolaenensis and S. hiloensis sp. nov. (Leptolyngbyaceae, Cyanobacteria) with an emendation of the genus. Phytotaxa 2020, 440, 108–128. [Google Scholar] [CrossRef]
- Schwabe, G.H. Zur autotrophen vegetation in ariden Böden, Blaualgen und Lebensraum IV. Osterr. Bot. Z. 1960, 107, 281–309. [Google Scholar] [CrossRef]
- Mühlsteinová, R.; Johansen, J.R.; Pietrasiak, N.; Martin, M.P. Polyphasic characterization of Kastovskya adunca gen. nov. et comb. nov. (Cyanobacteria: Oscillatoriales), from the desert soils of the Atacama Desert, Chile. Phytotaxa 2014, 163, 216–228. [Google Scholar] [CrossRef]
- Flechtner, V.R.; Johansen, J.R.; Belnap, J. The biological soil crusts of the San Nicholas Island: Enigmatic algae from a geographically isolated ecosystem. West. N. Am. Nat. 2008, 68, 405–436. [Google Scholar] [CrossRef]
- Alvarenga, D.O.; Andreote, A.P.D.; Branco, L.H.Z.; Delbaje, E.; Cruz, R.B.; de Mello Varani, A.; Fiore, M.F. Amazonocrinis nigriterrae gen. nov., sp. nov., Atlanticothrix silvestris gen. nov., sp. nov. and Denronalium phyllosphericum gen. nov., nostocacean cyanobacteria from Brazilian environments. Int. J. Syst. Evol. Microbiol. 2021, 71, 004811. [Google Scholar] [CrossRef]
- Machado de Lima, N.M.; Branco, L.H.Z. Biological soil crusts: New genera and species of cyanobacteria from Brazilian semi-arid regions. Phytotaxa 2020, 470, 263–281. [Google Scholar] [CrossRef]
- Johansen, J.R.; Jusko, B.M.; Mesfin, M.; Luknis, M.A.; Wain, A.; Hoyer, W.F.; Hasenstab-Lehman, K. Pseudoacaryochloris (Acaryochloridaceae, Cyanobacteria) species from Africa and North America: A disjunct distribution suggesting transatlantic wind dispersal. West. N. Am. Nat. 2024; 84, in press. [Google Scholar]
- Mehda, S.; Muñoz-Martín, M.Á.; Oustani, M.; Hamdi-Aïssa, B.; Perona, E.; Mateo, P. Lithic cyanobacterial communities in the polyextreme Sahara Desert: Implications for the search for the limits of life. Environ. Microbiol. 2022, 24, 451–474. [Google Scholar] [CrossRef]
- Mehda, S.; Perona, E.; Mateo, P.; Muñoz-Martín, M.Á. Validation of “Pseudacaryochloris sahariensis” nom. inval. (Acaryochloridaceae, Cyanophyceae) isolated from desert rocks in the Sahara. Not. Algarum 2023, 308, 2, ISSN: 2009-8987. [Google Scholar]
- Osorio-Santos, K.; Pietrasiak, N.; Bohunická, M.; Miscoe, L.H.; Kováčik, L.; Martin, M.P.; Johansen, J.R. Seven new species of Oculatella (Pseudanabaenales, Cyanobacteria): Taxonomically recognizing cryptic diversification. Eur. J. Phycol. 2014, 49, 450–470. [Google Scholar] [CrossRef]
- Pietrasiak, N.; Osorio-Santos, K.; Shalygin, S.; Martin, M.P.; Johansen, J.R. When is a lineage a species? A case study in Myxacorys gen. nov. (Synechococcales: Cyanobacteria) with the description of two new species from the Americas. J. Phycol. 2019, 55, 976–996. [Google Scholar] [CrossRef]
- Pietrasiak, N.; Reeve, S.; Osorio-Santos, K.; Lipson, D.A.; Johansen, J.R. Trichotorquatus gen. nov.—A new genus of soil cyanobacteria from American drylands. J. Phycol 2021, 57, 886–902. [Google Scholar] [CrossRef] [PubMed]
- Mühlsteinová, R.; Johansen, J.R.; Pietrasiak, N.; Martin, M.P.; Osorio-Santos, K.; Warren, S.D. Polyphasic characterization of Trichocoleus desertorum sp. nov. (Pseudanabaenales, Cyanobacteria) from desert soils and phylogenetic placement of the genus Trichocoleus. Phytotaxa 2014, 163, 241–261. [Google Scholar] [CrossRef]
- Moreira-Fernandes, V.; Giraldo-Silva, A.; Roush, D.; GarciaPichel, F. Coleofasciculaceae, a monophyletic home for the Microcoleus steenstrupii complex and other desiccation tolerant filamentous cyanobacteria. J. Phycol. 2021, 57, 1563–1579. [Google Scholar] [CrossRef]
- Kirchner, O. Schizophyta: Schizophyceae. In Die Natürlichen Pflanzenfamilien; Engler, A., Prantl, K., Eds.; Figures 48–62; Wilhelm Engelmann: Leipzig, Germany, 1898; Volume 1, pp. 45–92. [Google Scholar]
- Pietrasiak, N.; Mühlsteinová, R.; Siegesmund, M.; Johansen, J.R. Phylogenetic placement of Symplocastrum (Phormidiaceae, Cyanobacteria) with description of two new species: S. flechtnerae and S. torsivum. Phycologia 2014, 53, 529–541. [Google Scholar] [CrossRef]
- Patzelt, D.J.; Hodač, L.; Friedl, T.; Pietrasiak, N.; Johansen, J.R. Biodiversity of soil cyanobacteria in the hyper-arid Atacama Desert, Chile. J. Phycol. 2014, 50, 698–710. [Google Scholar] [CrossRef]
- Mehda, S.; Muñoz-Martin, M.A.; Oustani, M.; Hamdi-Aïssa, B.; Perona, E.; Mateo, P. Microenvironmental conditions drive the differential cyanobacterial community composition of biocrusts from the Sahara Desert. Microorganisms 2021, 9, 487. [Google Scholar] [CrossRef]
- Carmichael, W. Isolation, culture, and toxicity testing of toxic freshwater cyanobacteria (blue-green algae). In Fundamental Research in Homogenous Catalysis; Shilor, V., Ed.; Gordon & Breach Publ: New York, NY, USA, 1986; Volume 3, pp. 1249–1262. [Google Scholar]
- Rippka, R.; Deruelles, J.; Waterbury, J.B.; Herdman, M.; Stanier, R.Y. Generic assignments, stain histories and properties of pure cultures of cyanobacteria. Microbiology 1979, 111, 1–61. [Google Scholar] [CrossRef]
- Becerra-Absalón, I.; Muñoz-Martín, M.Á.; Montejano, G.; Mateo, P. Differences in the cyanobacterial community composition of biocrusts from the drylands of central Mexico. Are there endemic species? Front. Microbiol. 2019, 10, 937. [Google Scholar] [CrossRef]
- Loza, V.; Perona, E.; Mateo, P. Molecular fingerprinting of cyanobacteria from river biofilms as a water quality monitoring tool. Appl. Environ. Microbiol. 2013, 79, 1459–1472. [Google Scholar] [CrossRef]
- Wilmotte, A.; Van der Auwera, G.; De Wachter, R. Structure of the 16 S ribosomal RNA of the thermophilic cyanobacterium chlorogloeopsis HTF (‘mastigocladus laminosus HTF’) strain PCC7518, and phylogenetic analysis. FEBS Lett. 1993, 317, 96–100. [Google Scholar] [CrossRef] [PubMed]
- Flechtner, V.R.; Boyer, S.L.; Johansen, J.R.; De Noble, M.L. Spirirestis rafaelensis gen. et sp. nov. (Cyanophyceae), a new cyanobacterial genus from arid soils. Nova Hedwig. 2002, 74, 1–24. [Google Scholar] [CrossRef]
- Nübel, U.; Garcia-Pichel, F.; Muyzer, G. PCR primers to amplify 16S rRNA genes from cyanobacteria. Appl. Environ. Microbiol. 1997, 63, 3327–3332. [Google Scholar] [CrossRef]
- Larkin, M.A.; Blackshields, G.; Brown, N.P.; Chenna, R.; McGettigan, P.A.; McWilliam, H.; Valentin, F.; Wallace, I.M.; Wilm, A.; Lopez, R.; et al. Clustal W and Clustal X version 2.0. Bioinformatics 2007, 23, 2947–2948. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Li, D.; Liu, Y. Salt tolerance of Microcoleus vaginatus Gom., a cyanobacterium isolated from desert algal crust, was enhanced by exogenous carbohydrates. J. Arid. Environ. 2003, 55, 645–656. [Google Scholar] [CrossRef]
- Miller, M.A.; Pfeiffer, W.; Schwartz, T. Creating the CIPRES Science Gateway for inference of large phylogenetic trees. In Proceedings of the 2010 Gateway Computing Environments Workshop (GCE), New Orleans, LA, USA, 14 November 2010; pp. 1–8. [Google Scholar] [CrossRef]
- Ronquist, F.; Teslenko, M.; Van Der Mark, P.; Ayres, D.L.; Darling, A.; Hohna, S.; Larget, B.; Liu, L.; Suchard, M.A.; Huelsenbeck, J.P. MrBayes 3.2, efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 2012, 61, 539–542. [Google Scholar] [CrossRef]
- Stamatakis, A. RAxML version 8, a tool for phylogenetic analysis and post- analysis of large phylogenies. Bioinformatics 2014, 30, 1312–1313. [Google Scholar] [CrossRef]
- Drummond, A.J.; Ho, S.Y.W.; Phillips, M.J.; Rambaut, A. Relaxed phylogenetics and dating with confidence. PLoS Biol. 2006, 4, e88. [Google Scholar] [CrossRef]
- Gelman, A.; Rubin, D.B. Inference from iterative simulation using multiple sequences. Stat. Sci. 1992, 7, 157–511. [Google Scholar] [CrossRef]
- Rambaut, A. FigTree, Version 1.4.3. 2009. Available online: http://tree.bio.ed.ac.uk/software/figtree (accessed on 30 June 2024).
- Swofford, D.L. PAUP*. Phylogenetic Analysis Using Parsimony (*and Other Methods), Version 4.02b; Sinauer Associates: Sunderland, MA, USA, 1998. [Google Scholar]
- Zuker, M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003, 31, 3406–3415. [Google Scholar] [CrossRef] [PubMed]
- Johansen, J.R.; Casamatta, D.A. Recognizing cyanobacterial diversity through the adoption of a new species paradigm. Algol. Stud. 2005, 117, 71–93. [Google Scholar] [CrossRef]
- Fox, G.E.; Wisotzkey, J.D.; Jurtshuk, P. How close is close: 16S rRNA sequence identity may not be sufficient to guarantee species identity. Int. J. Syst. Bacteriol. 1992, 42, 166–170. [Google Scholar] [CrossRef]
- Casamatta, D.A.; Vis, M.L.; Sheath, R.G. Cryptic species in cyanobacterial systematics: A case study of Phormidium retzii (Oscillatoriales) using 16S rDNA and RAPD analyses. Aquat. Bot. 2003, 77L, 295–309. [Google Scholar] [CrossRef]
- Stanojkovic, A.; Skoupý, S.; Škaloud, P.; Dvořák, P. High genomic differentiation and limited gene flow indicate recent cryptic speciation within the genus Laspinema (cyanobacteria). Front. Microbiol. 2022, 13, 977454. [Google Scholar] [CrossRef]
- Yarza, P.; Yilmaz, P.; Pruesse, E.; Glöckner, F.O.; Ludwig, W.; Schleifer, K.H.; Whitman, W.B.; Euzéby, J.; Amann, R.; Roselló-Móra, R. Uniting the classification of cultured and uncultured bacteria and archaea using 16S rRNA gene sequences. Nat. Rev. Microbiol. 2014, 12, 635–645. [Google Scholar] [CrossRef]
- Casamatta, D.A.; Gomez, S.R.; Johansen, J.R. Rexia erecta gen. et sp. nov. and Capsosira lowei sp. nov., two newly described cyanobacterial taxa from the Great Smoky Mountain National Park (USA). Hydrobiologia 2006, 561, 13–26. [Google Scholar] [CrossRef]
- Johansen, J.R.; Bohunická, M.; Lukešová, A.; Hrčková, K.; Vaccarino, M.A.; Chesarino, N.M. Morphological and molecular characterization within 26 strains of the genus Cylindrospermum (Nostocaceae, Cyanobacteria), with descriptions of three new species. J. Phycol. 2014, 50, 187–202. [Google Scholar] [CrossRef]
- Řeháková, K.; Mareš, J.; Lukešová, A.; Zapomělová, E.; Bernardová, K.; Hrouzek, P. Nodularia (Cyanobacteria, Nostocaceae): A phylogenetically uniform genus with variable phenotypes. Phytotaxa 2014, 172, 235–246. [Google Scholar] [CrossRef]
- Hassler, H.B.; Probert, B.; Moore, C.; Lawson, E.; Jackson, R.W.; Russell, B.T.; Richards, V.P. Phylogenies of the 16S rRNA gene and its hypervariable regions lack concordance with core genome phylogenies. Microbiome 2022, 10, 104. [Google Scholar] [CrossRef] [PubMed]
- Bartoš, O.; Chmel, M.; Swierczková, I. The overlooked evolutionary dynamics of the 16S rRNA revises its role as the “gold standard” for bacterial species identification. Sci. Rep. 2024, 14, 9067. [Google Scholar] [CrossRef]
- Boyer, S.L.; Flechtner, V.R.; Johansen, J.R. Is the 16S–23S rRNA internal transcribed spacer region a good tool for use in molecular systematics and population genetics? A case study in cyanobacteria. Mol. Biol. Evol. 2001, 18, 1057–1069, ISSN: 0737-4038. [Google Scholar] [CrossRef] [PubMed]
- Johansen, J.R.; Kovacik, L.; Casamatta, D.A.; Fučíková, K.; Kaštovský, J. Utility of 16S–23S ITS sequence and secondary structure for recognition of intrageneric and intergeneric limits within cyanobacterial taxa: Leptolyngbya corticola sp. nov. (Pseudanabaenaceae, Cyanobacteria). Nova Hedwig. 2011, 92, 283–302. [Google Scholar] [CrossRef]
- Siegesmund, M.A.; Johansen, J.R.; Karsten, U.; Friedl, T. Coleofasciculus gen. nov. (Cyanobacteria): Morphological and molecular criteria for revision of the genus Microcoleus Gomont. J. Phycol. 2008, 44, 1572–1585. [Google Scholar] [CrossRef] [PubMed]
- Erwin, P.; Thacker, R. Cryptic diversity of the symbiotic cyanobacterium Synechococcus spongiarium among sponge hosts. Mol. Ecol. 2008, 17, 2937–2947. [Google Scholar] [CrossRef]
- Becerra-Absalón, I.; Johansen, J.R.; Muñoz-Martín, M.A.; Montejano, G. Chroakolemma gen. nov. (Leptolyngbyaceae, Cyanobacteria) from soil biocrusts in the semi-desert Central Region of Mexico. Phytotaxa 2018, 367, 201–218. [Google Scholar] [CrossRef]
- González-Resendiz, L.; Johansen, J.R.; Escobar-Sánchez, V.; Segal-Kischinevzky, C.; Jiménez-García, L.F.; León-Tejera, H. Two new species of Phyllonema (Rivulariaceae, Cyanobacteria) with an emendation of the genus. J. Phycol. 2018, 54, 638–652. [Google Scholar] [CrossRef]
- Bohunická, M.; Johansen, J.R.; Villanueva, C.D.; Mareš, J.; Štenclová, L.; Becerra-Absalón, I.; Hauer, T.; Kaštovský, J. Revision of the pantropical genus Brasilonema (Nostocales, Cyanobacteria), with the description of 24 species new to science. Fottea 2024, 24, 137–184. [Google Scholar] [CrossRef]
- Anagnostidis, K.; Komárek, J. Modern approach to the classification system of cyanophytes. 3-Oscillatoriales. Algol. Stud. 1988, 50–53, 327–472. [Google Scholar]
- Hernández-Mariné, M.; Roldán, M. Adherence of hormogonia to substrata is mediated by polysaccharides produced by necridic cells. Algol. Stud. 2005, 117, 239–249. [Google Scholar] [CrossRef]
- Berrendero, E.G.; Johansen, J.R.; Kaštovský, J.; Bohunická, M.; Čapková, K. Macrochaete gen. nov. (Cyanobacteria), a taxon morphologically and molecularly distinct from Calothrix. J. Phycol. 2016, 52, 638–655. [Google Scholar] [CrossRef] [PubMed]
- Hentschke, G.S.; Sant’Anna, C.L. Current trends and prospects for cyanobacterial taxonomy—Are only cultured populations. Algol. Stud. 2014, 147, 3–6. [Google Scholar] [CrossRef]
- James, I.N. Hadley Circulation. In Encyclopedia of Atmospheric Sciences, 1st ed.; Holton, J.R., Curry, J.A., Pyle, J.A., Eds.; Academic Press: Amsterdam, The Netherlands, 2022; pp. 919–924. ISBN 9780122270901. [Google Scholar] [CrossRef]
- Schneider, T. The general circulation of the atmosphere. Annu. Rev. Earth Planet. Sci. 2006, 34, 655–688. [Google Scholar] [CrossRef]
- Houston, J.; Hartley, A.J. The Central Andean west-slope rainshadow and its potential contribution to the origin of hyper-aridity in the Atacama Desert. Int. J. Climatol. 2003, 23, 1453–1464. [Google Scholar] [CrossRef]
- Dvořák, P.; Hašler, P.; Casamatta, D.A.; Poulíčková, A. Underestimated cyanobacterial diversity: Trends and perspectives of research in tropical environments. Fottea 2021, 21, 110–127. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).