Genetic Population Structure of Lane Snapper Lutjanus synagris (Linnaeus, 1758) in Western Atlantic: Implications for Conservation
Abstract
1. Introduction
2. Materials and Methods
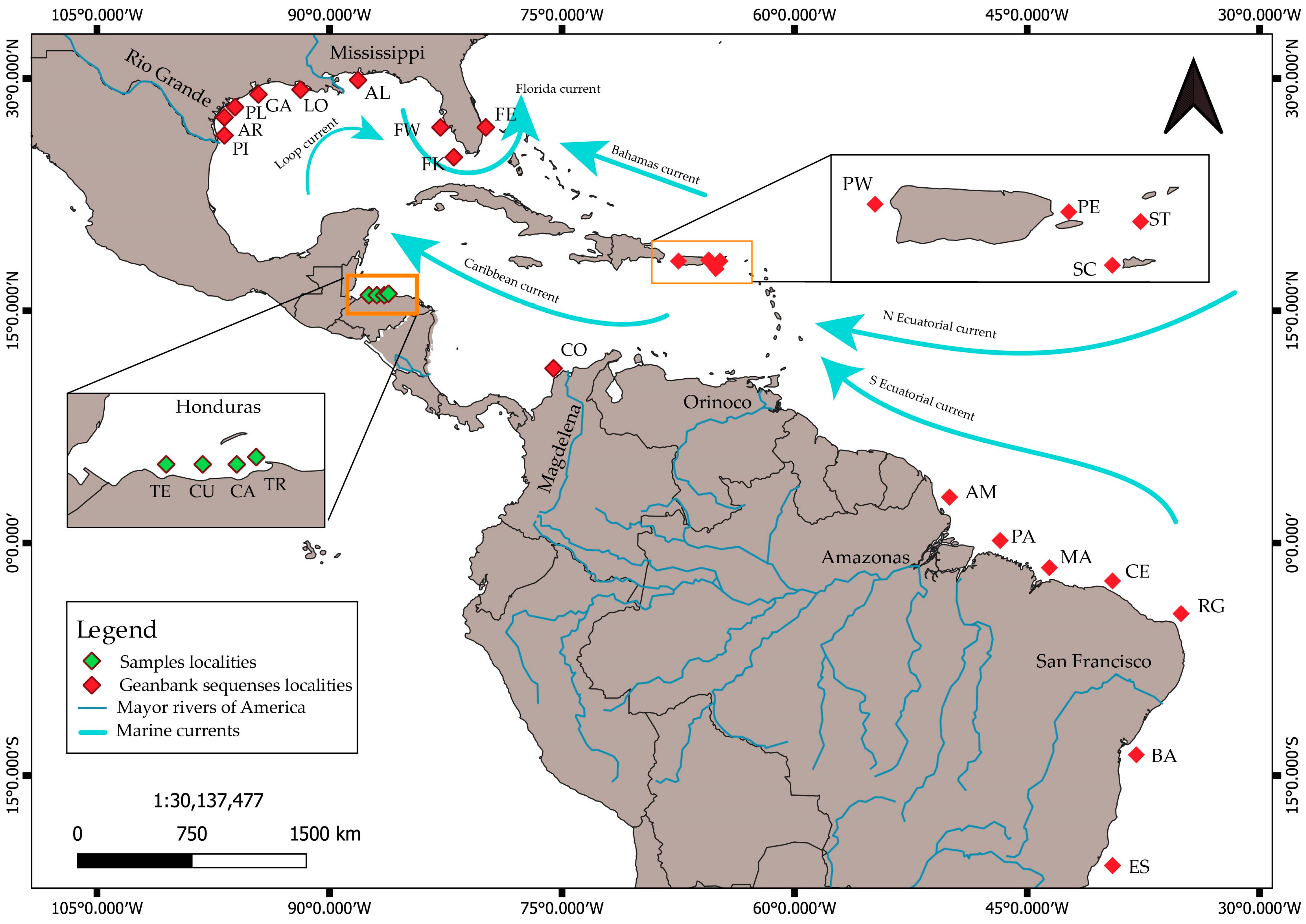
2.1. Fish Sampling
2.2. Laboratory Procedure
2.3. A Compilation of Sequences across the Western Atlantic
2.4. Genetic Structure
2.5. Demographic History in the Western Atlantic
3. Results
3.1. Genetic Diversity in Honduras
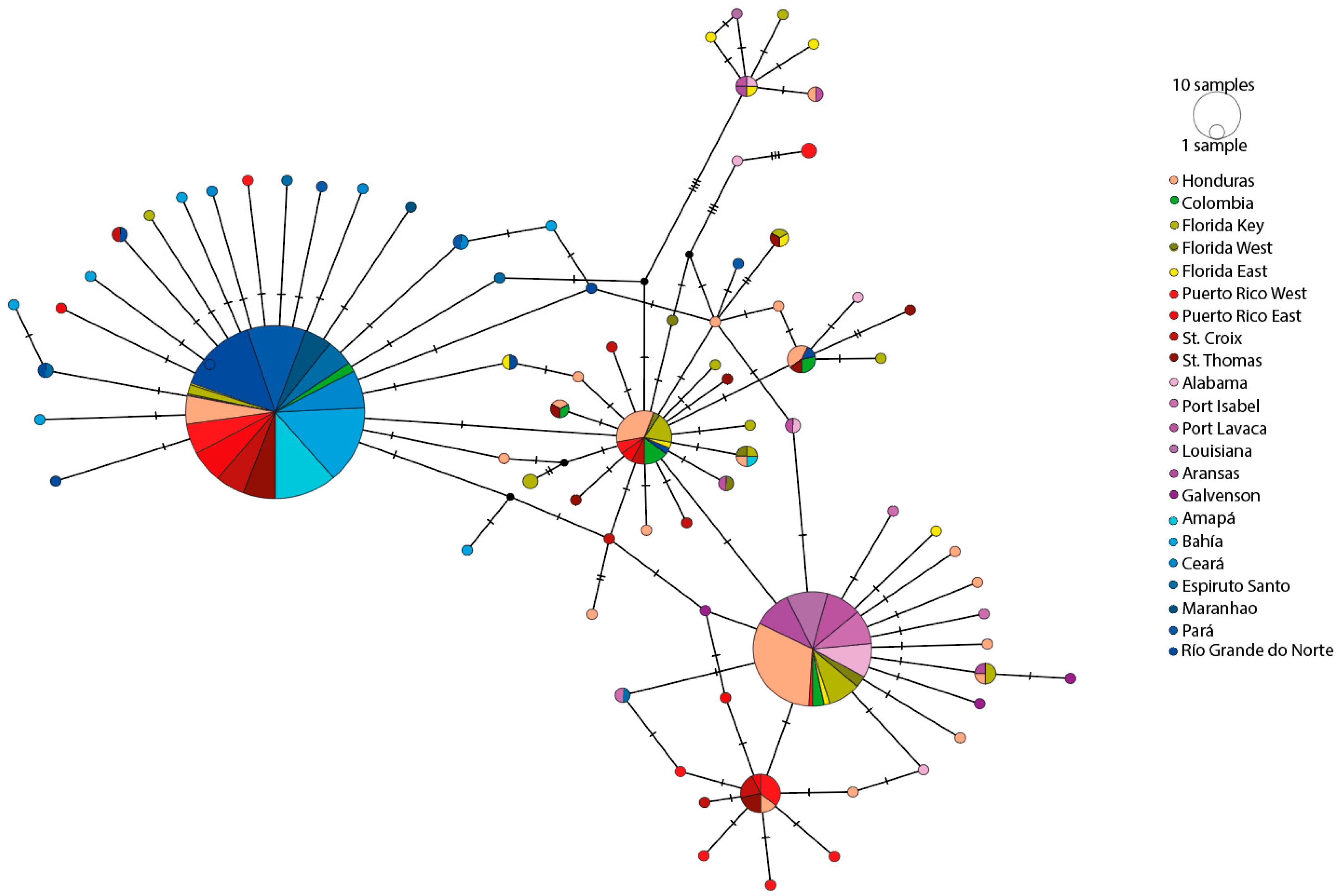
3.2. Genetic Variation and Population Structure in Honduras
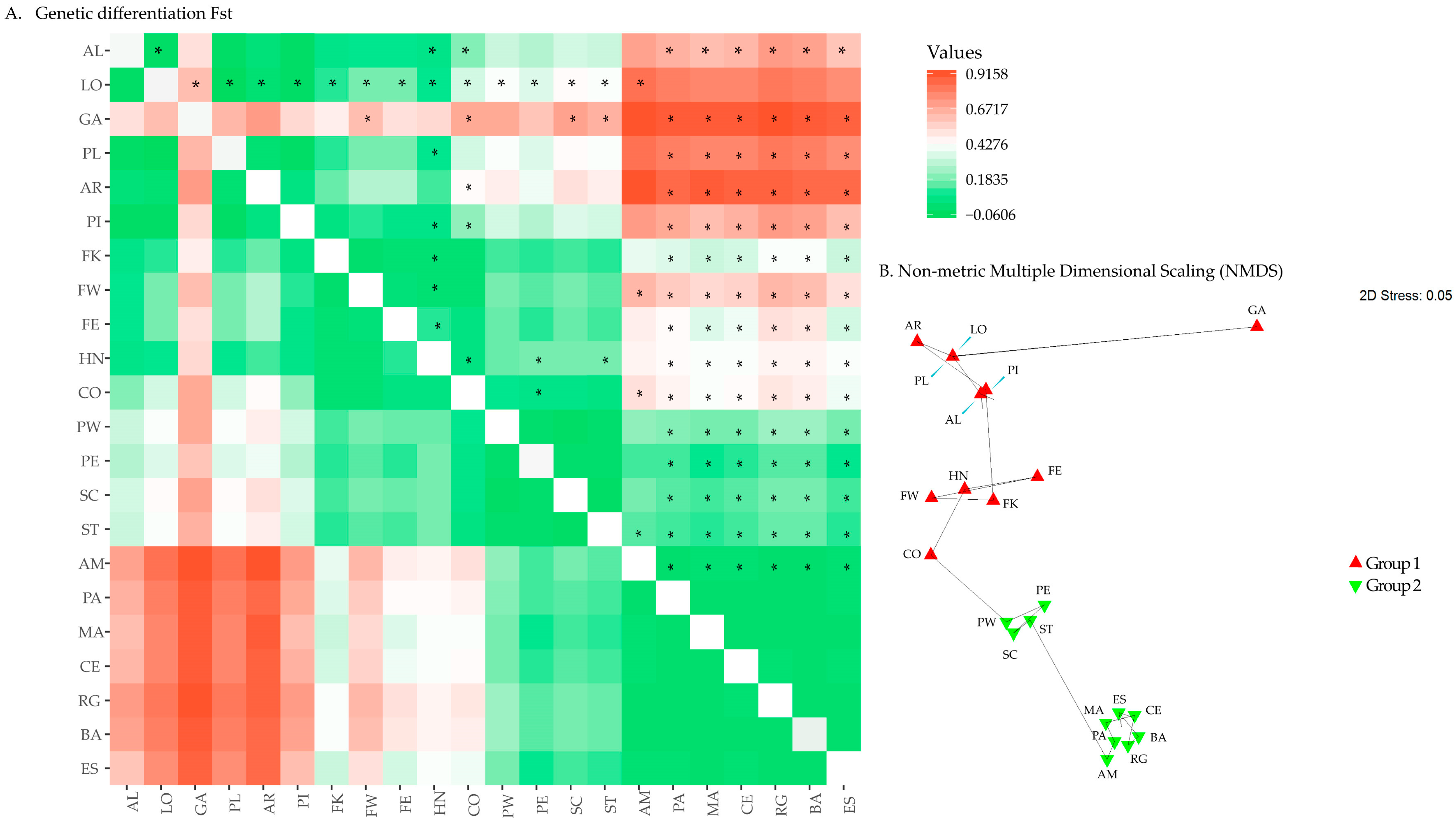
3.3. Genetic Variation and Population Structure in the Western Atlantic
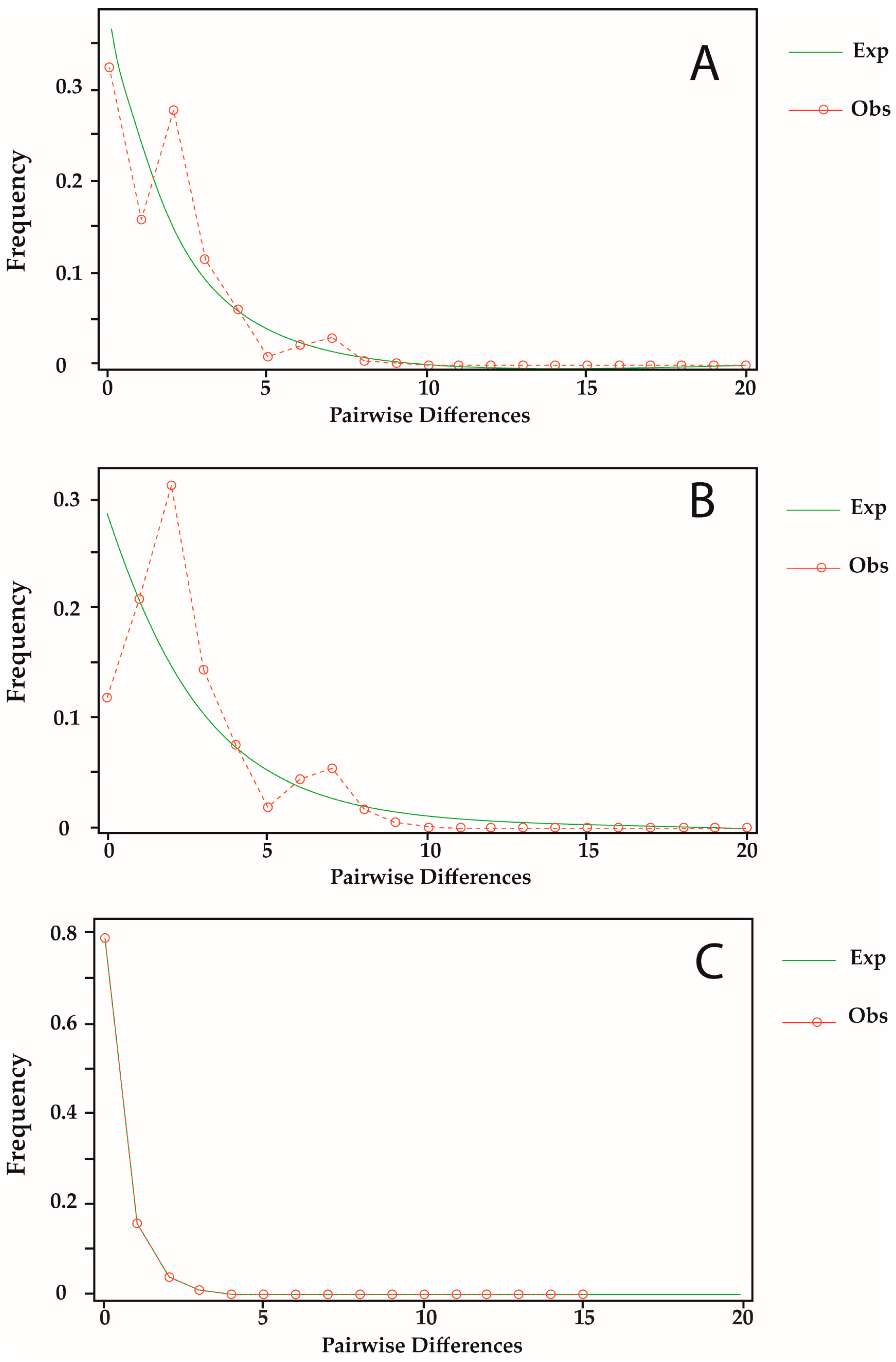
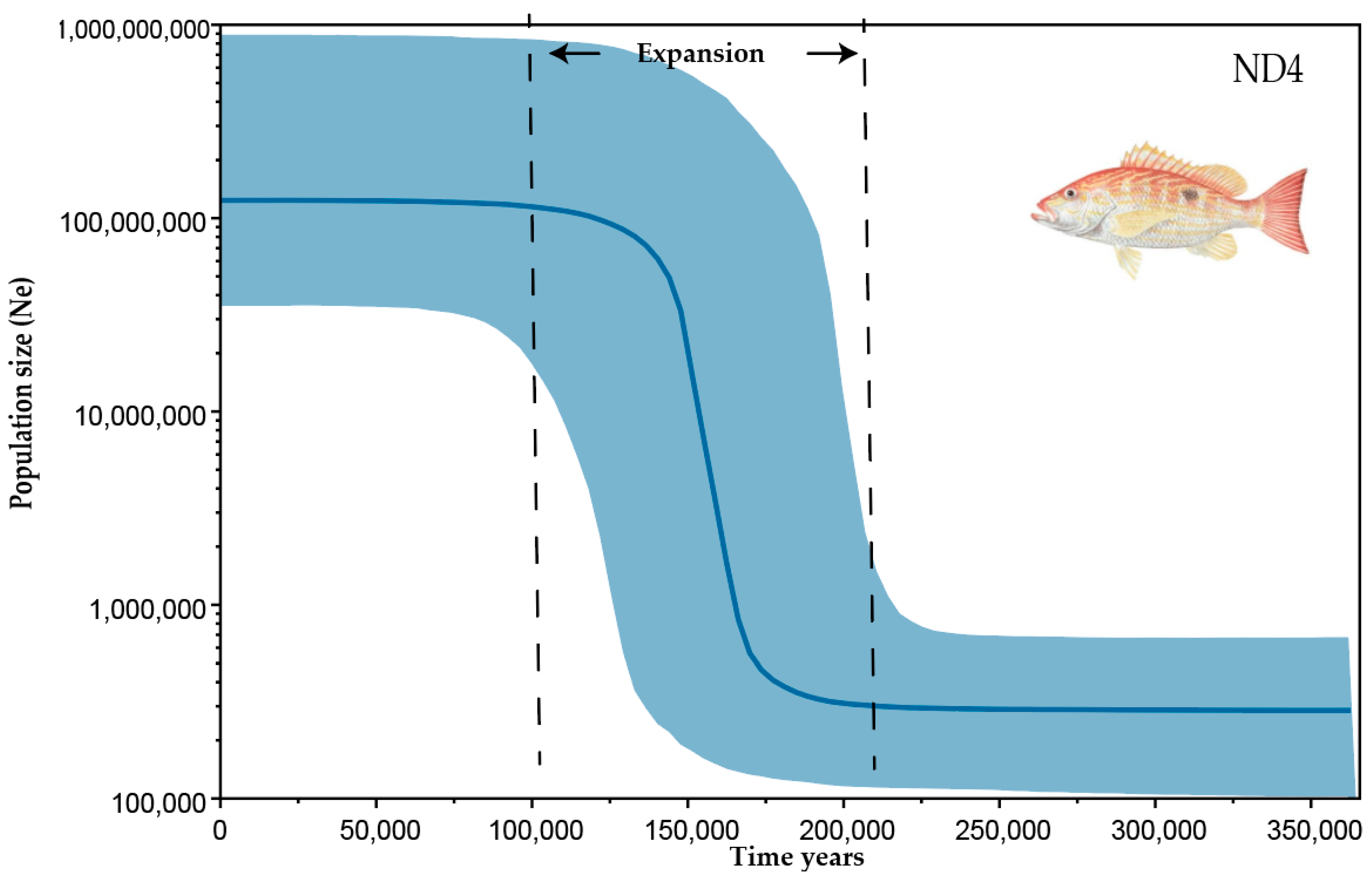
3.4. Demographic History
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Draheim, H.M.; Moore, J.A.; Etter, D.; Winterstein, S.R.; Scribner, K.T. Detecting black bear source–sink dynamics using individual-based genetic graphs. Proc. R. Soc. B Biol. Sci. 2016, 283, 20161002. [Google Scholar] [CrossRef] [PubMed]
- Gaggiotti, O.E. Population genetic models of source-sink metapopulations. Theor. Popul. Biol. 1996, 50, 178–208. [Google Scholar] [CrossRef] [PubMed]
- Green, A.; Fernandes, L.; Almany, G.; Abesamis, R.; McLeod, E.; Aliño, P.; White, A.T.; Salm, R.; Tanzer, J.; Pressey, R.L. Designing Marine Reserves for Fisheries Management, Biodiversity Conservation, and Climate Change Adaptation. Coast. Manag. 2014, 42, 143–159. [Google Scholar] [CrossRef]
- Canty, S.W.J.; Preziosi, R.F.; Rowntree, J.K. Dichotomy of mangrove management: A review of research and policy in the Mesoamerican reef region. Ocean Coast. Manag. 2018, 157, 40–49. [Google Scholar] [CrossRef]
- Green, A.; Maypa, A.; Almany, G.; Rhodes, K.; Weeks, R.; Abesamis, R.; Gleason, M.G.; Mumby, P.J.; White, A.T. Larval dispersal and movement patterns of coral reef fishes, and implications for marine reserve network design. Biol. Rev. 2015, 90, 1215–1247. [Google Scholar] [CrossRef]
- Green, A.; Chollett, I.; Suárez, A.; Dahlgren, C.; Cruz, S.; Zepeda, C.; Andino, J.; Robinson, J.; McField, M.; Fulton, S.; et al. Biophysical Principles for Designing a Network of Replenishment Zones for the Mesoamerican Reef System. The Nature Conservancy-México, 30 October 2017. [Google Scholar]
- Beltrán, D.M.; Schizas, N.V.; Appeldoorn, R.S.; Prada, C. Effective Dispersal of Caribbean Reef Fish is Smaller than Current Spacing among Marine Protected Areas. Sci. Rep. 2017, 7, 4689. [Google Scholar] [CrossRef]
- Christie, M.; Tissot, B.; Albins, M.; Beets, J.; Jia, Y.; Ortiz, D.; Thompson, S.E.; Hixon, M.A. Larval connectivity in an effective network of marine protected areas. PLoS ONE 2010, 5, e15715. [Google Scholar] [CrossRef] [PubMed]
- Cowen, R.; Paris, C.; Srinivasan, A. Scaling of connectivity in marine populations. Science 2006, 311, 522–527. [Google Scholar] [CrossRef]
- Soto, I.; Andréfouët, S.; Hu, C.; Muller-Karger, F.; Wall, C.; Sheng, J.; Hatcher, B.G. Physical connectivity in the Mesoamerican Barrier Reef System inferred from 9 years of ocean color observations. Coral Reefs 2009, 28, 415–425. [Google Scholar] [CrossRef]
- Green, B.S.; Mapstone, B.D.; Carlos, G.; Begg, G.A. Tropical Fish Otoliths: Information for Assessment, Management and Ecology. In Reviews: Methods and Technologies in Fish Biology and Fisheries; Springer: New York, NY, USA, 2009; Volume 11, 328p. [Google Scholar]
- Chollett, I.; Canty, S.W.J.; Box, S.J.; Mumby, P.J. Adapting to the impacts of global change on an artisanal coral reef fishery. Ecol. Econ. 2014, 102, 118–125. [Google Scholar] [CrossRef]
- Mech, S.G.; Hallett, J.G. Evaluating the Effectiveness of Corridors: A Genetic Approach. Conserv. Biol. 2001, 15, 467–474. [Google Scholar] [CrossRef]
- Balbar, A.C.; Metaxas, A. The current application of ecological connectivity in the design of marine protected areas. Glob. Ecol. Conserv. 2019, 17, e00569. [Google Scholar] [CrossRef]
- Pujolar, J.M.; Schiavina, M.; Di Franco, A.; Melia, P.; Guidetti, P.; Gatto, M.; De Leo, G.A.; Zane, L. Understanding the effectiveness of marine protected areas using genetic connectivity patterns and Lagrangian simulations. J. Conserv. Biogeogr. 2013, 19, 1531–1542. [Google Scholar] [CrossRef]
- White, J.W.; Botsford, L.W.; Hastings, A.; Largier, J.L. Population persistence in marine reserve networks: Incorporating spatial heterogeneities in larval dispersal. Mar. Ecol. Prog. Ser. 2009, 398, 49–67. [Google Scholar] [CrossRef]
- Miller, K.J.; Ayre, D.J. Protection of Genetic Diversity and Maintenance of Connectivity among Reef Corals within Marine Protected Areas. Conserv. Biol. 2008, 22, 1245–1254. [Google Scholar]
- Claudet, J.; Osenberg, C.W.; Domenici, P.; Badalamenti, F.; Milazzo, M.; Falcón, J.M.; Bertocci, I.; Benedetti-Cecchi, L.; Garcia-Charton, J.-A.; Goni, R.; et al. Marine reserves: Fish life history and ecological traits matter. Ecol. Appl. 2010, 20, 830–839. [Google Scholar] [CrossRef]
- Apostolaki, P.; Milner-Gulland, E.J.; McAllister, M.K.; Kirkwood, G.P. Modelling the effects of establishing a marine reserve for mobile fish species. Can. J. Fish. Aquat. Sci. 2002, 59, 405–415. [Google Scholar] [CrossRef]
- Guzman, H.M.; Benfield, S.; Breedy, O.; Mair, J.M. Broadening reef protection across the Marine Conservation Corridor of the Eastern Tropical Pacific: Distribution and diversity of reefs in Las Perlas Archipelago, Panama. Environ. Conserv. 2008, 35, 46–54. [Google Scholar] [CrossRef]
- Pendoley, K.L.; Schofield, G.; Whittock, P.A.; Ierodiaconou, D.; Hays, G.C. Protected species use of a coastal marine migratory corridor connecting marine protected areas. Mar. Biol. 2014, 161, 1455–1466. [Google Scholar] [CrossRef]
- Gell, F.R.; Roberts, C.M. Benefits beyond boundaries: The fishery effects of marine reserves. Trends Ecol. Evol. 2003, 18, 448–455. [Google Scholar] [CrossRef]
- Rakitin, A.; Kramer, D.L. Effect of a marine reserve on the distribution of coral reef fishes in Barbados. Mar. Ecol. Prog. Ser. 1996, 131, 97–113. [Google Scholar] [CrossRef]
- Diamond, S.L.; Kleisner, K.M.; Duursma, D.E.; Wang, Y. Designing marine reserves to reduce bycatch of mobile species: A case study using juvenile red snapper (Lutjanus campechanus). Can. J. Fish. Aquat. Sci. 2010, 67, 1335–1349. [Google Scholar] [CrossRef]
- McCook, L.J.; Ayling, T.; Cappo, M.; Choat, J.H.; Evans, R.D.; De Freitas, D.M.; Heupel, M.; Hughes, T.P.; Jones, G.P.; Mapstone, B.; et al. Adaptive management of the Great Barrier Reef: A globally significant demonstration of the benefits of networks of marine reserves. Proc. Natl. Acad. Sci. USA 2010, 107, 18278–18285. [Google Scholar] [CrossRef] [PubMed]
- Hilborn, R.; Stokes, K.; Maguire, J.J.; Smith, T.; Botsford, L.W.; Mangel, M.; Orensanz, J.; Parma, A.; Rice, J.; Bell, J.; et al. When can marine reserves improve fisheries management? Ocean Coast. Manag. 2004, 47, 197–205. [Google Scholar] [CrossRef]
- Fernandes, L.; Day, J.; Lewis, A.; Slegers, S.; Kerrigan, B.; Breen, D.; Cameron, D.; Jago, B.; Hall, J.; Lowe, D.; et al. Establishing representative no-take areas in the great barrier reef: Large-scale implementation of theory on marine protected areas. Conserv. Biol. 2005, 19, 1733–1744. [Google Scholar] [CrossRef]
- Isaza-Toro, E.; Selvaraj, J.J.; Giraldo-Lopez, A.; Ortíz-Ferrín, O.O.Y. Fish aggregating devices in the eastern tropical Pacific marine corridor, according to the Colombian Fisheries Observer Program. Aquat. Conserv. 2021, 31, 3311–3318. [Google Scholar] [CrossRef]
- Trejo-Martínez, J.; Brulé, T.; Morales-López, N.; Colás-Marrufo, T.; Sánchez-Crespo, M. Reproductive Strategy of a Continental Shelf Lane Snapper Population from the Southern Gulf of Mexico. Mar. Coast. Ficheries Manag. Ecosyst. Sci. 2021, 13, 140–156. [Google Scholar] [CrossRef]
- Viana, D.F.; Vieira Hazin, F.H.; Oliveira, P.G. Reproductive Biology of Lane Snapper, Lutjanus synagris (Perciformes: Lutjanidae), of Northern Pernambuco State, Brazil. Arq. Cienc. Mar. 2015, 48, 67–73. [Google Scholar]
- Rivera, A.; San Martín, J.; Guardiola, P. Diagnóstico Situacional de la Pesca Artesanal en los Municipios de Trujillo y Santa Fe; CORAL-GOAL: Colón, Honduras, 2019. [Google Scholar]
- Manickchand-Dass, S. Reproduction, age and growth of the lane snapper, Lutjanus synagris (Linnaeus), in Trinidad, West Indies. Bull. Mar. Sci. 1987, 40, 22–28. [Google Scholar]
- Carpenter, K.E. The Living Marine Resources of The Western Central Aatlantic; Smith, J.F., Whithaus, S., Askew, S., Kautenberger-Longo, M., DeAngelis, N., Eds.; FAO: Rome, Italy, 2002; pp. 1375–2127. [Google Scholar]
- Allen, G.R. Snappers of the world. Annotated and illustrated catalogue of lutjanid species known to date. In FAO Species Catalogue; FAO Fisheries Synopsis 6 (125), 2008; FAO: Rome, Italy, 1986; pp. 119–121. [Google Scholar]
- Karlsson, S.; Saillant, E.; Gold, J. Population structure and genetic variation of lane snapper (Lutjanus synagris) in the northern Gulf of Mexico. Mar. Biol. 2009, 156, 1841–1855. [Google Scholar] [CrossRef]
- Luckhurst, B.E.; Dean, J.M.; Reichert, M. Age, growth and reproduction of the lane snapper Lutjanus synagris (pisces: Lutjanidae) at Bermuda. Mar. Ecol. Prog. Ser. 2000, 203, 255–261. [Google Scholar] [CrossRef]
- Silva, D.; Martins, K.; Oliveira, J.; Da Silva, R.; Sampaio, I.; Schneider, H.; Gomes, G. Genetic differentiation in populations of lane snapper (Lutjanus synagris–Lutjanidae) from Western Atlantic as revealed by multilocus analysis. Fish Res. 2018, 198, 138–149. [Google Scholar] [CrossRef]
- Gold, J.R.; Saillant, E.; Cummings, N.J.; Renshaw, M.A. Genetic divergence and effective size among lane snapper in U.S. waters of the western atlantic ocean. N. Am. J. Fish Manag. 2011, 31, 209–223. [Google Scholar] [CrossRef]
- Gold, J.R.; Voelker, G.; Renshaw, M.A. Phylogenetic relationships of tropical western Atlantic snappers in subfamily Lutjaninae (Lutjanidae: Perciformes) inferred from mitochondrial DNA sequences. Biol. J. Linn. Soc. 2011, 102, 915–929. [Google Scholar] [CrossRef]
- Lee, W.J.; Conroy, J.; Howell, W.H.; Kocher, T.D. Structure and Evolution of Teleost Mitochondrial Control Regions. Mol. Evol. 1995, 41, 54–66. [Google Scholar] [CrossRef]
- Arevalo, E.; Scott, D.; Sites, J. Mitochondrial DNA Sequence Ddivergence and Phylogenetic Relationships among Eighte Chromosome Races of the Sceloporus grammicus Complex (Phrinosomatidae) in Central Mexico. Syst. Biol. 1994, 43, 387–418. [Google Scholar] [CrossRef]
- Bielawski, J.P.; Gold, J.R. Mutation patterns of mitochondrial H- and L-strand DNA in closely related cyprinid fishes. Genetics 2002, 161, 1589–1597. [Google Scholar] [CrossRef] [PubMed]
- Chow, S.; Hazama, K. Universal PCR primers for S7 ribosomal protein gene introns in fish. Mol. Ecol. 1998, 7, 1255–1256. [Google Scholar]
- Kearse, M.; Moir, R.; Wilson, A.; Stones-Havas, S.; Cheung, M.; Sturrock, S.; Buxton, S.; Cooper, A.; Markowitz, S.; Duran, C.; et al. Geneious Basic: An integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 2012, 28, 1647–1649. [Google Scholar] [CrossRef]
- Gold, J.R.; Saillant, E.; Ebelt, N.D.; Lem, S. Conservation Genetics of Gray Snapper (Lutjanus griseus) in U.S. Waters of the Northern Gulf of Mexico and Western Atlantic Ocean. Copeia 2009, 2009, 277–286. [Google Scholar] [CrossRef]
- Kruskal, J.B. Multidimiensional Scalin by Optimizing Goodness of Fit to Nonmetric Hypothesis. Pshychometrika 1964, 29, 1–27. [Google Scholar] [CrossRef]
- Excoffier, L.; Lischer, H.E.L. Arlequin suite ver 3.5: A new series of programs to perform population genetics analyses under Linux and Windows. Mol. Ecol. Resour. 2010, 10, 564–567. [Google Scholar] [CrossRef] [PubMed]
- Librado, P.; Rozas, J. DnaSp v5: A Software for comprehensive analysis of DNA polymorphism data. Bioinformatics 2009, 25, 1451–1452. [Google Scholar] [CrossRef] [PubMed]
- Dupanloup, I.; Schneider, S.; Excoffier, L. A simulated annealing approach to define the genetic structure of populations. Mol. Ecol. 2002, 11, 2571–2581. [Google Scholar] [CrossRef] [PubMed]
- Bandelt, H.; Forster, P.; Röhl, A. Median-joining networks for inferring intraspecific phylogenies. Mol. Biol. Evol. 1999, 16, 37–48. [Google Scholar] [CrossRef] [PubMed]
- Leigh, J.W.; Bryant, D.; Nakagawa, S. POPART: Full-feature software for haplotype network construction. Methods Ecol. Evol. 2015, 6, 1110–1116. [Google Scholar] [CrossRef]
- Tajima, F. Statistical Method for Testing the Neutral Mutation Hypothesis by DNA Polimorphism. Genet. Soc. Am. 1989, 3, 607–612. [Google Scholar]
- Fu, Y.X. Statistical tests of neutrality of mutations against population growth, hitchhiking and background selection. Genetics 1997, 147, 915–925. [Google Scholar] [CrossRef] [PubMed]
- Palumbi, S.R.; Cipriano, F.; Hare, M.P. Predicting nuclear gene coalescence from mitochondrial data: The three-times rule. Evolution 2001, 55, 859–868. [Google Scholar] [CrossRef]
- Cazes, M.H.; Hartl, D.; Clark, A. Principles of Population Genetics. Population (French Edition) 1999, 54, 1042. [Google Scholar] [CrossRef]
- Bouckaert, R.; Heled, J.; Kühnert, D.; Vaughan, T.; Wu, C.H.; Xie, D.; Suchard, M.A.; Rambaut, A.; Drummond, A.J. BEAST 2: A Software Platform for Bayesian Evolutionary Analysis. PLoS Comput. Biol. 2014, 10, e1003537. [Google Scholar] [CrossRef] [PubMed]
- Barido-Sottani, J.; Bošková, V.; Plessis LDu Kühnert, D.; Magnus, C.; Mitov, V.; Müller, N.F.; PečErska, J.; Rasmussen, D.A.; Zhang, C.; Drummond, A.J.; et al. Taming the BEAST—A Community Teaching Material Resource for BEAST 2. Syst. Biol. 2018, 67, 170–174. [Google Scholar] [CrossRef] [PubMed]
- Guindon, S.; Dufayard, J.F.; Lefort, V.; Anisimova, M.; Hordijk, W.; Gascuel, O. New Algorithms and Methods to Estimate Maximim-Likelihood Phylogenies Assessing the Performance of PhyML 3.0. Syst. Biol. 2010, 59, 307–321. [Google Scholar] [CrossRef] [PubMed]
- Lefort, V.; Longueville, J.E.; Gascuel, O. SMS: Smart Model Selection in PhyML. Mol. Biol. Evol. 2017, 34, 2422–2424. [Google Scholar] [CrossRef] [PubMed]
- Drummond, A.J.; Rambaut, A.; Shapiro, B.; Pybus, O.G. Bayesian coalescent inference of past population dynamics from molecular sequences. Mol. Biol. Evol. 2005, 22, 1185–1192. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Cai, Z.; Huang, L. Population genetic structure of crimson snapper Lutjanus erythropterus in East Asia, revealed by analysis of the mitochondrial control region. ICES J. Mar. Sci. 2006, 63, 693–704. [Google Scholar] [CrossRef]
- Rambaut, A.; Drummond, A.J.; Xie, D.; Baele, G.; Suchard, M.A. Posterior summarization in Bayesian phylogenetics using Tracer 1.7. Syst. Biol. 2018, 67, 901–904. [Google Scholar] [CrossRef] [PubMed]
- Teacher, A.G.; André, C.; Jonsson, P.R.; Merilä, J. Oceanographic connectivity and environmental correlates of genetic structuring in Atlantic herring in the Baltic Sea. Evol. Appl. 2013, 6, 549–567. [Google Scholar] [CrossRef] [PubMed]
- Fauvelot, C.; Bernardi, G.; Planes, A.S. Reductions in the mitochondrial DNA diversity of coral reef fish provide evidence of population bottlenecks resultin from Holocene sea-level change. Evolution 2003, 57, 1571–1583. [Google Scholar]
- Harborne, A.R.; Afzal, D.C.; Andrews, M.J. Honduras: Caribbean Coast. Mar. Pollut. Bull. 2001, 42, 1221–1235. [Google Scholar] [CrossRef]
- Truelove, N.K.; Griffiths, S.; Ley-Cooper, K.; Azueta, J.; Majil, I.; Box, S.; Behringer, D.C.; Butler, M.J.; Preziosi, R.F. Genetic evidence from the spiny lobster fishery supports international cooperation among Central American marine protected areas. Conserv. Genet. 2015, 16, 347–358. [Google Scholar] [CrossRef]
- Garber, A.F.; Tringali, M.D.; Stuck, K.C. Population structure and variation in red snapper (Lutjanus campechanus) from the Gulf of Mexico and Atlantic coast of Florida as determined from mitochondrial DNA control region sequence. Mar. Biotechnol. 2004, 6, 175–185. [Google Scholar] [CrossRef] [PubMed]
- Vasconcellos, A.V.; Vianna, P.; Paiva, P.C.; Schama, R.; Solé-Cava, A. Genetic and morphometric differences between yellowtail snapper (Ocyurus chrysurus, Lutjanidae) populations of the tropical West Atlantic. Genet. Mol. Biol. 2008, 31 (Suppl. S1), 308–316. [Google Scholar] [CrossRef]
- Truelove, N.K.; Kough, A.S.; Behringer, D.C.; Paris, C.B.; Box, S.J.; Preziosi, R.F.; Butler, M.J. Biophysical connectivity explains population genetic structure in a highly dispersive marine species. Coral Reefs 2017, 36, 233–244. [Google Scholar] [CrossRef]
- Taylor, M.S.; Hellberg, M.E. Comparative phylogeography in a genus of coral reef fishes: Biogeographic and genetic concordance in the Caribbean. Mol. Ecol. 2006, 15, 695–707. [Google Scholar] [CrossRef] [PubMed]
- Saillant, E.A.; Renshaw, M.A.; Cummings, N.J.; Gold, J.R. Conservation genetics and management of yellowtail snapper, Ocyurus chrysurus, in the US Caribbean and South Florida. Fish. Manag. Ecol. 2012, 19, 301–312. [Google Scholar] [CrossRef]
- Da Silva, R.; Veneza, I.; Sampaio, I.; Araripe, J.; Schneider, H.; Gomes, G. High Levels of Genetic Connectivity among Populations of Yellowtail Snapper, Ocyurus chrysurus (Lutjanidae–Perciformes), in the Western South Atlantic Revealed through Multilocus Analysis. PLoS ONE 2015, 10, e0122173. [Google Scholar] [CrossRef] [PubMed]
- Coimbra, M.R.M.; Benevides, E.; Farias, R.d.S.; da Silva, B.C.N.R.; Cloux, S.; Pérez-Muñuzuri, V.; Vera, M.; Torres, R. Restricted connectivity for cobia Rachycentron canadum (Perciformes: Rachycentridae) in the Western Atlantic Ocean. Fish. Oceanogr. 2023, 32, 495–508. [Google Scholar] [CrossRef]
- Heyman, W.D.; Kjerfve, B.; Ezer, T. Mesoamerican Reef Spawning Aggregations Help Maintain Fish Populations: A Review of Connectivity Research and Priorities for Science and Management Mexico Belize Honduras Guatemala. Caribbean Connectivity: Implications for Marine Protected Area Management. In Proceedings of the 59th Annual Meeting of the Gulf and Caribbean Fisheries Institute, Belize City, Belize, 9–11 November 2006; pp. 150–169. [Google Scholar]
- Taylor, M.S.; Hellberg, M.E. Genetic Evidence for Local Retention of Pelagic Larvae in a Caribbean Reef Fish. Science 2003, 299, 107–109. [Google Scholar] [CrossRef]
- Baums, I.B.; Miller, M.W.; Hellberg Michael, E. Regionally isolated populations of an imperiled Caribbean coral, Acropora palmata. Mol. Ecol. 2005, 14, 1377–1390. [Google Scholar] [CrossRef]
- Baums, I.B.; Paris, C.B.; Chérubin, L.M. A bio-oceanographic filter to larval dispersal in a reef-building coral. Limnol. Oceanogr. 2006, 51, 1969–1981. [Google Scholar] [CrossRef]
- Orr, M.R.; Smith, T.B. Ecology ans speciation. Trends Ecol. Evol. 1998, 13, 502–506. [Google Scholar] [CrossRef] [PubMed]
- Barreto, A.M.B.; Bezerra, F.H.R.; Suguio, K.; Tatumi, S.H.; Yee, M.; Paiva, R.P.; Munita, C.S. Late Pleistocene marine terrace deposits in northeastern Brazil: Sea-level change and tectonic implications. Palaleo 2002, 179, 57–69. [Google Scholar] [CrossRef]
- Carvalho, G.R.; Hauser, L. Molecular genetics and the stock concept in fisheries. Rev. Fish Biol. Fish. 2004, 4, 326–350. [Google Scholar] [CrossRef]
- Ludt, W.B.; Rocha, L.A. Shifting seas: The impacts of Pleistocene sea-level fluctuations on the evolution of tropical marine taxa. J. Biogeogr. 2015, 42, 25–38. [Google Scholar] [CrossRef]
- Berumen, M.L.; Almany, G.R.; Planes, S.; Jones, G.P.; Saenz-Agudelo, P.; Thorrold, S.R. Persistence of self-recruitment and patterns of larval connectivity in a marine protected area network. Ecol. Evol. 2012, 2, 444–452. [Google Scholar] [CrossRef] [PubMed]
- Chapman, M.R.; Kramer, D.L. Movements of fishes within and among fringing coral reefs in Barbados. Environ. Biol. Fishes 2000, 57, 11–24. [Google Scholar] [CrossRef]
- Foster, N.; Paris, C.; Kool, J.; Baums, I.; Stevens, J.; Sanchez, J.; Bastidas, C.; Agudelo, C.; Bush, P.; Day, O.; et al. Connectivity of Caribbean coral populations: Complementary insights from empirical and modelled gene flow. Mol. Ecol. 2012, 21, 1143–1157. [Google Scholar] [CrossRef]
| Indices | Sample Localities | ||||
|---|---|---|---|---|---|
| n | S | K | H | π | |
| Honduras localities | |||||
| D-loop | |||||
| TE | 28 | 35 | 21 | 0.963 | 0.00923 |
| CU | 30 | 57 | 25 | 0.982 | 0.01273 |
| CA | 30 | 46 | 26 | 0.989 | 0.01142 |
| TR | 15 | 38 | 14 | 0.990 | 0.01088 |
| Total | 103 | 76 | 72 | 0.977 | 0.01106 |
| ND4 | |||||
| TE | 27 | 11 | 10 | 0.766 | 0.00298 |
| CU | 15 | 4 | 5 | 0.695 | 0.00212 |
| CA | 27 | 10 | 11 | 0.786 | 0.00274 |
| TR | 14 | 10 | 6 | 0.747 | 0.00359 |
| Total | 83 | 25 | 20 | 0.740 | 0.00279 |
| S7-1 | |||||
| TE | 29 | 8 | 11 | 0.938 | 0.00391 |
| CU | 25 | 11 | 21 | 0.942 | 0.00513 |
| CA | 25 | 11 | 20 | 0.960 | 0.00502 |
| TR | 15 | 9 | 15 | 0.952 | 0.00549 |
| Total | 94 | 13 | 53 | 0.963 | 0.00496 |
| ND4 Atlantic Ocean localities | |||||
| Gulf of Mexico [35] | 93 | 43 | 16 | 0.346 | 0.00372 |
| Florida [38] | 77 | 40 | 21 | 0.845 | 0.00461 |
| Honduras [present study] | 83 | 35 | 20 | 0.740 | 0.00279 |
| Colombia [37] | 16 | 4 | 5 | 0.808 | 0.00226 |
| Puerto Rico [38] | 101 | 41 | 25 | 0.590 | 0.00291 |
| Brazil [37] | 216 | 22 | 23 | 0.210 | 0.00051 |
| Total | 586 | 66 | 79 | 0.676 | 0.00321 |
| SAMOVA Fixation Index | ||||
| K Value | FCT | FST | p Value | |
| 2 | 0.427 | 0.479 | p < 0.001 | |
| 3 | 0.425 | 0.475 | p < 0.001 | |
| 4 | 0.423 | 0.449 | p < 0.001 | |
| AMOVA | ||||
| Source of Variation | d.f. | Sum of Squares | Variance Components | % of Variation |
| Among groups (va) | 1 | 126.511 | 0.448 va | 42.67 |
| Among populations (vb) | 20 | 37.996 | 0.055 vb | 5.25 |
| Within populations (vc) | 541 | 295.919 | 0.547 vc | 52.08 |
| Tota | 562 | 460.426 | 1.050 | |
| Indices | n | S | K | H | π | D | Fu’s Fs | p | |
|---|---|---|---|---|---|---|---|---|---|
| Localities | |||||||||
| Sequences ND4 Atlantic | |||||||||
| Group 1: Gulf of Mexico, Florida, Honduras, and Colombia | 150 | 44 | 45 | 0.846 | 0.00440 | −2.13534 | −34.532 | <0.01 | |
| Group 2: Puerto Rico and Brazil | 249 | 42 | 43 | 0.374 | 0.00149 | −2.55341 | −33.830 | <0.01 | |
| Total | 399 | 66 | 79 | 0.676 | 0.00321 | −2.39976 | −129.275 | <0.01 | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Núñez-Vallecillo, M.; Vera-Escalona, I.; Rivera, A.; Górski, K.; Brante, A. Genetic Population Structure of Lane Snapper Lutjanus synagris (Linnaeus, 1758) in Western Atlantic: Implications for Conservation. Diversity 2024, 16, 336. https://doi.org/10.3390/d16060336
Núñez-Vallecillo M, Vera-Escalona I, Rivera A, Górski K, Brante A. Genetic Population Structure of Lane Snapper Lutjanus synagris (Linnaeus, 1758) in Western Atlantic: Implications for Conservation. Diversity. 2024; 16(6):336. https://doi.org/10.3390/d16060336
Chicago/Turabian StyleNúñez-Vallecillo, Mayra, Iván Vera-Escalona, Antonella Rivera, Konrad Górski, and Antonio Brante. 2024. "Genetic Population Structure of Lane Snapper Lutjanus synagris (Linnaeus, 1758) in Western Atlantic: Implications for Conservation" Diversity 16, no. 6: 336. https://doi.org/10.3390/d16060336
APA StyleNúñez-Vallecillo, M., Vera-Escalona, I., Rivera, A., Górski, K., & Brante, A. (2024). Genetic Population Structure of Lane Snapper Lutjanus synagris (Linnaeus, 1758) in Western Atlantic: Implications for Conservation. Diversity, 16(6), 336. https://doi.org/10.3390/d16060336








