Identifying Morphs of the Yellow-Legged Hornet (Vespa velutina) and Other Pests of Quarantine Importance with Geometric Morphometrics
Abstract
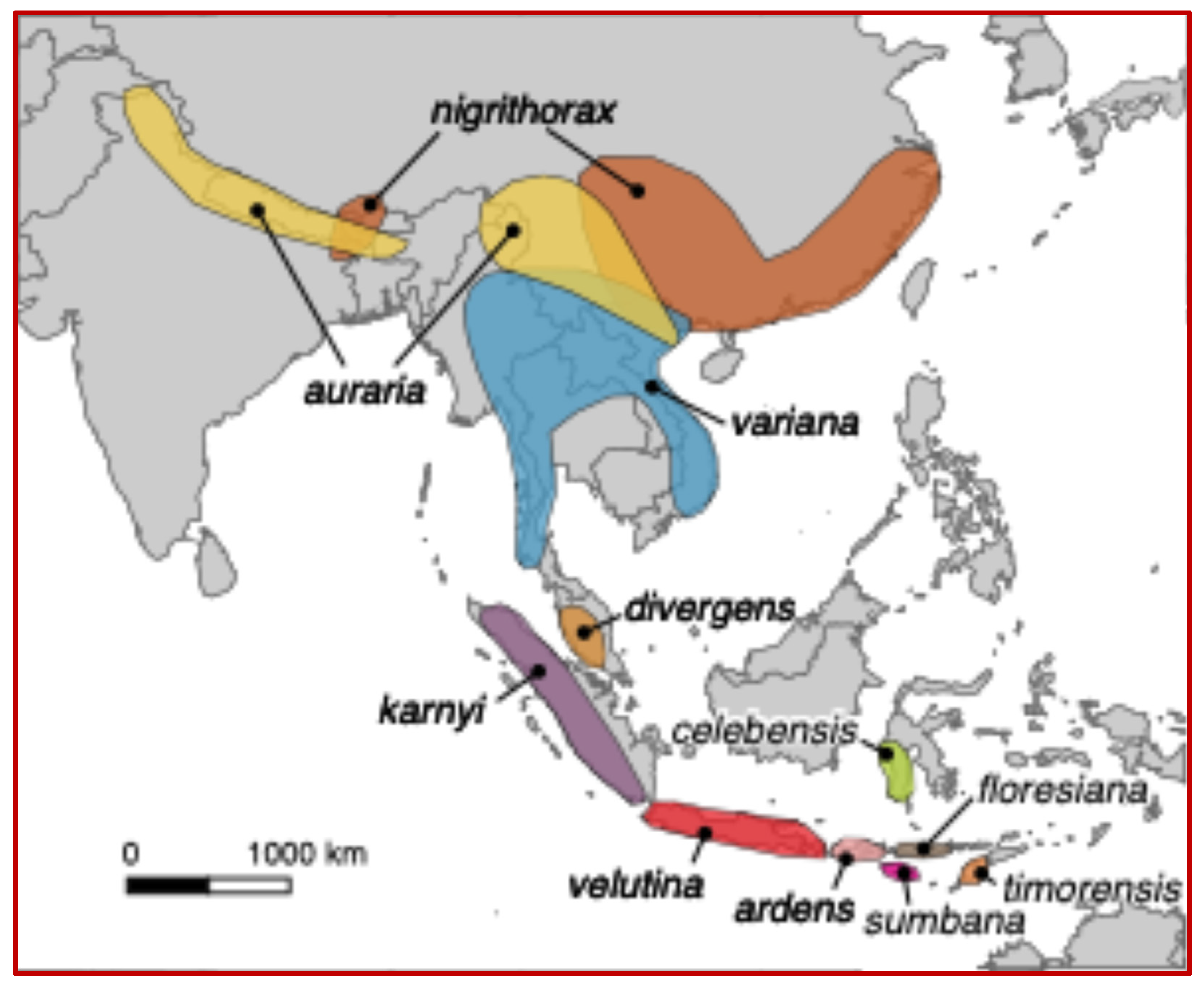
1. Introduction
2. Materials and Methods
2.1. Geometric Morphometrics
2.2. Molecular Sequencing and Analysis
3. Results
4. Discussion
4.1. Accuracy of GMM Identifications
4.2. Strengths and Weaknesses of the DNA Barcode Analysis
4.3. The Provenance of the Specimen Intercepted in Utah
4.4. Using GMM to Determine the Provenance of Invasive Insects
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Disclaimer
References
- Perrard, A.; Baylac, M.; Carpenter, J.M.; Villemant, C. Evolution of wing shape in hornets: Why is the wing venation efficient for species identification? J. Evol. Biol. 2014, 27, 2665–2675. [Google Scholar] [CrossRef] [PubMed]
- Smith-Pardo, A.H.; Carpenter, J.M.; Kimsey, L. The diversity of hornets in the Genus Vespa (Hymenoptera: Vespidae; Vespinae), their importance, and interceptions in the United States. Insect Syst. Divers. 2020, 4, 2. [Google Scholar] [CrossRef]
- Monceau, K.; Bonnard, O.; Thiéry, D. Vespa velutina: A new invasive predator of honeybees in Europe. J. Pest Sci. 2014, 87, 1–16. [Google Scholar] [CrossRef]
- Fedele, E.; Gervasini, E.; Cardoso, A.C.; La Notte, A.; Vallecillo, S.; Tsiamis, K.; Maes, J. Invasive Alien Species Impact on Ecosystem Services–Asian Hornet (Vespa velutina nigrithorax) Case Study; EUR 29827 ΕΝ; Publications Office of the European Union: Luxembourg, 2019; ISBN 978-92-76-09511-8. [Google Scholar]
- Laborde-Castérot, H.; Darrouzet, E.; Le Roux, G.; Labadie, M.; Delcourt, N.; de Haro, L.; Vodovar, D.; Langrand, J.; French Poison Control Centers Research Group. Ocular lesions other than stings following yellow-legged hornet (Vespa velutina nigrithorax) projections, as reported to French Poison control centers. JAMA Ophthalmol. 2021, 139, 105–108. [Google Scholar] [CrossRef]
- Archer, M.E. Vespine Wasps of the World: Behavior, Ecology & Taxonomy of the Vespinae; SSP-Siri Scientific Press: Manchester, UK, 2012; 352p. [Google Scholar]
- Perrard, A.; Arca, M.; Rome, Q.; Muller, F.; Tan, J.; Bista, S.; Nugroho, H.; Baudoin, R.; Baylac, M.; Silvain, J.-F.; et al. Geographic variation of melanization patterns in a hornet species: Genetic differences, climatic pressures or aposematic constraints? PLoS ONE 2014, 9, e94162. [Google Scholar] [CrossRef] [PubMed]
- Choi, M.B.; Martin, S.J.; Jong Wook, L. Distribution, spread, and impact of the invasive hornet Vespa velutina in South Korea. J. Asia-Pac. Entomol. 2012, 15, 473–477. [Google Scholar] [CrossRef]
- Jung, C.; Kang, M.S.; Kim, D.W.; Lee, H.S. Vespid wasps (Hymenoptera: Vespidae) occurring around apiaries in Andong, Korea. I. Taxonomy and life history. Korean J. Apic. 2007, 22, 53–62. [Google Scholar]
- Kishi, S.; Goka, K. Review of the invasive yellow-legged hornet, Vespa velutina nigrithorax (Hymenoptera: Vespidae), in Japan and its possible chemical control. Appl. Entomol. Zool. 2017, 52, 361–368. [Google Scholar] [CrossRef]
- Minoshima, Y.N.; Yamane, S.; Ueno, T. An invasive alien hornet, Vespa velutina nigrithorax du Buysson (Hymenoptera, Vespidae), found in Kitakyushu, Kyushu Island: A first record of the species from mainland Japan. Jpn. J. Syst. Entomol. 2015, 21, 259–261. [Google Scholar]
- Bertolino, S.; Lioy, S.; Laurino, D.; Manino, A.; Porporato, M. Spread of the invasive yellow-legged hornet Vespa velutina (Hymenoptera: Vespidae) in Italy. Appl. Entomol. Zool. 2016, 5, 589–597. [Google Scholar] [CrossRef]
- Arca, M.; Mougel, F.; Guillemaud, T.; Dupas, S.; Rome, Q.; Perrard, A.; Muller, F.; Fossoud, A.; Capdevielle-Dulac, C.; Torres-Leguizamon, M.; et al. Reconstructing the invasion and the demographic history of the yellow-legged hornet, Vespa velutina, in Europe. Biol. Invasions 2015, 17, 2357–2371. [Google Scholar] [CrossRef]
- Robinet, C.; Suppo, C.; Darrouzet, E. Rapid spread of the invasive yellow-legged hornet in France: The role of human-mediated dispersal and the effects of control measures. J. Appl. Ecol. 2017, 54, 205–215. [Google Scholar] [CrossRef]
- Keeling, M.J.; Franklin, D.N.; Datta, S.; Brown, M.A.; Budge, G.E. Predicting the spread of the Asian hornet (Vespa velutina) following its incursion into Great Britain. Sci. Rep. 2017, 7, 6240. [Google Scholar] [CrossRef] [PubMed]
- CABI. Vespa velutina (Asian Hornet)–Invasive Species Compendium. 2020. Available online: https://www.cabi.org/isc/datasheet/109164 (accessed on 15 December 2020).
- Leza, M.; Miranda, M.A.; Colomar, V. First detection of Vespa velutina nigrithorax (Hymenoptera: Vespidae) in the Balearic Islands (Western Mediterranean): A challenging study case. Biol. Invasions 2017, 20, 1643–1649. [Google Scholar] [CrossRef]
- Husemann, M.; Sterr, A.; Maack, S.; Abraham, R. The northernmost record of the Asian hornet Vespa velutina nigrithorax (Hymenoptera, Vespidae). Evol. Syst. 2020, 4, 1–4. [Google Scholar] [CrossRef]
- du Buysson, R. Monographie des guêpes ou Vespa. Ann. Société Entomol. Fr. 1905, 72, 260–288. [Google Scholar]
- van der Vecht, J. The Vespinae of the Indo-Malayan and Papuan areas (Hymenoptera, Vespidae). Zool. Verh. 1957, 34, 1–82. [Google Scholar]
- Archer, M.E. Taxonomy, distribution, and nesting biology of the Vespa bicolor group (Hym., Vespinae). Entomol. Mon. Mag. 1994, 130, 149–158. [Google Scholar]
- van der Vecht, J. Notes on Oriental Vespinae, including some species from China and Japan (Hymenoptera, Vespidae). Zool. Meded. 1959, 13, 205–232. [Google Scholar]
- Rohlf, F.J. Relative-warp analysis and an example of its application to mosquito wings. In Contributions to Morphometrics; Marcus, L.F., Bellow, E., Garcia-Valdecasas, A., Eds.; Museo Nacional de Ciencias Naturales: Madrid, Spain, 1993; pp. 131–159. [Google Scholar]
- Weeks, P.J.D.; Gauld, I.D.; Gaston, K.J.; O’Neill, M.A. Automating the identification of insects: A new solution to an old problem. Bull. Entomol. Res. 1997, 87, 203–211. [Google Scholar] [CrossRef]
- Baylac, M.; Villemant, C.; Simbolotti, G. Combining geometric morphometrics with pattern recognition for the investigation of species complexes. Biol. J. Linn. Soc. 2003, 80, 89–98. [Google Scholar] [CrossRef]
- Houle, D.; Mezey, J.; Galpern, P.; Carter, A. Automated measurement of Drosophila wings. BMC Evol. Biol. 2003, 3, 25. [Google Scholar] [CrossRef] [PubMed]
- Sadeghi, S.; Adriaens, D.; Dumont, H.J. Geometric morphometric analysis of wing shape variation in ten European populations of Calopteryx splendens (Harris, 1782). Odonatologica 2009, 38, 343–360. [Google Scholar]
- Perrard, A. Systématique et Morphométrie Géométrique: L’Évolution de la Nervation Alaire au Sein du Genre Vespa (Hyménoptères, Vespidés). Ph.D. Dissertation, Muséum National d’Histoire Naturelle, Paris, France, 2012. [Google Scholar]
- Carpenter, J.M.; Kojima, J.I. Checklist of the species in the subfamily Vespinae (Insecta: Hymenoptera: Vespidae). Nat. Hist. Bull. Ibaraki Univ. 1997, 1, 51–92. [Google Scholar]
- Nguyen, L.T.P.; Saito, F.; Kojima, J.I.; Carpenter, J.M. Vespidae of Viet Nam (Insecta: Hymenoptera). 2. Taxonomic notes on Vespinae. Zool. Sci. 2006, 23, 95–104. [Google Scholar] [CrossRef]
- Do, Y.; Park, W.-B.; Park, J.-K.; Kim, C.-J.; Choi, M.B. Genetic and morphological variation of Vespa velutina nigrithorax which is an invasive species in a mountainous area. Sci. Rep. 2022, 12, 4737. [Google Scholar] [CrossRef]
- Rohlf, F.J. tpsDig, Digitize Landmarks and Outlines, Version 2.31; Stony Brook; Department of Ecology and Evolution, State University of New York: New York, NY, USA, 2006. [Google Scholar]
- Bookstein, F.L. Morphometric Tools for Landmark Data; Cambridge University Press: Cambridge, UK, 1991; 435p. [Google Scholar]
- Dryden, I.L.; Mardia, K.V. Statistical Shape Analysis; John Wiley & Sons: Chichester, UK, 1998; 347p. [Google Scholar]
- Klingenberg, C.P. MorphoJ: An integrated software package for geometric morphometrics. Mol. Ecol. Resour. 2011, 11, 353–357. Available online: https://morphometrics.uk/MorphoJ_page.html (accessed on 10 June 2023). [CrossRef]
- Hammer, Ø.; Harper, D.A.T.; Ryan, P.D. PAST: Paleontological Statistics software package for education and analysis. Palaeontol. Electron. 2001, 4, 1–9. Available online: https://www.nhm.uio.no/english/research/resources/past/ (accessed on 20 June 2024).
- Polly, P.D. Geometric Morphometrics for Mathematica. Version 13.0. 2024. Available online: https://github.com/pdpolly/Morphometrics-for-Mathematica (accessed on 20 June 2024).
- Rohlf, F.J.; Slice, D. Extensions of the Procrustes method for the optimal superimposition of landmarks. Syst. Zool. 1990, 39, 40–59. [Google Scholar] [CrossRef]
- Polly, P.D.; Motz, G.J. Patterns and processes in morphospace: Geometric morphometrics of three-dimensional objects. Paleontol. Soc. Pap. 2016, 22, 71–99. [Google Scholar] [CrossRef]
- Zelditch, M.L.; Swiderski, D.L.; Fink, W.L. Geometric Morphometrics for Biologists: A Primer, 2nd ed.; Academic Press: San Diego, CA, USA, 2012; 488p. [Google Scholar]
- Pearson, K. LIII. On lines and planes of closest fit to systems of points in space. Lond. Edinb. Dublin Philos. Mag. J. Sci. 1901, 2, 559–572. [Google Scholar] [CrossRef]
- Wishart, J. The generalized product moment distribution in samples from a normal multivariate population. Biometrika 1928, 20A, 32–52. [Google Scholar] [CrossRef]
- Uyeda, J.C.; Caetano, D.S.; Pennell, M.W. Comparative analysis of principal components can be misleading. Syst. Biol. 2015, 64, 677–689. [Google Scholar] [CrossRef]
- Adams, D.C.; Collyer, M.L. Multivariate phylogenetic comparative methods: Evaluations, comparisons, and recommendations. Syst. Biol. 2018, 67, 14–31. [Google Scholar] [CrossRef] [PubMed]
- Rao, C.R. The utilization of multiple measurements in problems of biological classification. J. R. Stat. Soc. Ser. B 1948, 10, 159–203. [Google Scholar] [CrossRef]
- Mitteroecker, P.; Bookstein, F. Linear discrimination, ordination, and the visualization of selection gradients in modern morphometrics. Evol. Biol. 2011, 38, 100–114. [Google Scholar] [CrossRef]
- Cardini, A.; O’Higgins, P.; Rohlf, F.J. Seeing distinct groups where there are none: Spurious patterns from between-group PCA. Evol. Biol. 2019, 46, 303–316. [Google Scholar] [CrossRef]
- Cardini, A.; Polly, P.D. Cross-validated between group PCA scatterplots: A solution to spurious group separation? Evol. Biol. 2020, 47, 85–95. [Google Scholar] [CrossRef]
- Felsenstein, J. Maximum-likelihood estimation of evolutionary trees from continuous characters. Am. J. Hum. Genet. 1973, 25, 471–492. [Google Scholar]
- Felsenstein, J. Inferring Phylogenies; Sinauer Associates: Sunderland, MA, USA, 2004; 580p. [Google Scholar]
- Caumul, R.; Polly, P.D. Phylogenetic and environmental components of morphological variation: Skull, mandible and molar shape in marmots (Marmota, Rodentia). Evolution 2005, 59, 2460–2472. [Google Scholar]
- Rohlf, F.J. Geometric morphometrics and phylogeny. In Morphology, Shape, and Phylogenetics; MacLeod, N., Forey, P., Eds.; Taylor and Francis: Abingdon, UK, 2002; pp. 175–193. [Google Scholar]
- Felsenstein, J. PHYLIP–Phylogeny Inference Package (Version 3.2). Cladistics 1989, 5, 164–166. [Google Scholar]
- Folmer, O.; Black, M.; Hoeh, W.; Lutz, R.; Vrijenhoek, R. DNA primers for amplification of mitochondrial cytochrome coxidase subunit I from diverse metazoan invertebrates. Mol. Mar. Biol. Biotechnol. 1994, 3, 294–299. [Google Scholar] [PubMed]
- Ratnasingham, S.; Hebert, P.D.N. BOLD: The Barcode of Life Data System (http://www.barcodinglife.org). Mol. Ecol. Notes 2007, 7, 355–364. [Google Scholar] [CrossRef] [PubMed]
- Katoh, K.; Misawa, K.; Kuma, K.; Miyata, T. MAFFT: A novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acid Res. 2002, 30, 3059–3066. [Google Scholar] [CrossRef] [PubMed]
- Swofford, D.L. PAUP*: Phylogenetic Analysis Using Parsimony (*and Other Methods). Version 4; Sinauer Associates: Sunderland, MA, USA, 2003. [Google Scholar]
- Allendorf, F.W.; Lundquist, L.L. Introduction: Population biology, evolution, and control of invasive species. Conserv. Biol. 2003, 17, 24–30. [Google Scholar] [CrossRef]
- Hebert, P.D.N.; Cywinska, A.; Ball, S.L.; deWaard, J.R. Biological identifications through DNA barcodes. Proc. R. Soc. Lond. 2003, 270, 313–321. [Google Scholar] [CrossRef] [PubMed]
- Klopfstein, S.; Kropf, C.; Bauri, H. Wolbachia endosymbionts distort DNA barcoding in the parasitoid wasp genus Diplazon (Hymenoptera: Ichneumonidae). Zool. J. Linn. Soc. 2015, 177, 541–557. [Google Scholar] [CrossRef]
- Bleidorn, C.; Henze, K. A new primer pair for barcoding of bees (Hymenoptera: Anthophila) without amplifying the orthologous coxA gene of Wolbachia bacteria. BMC Res. Notes 2021, 14, 427. [Google Scholar] [CrossRef] [PubMed]
- Sherpa, S.; Renaud, J.; Guéguen, M.; Besnard, G.; Mouyon, L.; Rey, D.; Després, L. Landscape does matter: Disentangling founder effects from natural and human-aided post-introduction dispersal during an ongoing biological invasion. J. Anim. Ecol. 2020, 89, 2027–2042. [Google Scholar] [CrossRef]
- Namin, S.M.; Jung, C. Genetic diversity of genus Vespa including an invaded species of V. velutina (Hymenoptera: Vespidae) in Korea inferred from DNA barcoding data. J. Asian-Pac. Entomol. 2020, 23, 540–545. [Google Scholar] [CrossRef]
- Darrouzet, E.; Gévar, J.; Guignard, Q.; Aron, S. Production of early diploid males by European colonies of the invasive hornet Vespa velutina nigrithorax. PLoS ONE 2015, 10, e0136680. [Google Scholar] [CrossRef] [PubMed]
- Bouzar, C.; Bankhead-Dronnet, S.; Gévar, J.; Darrouzet, E. Chemical and genetic evidences that multiple hornet colonies attack honeybee colonies. Insectes Sociaux. 2022, 69, 159–168. [Google Scholar] [CrossRef]
- Villement, C.; Streito, J.-C.; Haxaire, J. Premier bilan de l’invasion de Vespa velutina Lepeletier en France. Bull. Société Entomol. Fr. 2006, 111, 535–538. [Google Scholar] [CrossRef]
- Polly, P.D. Paleophylogeography: The tempo of geographic differentiation in marmots (Marmota). J. Mammal. 2003, 84, 369–384. [Google Scholar] [CrossRef]
- Polly, P.D.; Head, J.J. Maximum-likelihood identification of fossils: Taxonomic identification of Quaternary marmots (Rodentia, Mammalia) and identification of vertebral position in the pipesnake Cylindrophis (Serpentes, Reptilia). In Morphometrics-Applications in Biology and Paleontology; Elewa, A.M.T., Ed.; Springer: Heidelberg, Germany, 2004; pp. 197–222. [Google Scholar]
- Mutanen, M.; Pretorius, E. Subjective visual evaluation vs. traditional and geometric morphometrics in species delimitation: A comparison of moth genitalia. Syst. Entomol. 2007, 32, 371–386. [Google Scholar] [CrossRef]
- Villemant, C.; Simbolotti, G.; Kenis, M. Discrimination of Eubazus (Hymenoptera, Braconidae) sibling species using geometric morphometrics analysis of wing venation. Syst. Entomol. 2007, 32, 625–634. [Google Scholar] [CrossRef]
- Jaramillo, N.; Dujardin, J.P.; Calle-Londoño, D.; Fonseca-González, I. Geometric morphometrics for the taxonomy of 11 species of Anopheles (Nyssorhynchus) mosquitoes. Med. Vet. Entomol. 2015, 29, 26–36. [Google Scholar] [CrossRef]
- Christodoulou, M.D.; Clark, J.Y.; Culham, A. The Cinderella discipline: Morphometrics and their use in botanical classification. Bot. J. Linn. Soc. 2020, 194, 385–396. [Google Scholar] [CrossRef]
- Cardini, A.; Elton, S.; Kovarovic, K.F.; Viðarsdóttir, U.S.; Polly, P.D. Impact of sampling error on the assessment of morphospecies using geometric morphometrics in primates and other mammals. Evol. Biol. 2021, 48, 190–220. [Google Scholar] [CrossRef]
- Rosa, V.G.D.; Torres, M.A.J.; Demayo, C.G. Geometric morphometric tools in the analysis of shell shape of twelve local populations of the invasive snail Achatina fulica Bowdich from the Philippines. In Proceedings of the 2010 International Conference on Environmental Engineering and Applications, Singapore, 10–12 September 2010; Volume 2010, pp. 91–95. [Google Scholar]
- Rama Rao, S.; Liew, T.S.; Yow, Y.Y.; Ratnayeke, S. Cryptic diversity: Two morphologically similar species of invasive apple snail in Peninsular Malaysia. PLoS ONE 2018, 13, e0196582. [Google Scholar] [CrossRef]
- Lemic, D.; Pajač Živković, I.; Šuliček, M.; Benítez, H.A. Exploratory Analysis of Color Forms’ Variability in the Invasive Asian Lady Beetle Harmonia axyridis (Pallas 1773). Animals 2021, 11, 2436. [Google Scholar] [CrossRef]
- Devine, J.; Aponte, J.D.; Katz, D.C.; Liu, W.; LVercio, D.L.; Forkert, N.D.; Marcucio, R.; Percival, C.J.; Hallgrímsson, B. A registration and deep learning approach to automated landmark detection for geometric morphometrics. Evol. Biol. 2020, 47, 246–259. [Google Scholar] [CrossRef] [PubMed]
- Porto, A.; Voje, K.L. ML-morph: A fast, accurate, and general approach for automated detection and landmarking of biological structures in images. Methods Ecol. Evol. 2020, 11, 500–512. [Google Scholar] [CrossRef]
- Porto, A.; Rolfe, S.; Maga, A.M. ALPACA: A fast and accurate computer vision approach for automated landmarking of three-dimensional biological structures. Methods Ecol. Evol. 2021, 12, 2129–2144. [Google Scholar] [CrossRef]
- Guisande, C.; Manjarrés-Hernández, A.; Pelayo-Villamil, P.; Granado-Lorencio, C.; Riveiro, I.; Acuña, A.; Prieto-Piraquive, E.; Janeiro, E.; Matías, J.M.; Patti, C.; et al. IPez: An expert system for the taxonomic identification of fishes based on machine learning techniques. Fish. Res. 2010, 102, 240–247. [Google Scholar] [CrossRef]
- MacLeod, N.; Benfield, M.; Culverhouse, P. Time to automate identification. Nature 2010, 467, 154–155. [Google Scholar] [CrossRef]
- Wäldchen, J.; Mäder, P. Machine learning for image-based species identification. Methods Ecol. Evol. 2018, 9, 2216–2225. [Google Scholar] [CrossRef]
- Wäldchen, J.; Rzanny, M.; Seeland, M.; Mäder, P. Automated plant species identification—Trends and future directions. PLoS Comput. Biol. 2018, 14, e1005993. [Google Scholar] [CrossRef]
- Wang, M.-M.; Yin, P.-K.; Tang, Y.-N.; Yang, Z.-Z.; Xiao, H.; Zhang, C.-G.; Yang, Y.-H.; Yang, D.-S. Utility of DNA barcoding for identification of common Vespa species (Hymenoptera: Vespidae) from Yunnan, China. Entomol. Res. 2022, 52, 111–117. [Google Scholar] [CrossRef]







| SS | df | MS | ||||||||
| auraria | 0.00 | Between | 0.023 | 10 | 0.0023 | |||||
| celebensis | 0.09 | 0.00 | Within | 0.035 | 221 | 0.0002 | ||||
| divergens | 0.00 | 0.00 | 0.00 | Total | 0.058 | 231 | 0.0003 | |||
| floresiana | 0.00 | 0.00 | 0.01 | 0.00 | F-ratio = 14.59 | |||||
| karnyi | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | P(non-parametric) < 0.001 | ||||
| nigrithorax | 0.00 | 0.01 | 0.00 | 0.00 | 0.00 | 0.00 | R2 = 0.40 | |||
| sumbana | 0.00 | 0.00 | 0.01 | 0.00 | 0.00 | 0.00 | 0.00 | |||
| timorensis | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | ||
| variana | 0.00 | 0.13 | 0.02 | 0.00 | 0.02 | 0.00 | 0.13 | 0.01 | 0.00 | |
| velutina | 0.00 | 0.00 | 0.01 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 |
| ardens | auraria | celebensis | divergens | floresiana | karnyi | nigrithorax | sumbana | timorensis | variana | |
| ardens | auraria | celebensis | divergens | floresiana | karnyi | nigrithorax | sumbana | timorensis | varians | velutina | Total | % Correctly Classified | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ardens | 11 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 11 | 100% |
| auraria | 0 | 13 | 0 | 0 | 0 | 1 | 1 | 1 | 0 | 2 | 0 | 18 | 72% |
| celebensis | 0 | 0 | 5 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 6 | 83% |
| divergens | 0 | 0 | 0 | 33 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 34 | 97% |
| floresiana | 0 | 2 | 0 | 0 | 15 | 0 | 0 | 0 | 0 | 0 | 0 | 17 | 88% |
| karnyi | 0 | 1 | 0 | 0 | 1 | 22 | 0 | 0 | 0 | 0 | 0 | 24 | 92% |
| nigrithorax | 0 | 1 | 0 | 0 | 0 | 2 | 27 | 0 | 0 | 2 | 0 | 32 | 84% |
| sumbana | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 25 | 0 | 0 | 0 | 25 | 100% |
| timorensis | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 7 | 0 | 0 | 8 | 88% |
| varians | 0 | 1 | 0 | 0 | 1 | 0 | 1 | 0 | 0 | 5 | 0 | 8 | 63% |
| velutina | 2 | 0 | 1 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 44 | 49 | 90% |
| Total | 13 | 18 | 6 | 33 | 20 | 26 | 29 | 27 | 7 | 9 | 44 | 232 | |
| % classifications correct | 85% | 72% | 83% | 100% | 75% | 85% | 93% | 93% | 100% | 56% | 100% |
| Taxon | D2 | P (same) |
|---|---|---|
| ardens | 62.8 | <0.0001 |
| auraria | 35.2 | <0.0001 |
| celebensis | 892.1 | <0.0001 |
| divergens | 66.4 | <0.0001 |
| floresiana | 79.7 | <0.0001 |
| karnyi | 28.1 | <0.0001 |
| nigrithorax | 12.7 | 0.01 |
| sumbana | 65.26 | <0.0001 |
| timorensis | 696.9 | <0.0001 |
| variana | 44.4 | <0.0001 |
| velutina | 24.7 | <0.0001 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Smith-Pardo, A.; Polly, P.D.; Gilligan, T. Identifying Morphs of the Yellow-Legged Hornet (Vespa velutina) and Other Pests of Quarantine Importance with Geometric Morphometrics. Diversity 2024, 16, 367. https://doi.org/10.3390/d16070367
Smith-Pardo A, Polly PD, Gilligan T. Identifying Morphs of the Yellow-Legged Hornet (Vespa velutina) and Other Pests of Quarantine Importance with Geometric Morphometrics. Diversity. 2024; 16(7):367. https://doi.org/10.3390/d16070367
Chicago/Turabian StyleSmith-Pardo, Allan, P. David Polly, and Todd Gilligan. 2024. "Identifying Morphs of the Yellow-Legged Hornet (Vespa velutina) and Other Pests of Quarantine Importance with Geometric Morphometrics" Diversity 16, no. 7: 367. https://doi.org/10.3390/d16070367
APA StyleSmith-Pardo, A., Polly, P. D., & Gilligan, T. (2024). Identifying Morphs of the Yellow-Legged Hornet (Vespa velutina) and Other Pests of Quarantine Importance with Geometric Morphometrics. Diversity, 16(7), 367. https://doi.org/10.3390/d16070367








