A New Record of Bufo gargarizans Complex (Bufonidae, Anura) from Truong Son Mounts, Ha Tinh and Ha Giang Provinces, Vietnam Based on Molecular Evidence with a Description of a New Species
Abstract
1. Introduction
2. Materials and Methods
2.1. Sampling
2.2. Molecular and Phylogenetic Analyses
2.3. Morphological Analysis
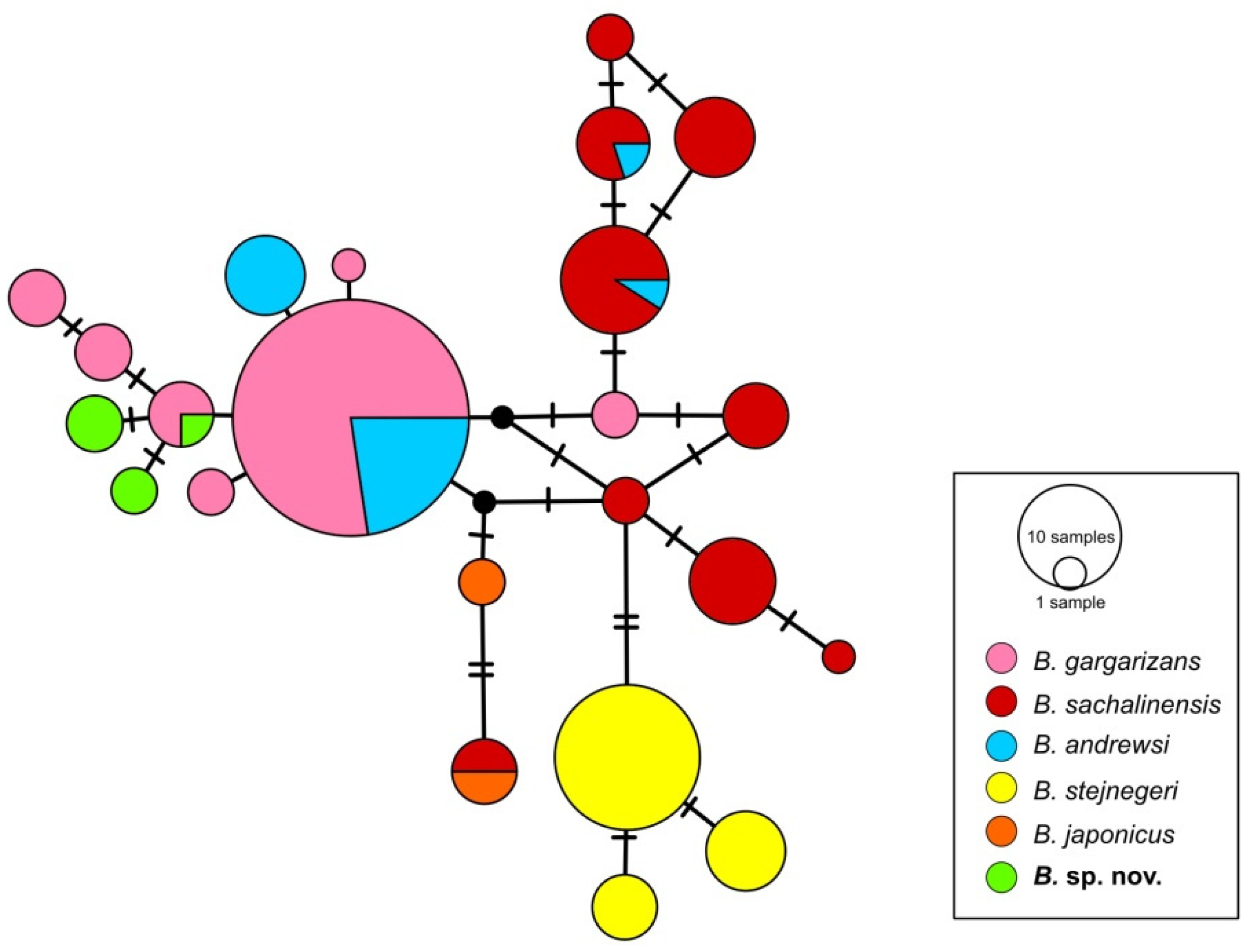
3. Results
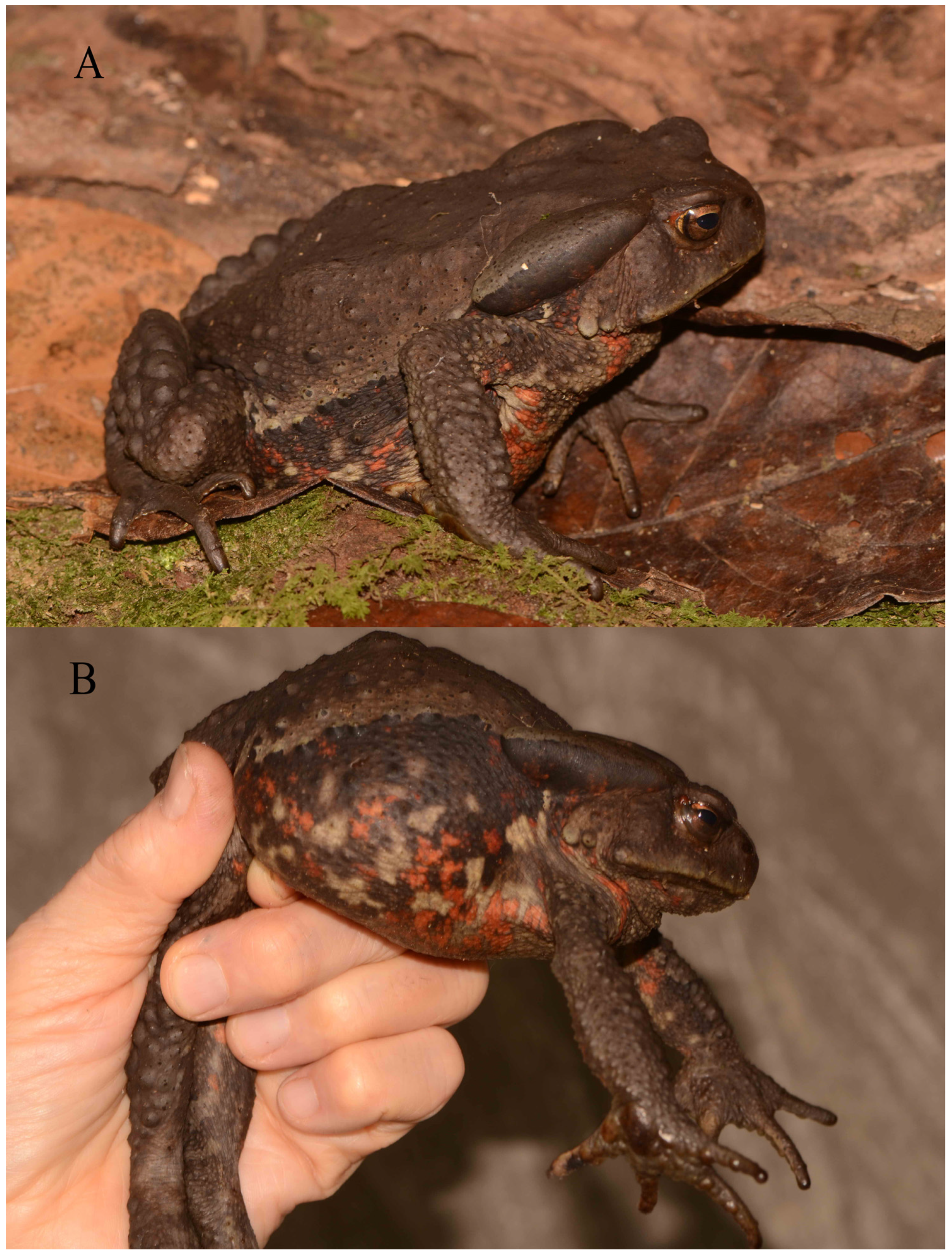
Taxonomic Account
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A. Accession Numbers of the GenBank of Used Samples
Appendix A.1. ND2 (n = 97)
Appendix A.2. RAG1 (n = 76)
References
- Frost, D.R. Amphibian Species of the World. Available online: https://amphibiansoftheworld.amnh.org/ (accessed on 17 December 2023).
- Qi, S.; Lyu, Z.-T.; Song, H.-M.; Wei, S.-C.; Zhong, Q.-F.; Wang, Y.-Y. A New Species of Stream-Living Toad (Anura: Bufonidae: Bufo) from Guangdong, China. Vertebr. Zool. 2023, 73, 677–689. [Google Scholar] [CrossRef]
- Pham, C.T.; Nguyen, T.Q.; Bernardes, M.; Nguyen, T.T.; Ziegler, T. First Records of Bufo gargarizans Cantor, 1842 and Odorrana lipuensis Mo, Chen, Wu, Zhang et Zhou, 2015 (Anura: Bufonidae, Ranidae) from Vietnam. Russ. J. Herpetol. 2016, 23, 103–107. [Google Scholar] [CrossRef]
- Othman, S.N.; Litvinchuk, S.N.; Maslova, I.; Dahn, H.; Messenger, K.R.; Andersen, D.; Jowers, M.J.; Kojima, Y.; Skorinov, D.V.; Yasumiba, K.; et al. From Gondwana to the Yellow Sea, Evolutionary Diversifications of True Toads Bufo sp. in the Eastern Palearctic and a Revisit of Species Boundaries for Asian Lineages. Elife 2022, 11, e70494. [Google Scholar] [CrossRef] [PubMed]
- Poyarkov, N.A.; Nguyen, T.V.; Popov, E.S.; Geissler, P.; Pawangkhanant, P.; Neang, T.; Suwannapoom, C.; Orlov, N.L. Recent Progress in Taxonomic Studies, Biogeographic Analysis, and Revised Checklist of Amphibians in Indochina. Russ. J. Herpetol. 2021, 28, 1–110. [Google Scholar] [CrossRef]
- Sambrook, J.; Fritsch, E.F.; Maniatis, T. Molecular Cloning: A Laboratory Manual, 2nd ed.; Cold Spring Harbor Lab.: New York, NY, USA, 1989; ISBN 978-0-87969-309-1. [Google Scholar]
- Fu, J.; Weadick, C.J.; Zeng, X.; Wang, Y.; Liu, Z.; Zheng, Y.; Li, C.; Hu, Y. Phylogeographic Analysis of the Bufo gargarizans species Complex: A Revisit. Mol. Phylogenet. Evol. 2005, 37, 202–213. [Google Scholar] [CrossRef] [PubMed]
- Macey, J.R.; Schulte, J.A., 2nd; Larson, A.; Fang, Z.; Wang, Y.; Tuniyev, B.S.; Papenfuss, T.J. Phylogenetic Relationships of Toads in the Bufo bufo species Group from the Eastern Escarpment of the Tibetan Plateau: A Case of Vicariance and Dispersal. Mol. Phylogenet. Evol. 1998, 9, 80–87. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, L.-T.; Schmidt, H.A.; von Haeseler, A.; Minh, B.Q. IQ-TREE: A Fast and Effective Stochastic Algorithm for Estimating Maximum-Likelihood Phylogenies. Mol. Biol. Evol. 2015, 32, 268–274. [Google Scholar] [CrossRef] [PubMed]
- Trifinopoulos, J.; Nguyen, L.-T.; von Haeseler, A.; Minh, B.Q. W-IQ-TREE: A Fast Online Phylogenetic Tool for Maximum Likelihood Analysis. Nucl. Acids Res. 2016, 44, W232–W235. [Google Scholar] [CrossRef] [PubMed]
- Kalyaanamoorthy, S.; Minh, B.Q.; Wong, T.K.F.; von Haeseler, A.; Jermiin, L.S. ModelFinder: Fast Model Selection for Accurate Phylogenetic Estimates. Nat. Methods 2017, 14, 587–589. [Google Scholar] [CrossRef]
- Guindon, S.; Dufayard, J.-F.; Lefort, V.; Anisimova, M.; Hordijk, W.; Gascuel, O. New Algorithms and Methods to Estimate Maximum-Likelihood Phylogenies: Assessing the Performance of PhyML 3.0. Syst. Biol. 2010, 59, 307–321. [Google Scholar] [CrossRef]
- Anisimova, M.; Gil, M.; Dufayard, J.-F.; Dessimoz, C.; Gascuel, O. Survey of Branch Support Methods Demonstrates Accuracy, Power, and Robustness of Fast Likelihood-Based Approximation Schemes. Syst. Biol. 2011, 60, 685–699. [Google Scholar] [CrossRef] [PubMed]
- Hoang, D.T.; Chernomor, O.; von Haeseler, A.; Minh, B.Q.; Vinh, L.S. UFBoot2: Improving the Ultrafast Bootstrap Approximation. Mol. Biol. Evol. 2018, 35, 518–522. [Google Scholar] [CrossRef] [PubMed]
- Clement, M.; Posada, D.; Crandall, K.A. TCS: A Computer Program to Estimate Gene Genealogies. Mol. Ecol. 2000, 9, 1657–1659. [Google Scholar] [CrossRef] [PubMed]
- Leigh, J.W.; Bryant, D. PopART: Full-Feature Software for Haplotype Network Construction. Methods Ecol. Evol. 2015, 6, 1110–1116. [Google Scholar] [CrossRef]
- Stephens, M.; Smith, N.J.; Donnelly, P. A New Statistical Method for Haplotype Reconstruction from Population Data. Am. J. Hum. Genet. 2001, 68, 978–989. [Google Scholar] [CrossRef] [PubMed]
- Librado, P.; Rozas, J. DnaSP v5: A Software for Comprehensive Analysis of DNA Polymorphism Data. Bioinformatics 2009, 25, 1451–1452. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef]
- Pham, C.T.; Le, M.D.; Hoang, C.V.; Pham, A.V.; Ziegler, T.; Nguyen, T.Q. First Records of Bufo luchunnicus (Yang et Rao, 2008) and Amolops wenshanensis Yuan, Jin, Li, Stuart et Wu, 2018 (Anura: Bufonidae, Ranidae) from Vietnam. Russ. J. Herpetol. 2020, 27, 81–86. [Google Scholar] [CrossRef]
- Ananjeva, N.B.; Borkin, L.Y.; Darevsky, I.S.; Orlov, N.L. Amphibia and Reptilia. Encyclopedia of the Nature of Russia. (Field Guide of Amphibians and Reptiles of Russia and Adjacent Countries); ABF Publishing Company: Moscow, Russia, 1998; ISBN 5-87484-066-4. (In Russian) [Google Scholar]
- Nguyen, S.V.; Ho, C.T.; Nguyen, T.Q. Herpetofauna of Vietnam; Edition Chimaira: Frankfurt am Main, Germany, 2009; ISBN 978-3-89973-462-1. [Google Scholar]
- Zhu, X.; Chen, C.; Jiang, Y.; Zhao, L.; Jin, L. Geographical Variation of Organ Size in Andrew’s Toad (Bufo andrewsi). Front. Ecol. Evol. 2022, 10, 972942. [Google Scholar] [CrossRef]
- AmphibiaWeb—Bufo gargarizans: Asiatic Toad. Available online: https://amphibiaweb.org/species/179/ (accessed on 17 February 2024).
- Fei, L.; Ye, C.Y.; Jiang, P. Colored Atlas of Chinese Amphibians and Their Distributions; Sichuan Science and Technology Press: Chengdu, China, 2012; ISBN 978-7-5364-7545-8. [Google Scholar]
- Borzée, A.; Santos, J.L.; Sánchez-RamÍrez, S.; Bae, Y.; Heo, K.; Jang, Y.; Jowers, M.J. Phylogeographic and Population Insights of the Asian Common Toad (Bufo gargarizans) in Korea and China: Population Isolation and Expansions as Response to the Ice Ages. PeerJ 2017, 5, e4044. [Google Scholar] [CrossRef]
- Lee, C.; Fong, J.J.; Jiang, J.-P.; Li, P.-P.; Waldman, B.; Chong, J.R.; Lee, H.; Min, M.-S. Phylogeographic Study of the Bufo gargarizans species Complex, with Emphasis on Northeast Asia. Anim. Cells Syst. 2021, 25, 434–444. [Google Scholar] [CrossRef]
- Zhan, A.; Fu, J. Past and Present: Phylogeography of the Bufo gargarizans species Complex Inferred from Multi-Loci Allele Sequence and Frequency Data. Mol. Phylogenet. Evol. 2011, 61, 136–148. [Google Scholar] [CrossRef] [PubMed]
- Kasatani, M. Phylogeography of the Miyako Toad, Bufo gargarizans Miyakonis (Anura: Bufonidae), Inferred from Mitochondrial Sequence Data. Curr. Herpetol. 2023, 42, 144–161. [Google Scholar] [CrossRef]
- Fu, J. Bufo gargarizans: What’s in a Name? Asian Herpetol. Res. 2023, 14, 246–255. [Google Scholar] [CrossRef]
- Dufresnes, C.; Litvinchuk, S.N. Diversity, Distribution and Molecular Species Delimitation in Frogs and Toads from the Eastern Palaearctic. Zool. J. Linn. Soc. 2022, 195, 695–760. [Google Scholar] [CrossRef]
- Ehl, S.; Vences, M.; Veith, M. Reconstructing evolution at the community level: A case study on Mediterranean amphibians. Mol. Phylogenet. Evol. 2019, 134, 211–225. [Google Scholar] [CrossRef]
- Fong, J.J.; Yang, B.-T.; Li, P.-P.; Waldman, B.; Min, M.-S. Phylogenetic systematics of the water toad (Bufo stejnegeri) elucidates the evolution of semi-aquatic toad ecology and Pleistocene glacial refugia. Front. Ecol. Evol. 2020, 7, 523. [Google Scholar] [CrossRef]
- Liedtke, H.C.; Müller, H.; Rödel, M.O.; Menegon, M.; Gonwouo, L.N.; Barej, M.F.; Gvoždík, V.; Schmitz, A.; Channing, A.; Nagel, P.; et al. No ecological opportunity signal on a continental scale? Diversification and life-history evolution of African true toads (Anura: Bufonidae). Evolution 2016, 70, 1717–1733. [Google Scholar] [CrossRef]
- Stöck, M.; Moritz, C.; Hickerson, M.; Frynta, D.; Dujsebayeva, T.; Eremchenko, V.; Macey, J.R.; Papenfuss, T.J.; Wake, D.B. Evolution of mitochondrial relationships and biogeography of Palearctic green toads (Bufo viridis subgroup) with insights in their genomic plasticity. Mol. Phylogenet. Evol. 2006, 41, 663–689. [Google Scholar] [CrossRef]
- Tong, H.; Wo, Y.; Liao, P.; Jin, Y. Phylogenetic, demographic and dating analyses of Bufo gargarizans populations from the Zhoushan archipelago and mainland China. Asian Herpetol. Res. 2017, 8, 165–173. [Google Scholar] [CrossRef]




| Collector | Date | Locality | Coordinates | Voucher Number | RAG-1 (nuDNA) GenBank Number | ND2 (mtDNA) GenBank Number |
|---|---|---|---|---|---|---|
| N. Orlov, Manh Van Le, Tao Thien Nguyen, L. Iogansen. | 16 October 2019 | Vietnam, Ha Tinh Province, Vu Quang NP. | 18.16 N 105 21 36 E | IGR 10497, female | OR113666 | OR113672 |
| N.Orlov | 11 June 2019 | Vietnam, Ha Giang, Quan Ba, Tung Vai | 23.06 N 104.92 E | ZISP:15117 male | OR113664 | OR113670 |
| N.Orlov | 11 June 2019 | Vietnam, Ha Giang, Quan Ba, Tung Vai | 23.06 N 104.92 E | ZISP:15118 female | OR113665 | OR113671 |
| N. Orlov, L. Iogansen | 18 July 2021 | Primorskiy kray, Bulyga-Fadeevo | 44.04 N 133.88 E | ZISP:15119 subad. | OR113661 | OR113667 |
| N. Orlov, L. Iogansen | 18 July 2021 | Primorskiy kray, Bulyga-Fadeevo | 44.04 N 133.88 E | ZISP:15120, female | OR113662 | OR113668 |
| N. Orlov, L. Iogansen | 18 July 2021 | Primorskiy kray, Bulyga-Fadeevo | 44.04 N 133.88 E | ZISP:15121, female | OR113663 | OR113669 |
| Species | sp. nov. | Andrewsi | Gargarizans | Sachalinensis | Stejnegeri | Japonicus |
|---|---|---|---|---|---|---|
| sp. nov. | – | 0.5 | 0.4 | 1.0 | 1.2 | 0.9 |
| Andrewsi | 4.3 | – | 0.2 | 0.7 | 0.9 | 0.7 |
| Gargarizans | 5.4 | 5.1 | – | 0.7 | 0.9 | 0.7 |
| Sachalinensis | 6.0 | 5.5 | 4.2 | – | 1.0 | 1.0 |
| Stejnegeri | 7.0 | 7.2 | 8.0 | 7.9 | – | 1.0 |
| Japonicus | 6.2 | 6.0 | 7.1 | 6.1 | 7.3 | – |
| Characters | ZISP 15117 Male Paratype | ZISP 15118 Female Paratype | IGR 10497 Female Holotype |
|---|---|---|---|
| SVL | 65.5 | 73.5 | 125.4 |
| HL | 23.3 | 25.2 | 65.4 |
| HW | 20.4 | 29.0 | 60.3 |
| MN | 15.1 | 16.5 | 50.3 |
| MFE | 12.3 | 14.0 | 43.4 |
| MBE | 12.0 | 13.4 | 35.7 |
| SNL | 10.7 | 11.8 | 13.7 |
| ED | 7.6 | 8.0 | 10.6 |
| ED/SNL | 0.7 | 0.7 | 0.8 |
| UEW | 6.5 | 7.0 | 8.6 |
| IN | 5.6 | 5.8 | 8.5 |
| IOD | 10.9 | 12.3 | 12.5 |
| DAE | 18.5 | 18.9 | 20.5 |
| DPE | 26.5 | 27.5 | 31.5 |
| NS | 5.16 | 5.4 | 6.7 |
| EN | 6.0 | 6.3 | 7.8 |
| FLL | 18.2 | 19.0 | 23.6 |
| HAL | 37.8 | 39.0 | 59.8 |
| FLL/HAL | 0.5 | 0.5 | 0.4 |
| FHL | 27.7 | 28.6 | 31.8 |
| IPT | 4.2 | 4.8 | 5.9 |
| OPT | 4.7 | 5.0 | 6.6 |
| Finger I | 8.6 | 9.0 | 11.6 |
| Finger II | 6.5 | 7.0 | 9.9 |
| Finger III | 12.7 | 12.9 | 15.7 |
| Finger IV | 9.5 | 10.0 | 11.5 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Orlov, N.L.; Ananjeva, N.B.; Ermakov, O.A.; Lukonina, S.A.; Ninh, H.T.; Nguyen, T.T. A New Record of Bufo gargarizans Complex (Bufonidae, Anura) from Truong Son Mounts, Ha Tinh and Ha Giang Provinces, Vietnam Based on Molecular Evidence with a Description of a New Species. Diversity 2024, 16, 361. https://doi.org/10.3390/d16070361
Orlov NL, Ananjeva NB, Ermakov OA, Lukonina SA, Ninh HT, Nguyen TT. A New Record of Bufo gargarizans Complex (Bufonidae, Anura) from Truong Son Mounts, Ha Tinh and Ha Giang Provinces, Vietnam Based on Molecular Evidence with a Description of a New Species. Diversity. 2024; 16(7):361. https://doi.org/10.3390/d16070361
Chicago/Turabian StyleOrlov, Nikolai L., Natalia B. Ananjeva, Oleg A. Ermakov, Svetlana A. Lukonina, Hoa Thi Ninh, and Tao Thien Nguyen. 2024. "A New Record of Bufo gargarizans Complex (Bufonidae, Anura) from Truong Son Mounts, Ha Tinh and Ha Giang Provinces, Vietnam Based on Molecular Evidence with a Description of a New Species" Diversity 16, no. 7: 361. https://doi.org/10.3390/d16070361
APA StyleOrlov, N. L., Ananjeva, N. B., Ermakov, O. A., Lukonina, S. A., Ninh, H. T., & Nguyen, T. T. (2024). A New Record of Bufo gargarizans Complex (Bufonidae, Anura) from Truong Son Mounts, Ha Tinh and Ha Giang Provinces, Vietnam Based on Molecular Evidence with a Description of a New Species. Diversity, 16(7), 361. https://doi.org/10.3390/d16070361







