Bears into the Niche-Space: Phylogeography and Phyloclimatic Model of the Family Ursidae
Abstract
1. Introduction
2. Materials and Methods
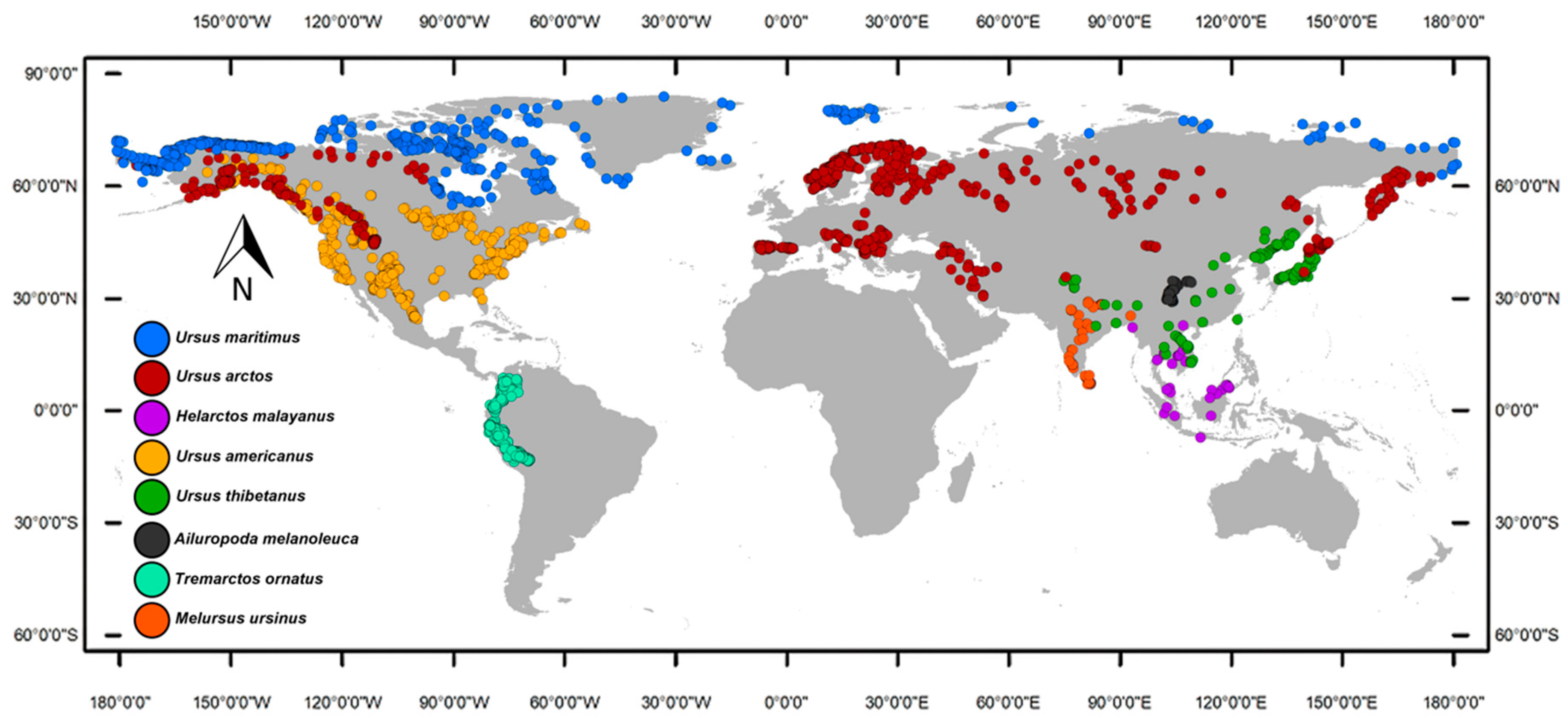
2.1. Occurrence Records
2.2. Bioclimatic Variables
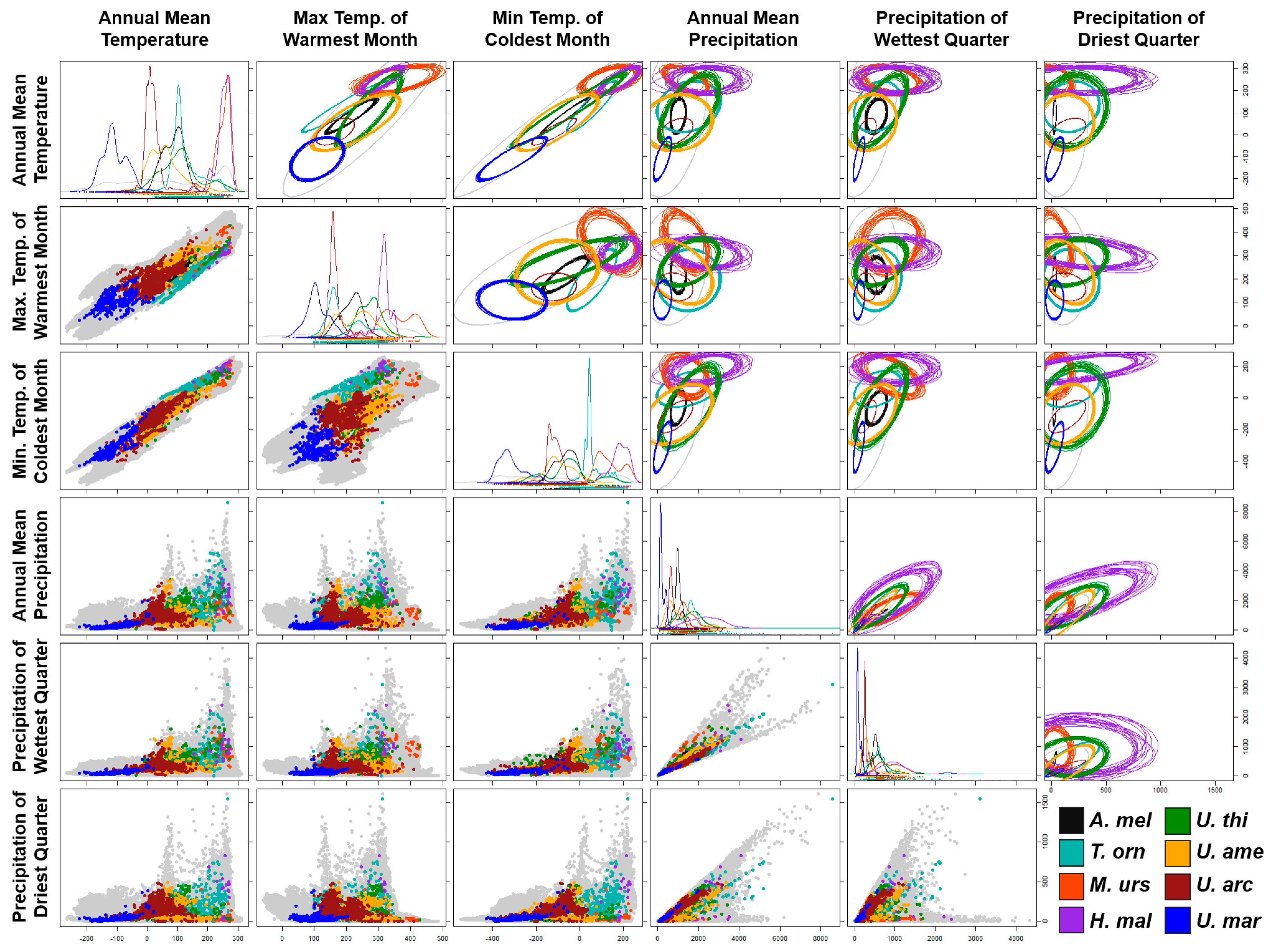
2.3. Environmental Characterization
2.4. Time-Calibrated Bayesian Ultrametric Tree
2.5. Phyloclimatic Analysis
3. Results
3.1. Niche Modeling Analysis
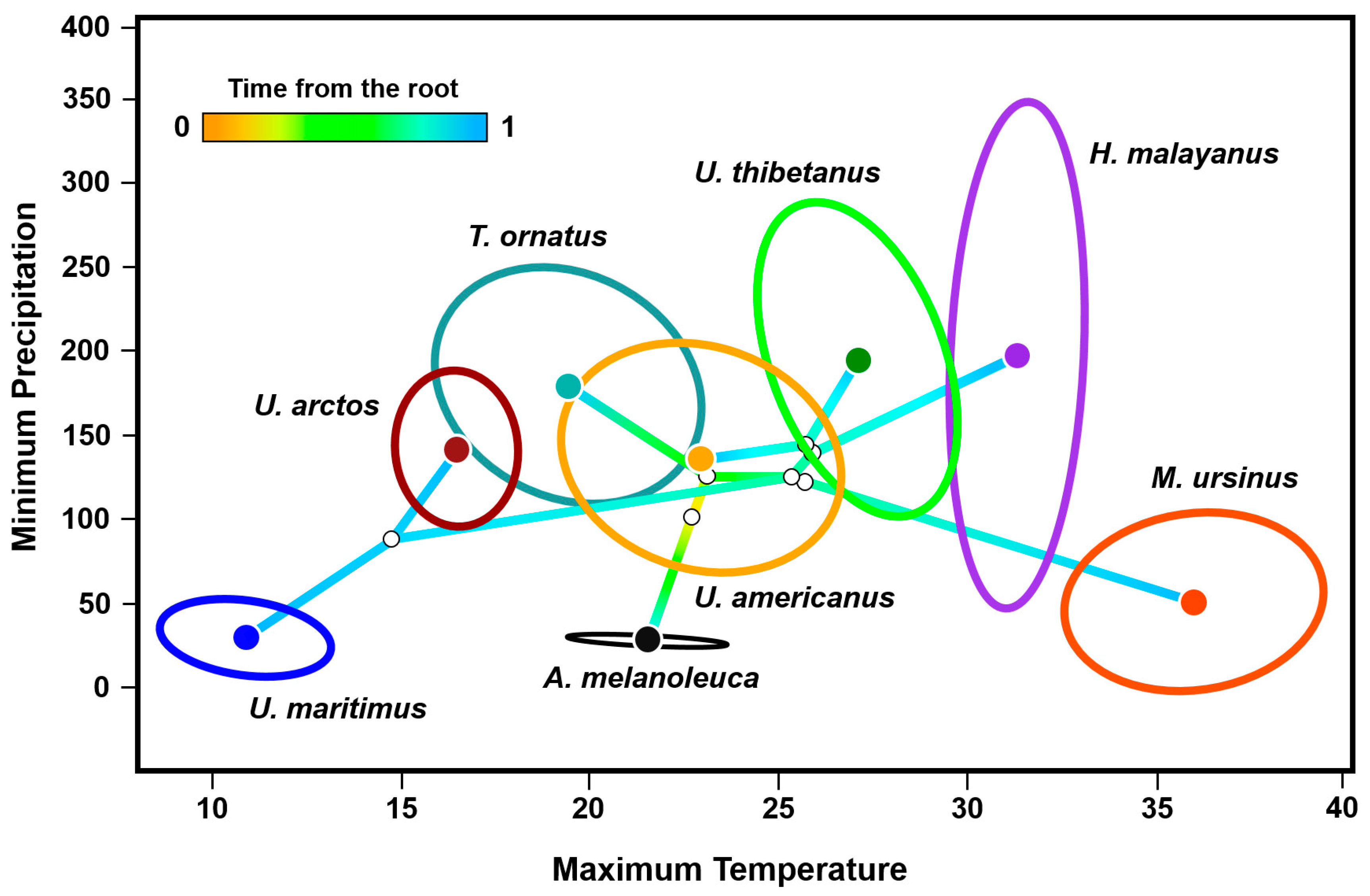
3.2. Phyloclimatic Analysis
4. Discussion
4.1. Ecological Niche and Phylogeography of the Extant Bear Species
4.2. Environmental Adaptation, Niche Conservatism, and Niche Evolution
4.3. Geographic Ranges, Environmental Preferences, and Niche Trajectories
4.4. Implications for Conservation and Management
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kalkvik, H.M.; Stout, I.J.; Doonan, T.J.; Parkinson, C.L. Investigating niche and lineage diversification in widely distributed taxa: Phylogeography and ecological niche modeling of the Peromyscus maniculatus species group. Ecography 2012, 35, 54–64. [Google Scholar] [CrossRef]
- Rolland, J.; Silvestro, D.; Schluter, D.; Guisan, A.; Broennimann, O.; Salamin, N. The impact of endothermy on the climatic niche evolution and the distribution of vertebrate diversity. Nat. Ecol. Evol. 2018, 2, 459–464. [Google Scholar] [CrossRef] [PubMed]
- Avise, J.C. Phylogeography: The History and Formation of Species; Harvard University Press: Cambridge, MA, USA, 2000. [Google Scholar]
- Carstens, B.C.; Richards, C.L. Integrating coalescent and ecological niche modeling in comparative phylogeography. Evolution 2007, 61, 1439–1454. [Google Scholar] [CrossRef] [PubMed]
- Soberón, J.; Nakamura, M. Niches and distributional areas: Concepts, methods, and assumptions. Proc. Natl. Acad. Sci. USA 2009, 106, 19644–19650. [Google Scholar] [CrossRef] [PubMed]
- Knowles, L.L.; Carstens, B.C.; Keat, M.L. Coupling genetic and ecological-niche models to examine how past population distributions contribute to divergence. Curr. Biol. 2007, 17, 940–946. [Google Scholar] [CrossRef] [PubMed]
- Alvarado-Serrano, D.F.; Knowles, L.L. Ecological niche models in phylogeographic studies: Applications, advances and precautions. Mol. Ecol. Resour. 2014, 14, 233–248. [Google Scholar] [CrossRef] [PubMed]
- Luna-Aranguré, C.; Vázquez-Domínguez, E. Analysis of the application of ecological niche modeling in phylogeographic studies: Contributions, challenges, and future. Therya 2020, 11, 47–55. [Google Scholar] [CrossRef]
- Suárez-Atilano, M.; Rojas-Soto, O.; Parra, J.L.; Vázquez-Domínguez, E. The role of the environment on the genetic divergence between two Boa imperator lineages. J. Biogeog. 2017, 44, 2045–2056. [Google Scholar] [CrossRef]
- Wiens, J.J.; Graham, C.H. Niche conservatism: Integrating evolution, ecology, and conservation biology. Ann. Rev. Ecol. Evol. Syst. 2005, 36, 519–539. [Google Scholar] [CrossRef]
- Eliosa-León, H.R.; Montes de Oca, A.N.; Navarro-Carbajal, M.C. Conservadurismo filogenético del nicho ecológico un enfoque integral de la evolución. Ciencias 2010, 98, 64–698. [Google Scholar]
- Losos, J.B.; Leal, M.; Glor, R.E.; De Queiroz, K.; Hertz, P.E.; Rodríguez Schettino, L.; Chamizo Lara, A.; Jackman, T.R.; Larson, A. Niche lability in the evolution of a Caribbean lizard community. Nature 2003, 424, 542–545. [Google Scholar] [CrossRef] [PubMed]
- Holt, R.D. Evolution of the ecological niche. In The Princeton Guide to Evolution; Losos, J.B., Baum, D.A., Futuyma, D.J., Hoekstra, H.E., Lenski, R.E., Moore, A.J., Peichel, C.L., Schluter, D., Whitlock, M.C., Eds.; Princeton University Press: Princeton, NJ, USA, 2014; pp. 288–297. [Google Scholar] [CrossRef]
- Pyron, R.A.; Costa, G.C.; Patten, M.A.; Burbrink, F.T. Phylogenetic niche conservatism and the evolutionary basis of ecological speciation. Biol. Rev. 2015, 90, 1248–1262. [Google Scholar] [CrossRef] [PubMed]
- Castro-Insua, A.; Gómez-Rodríguez, C.; Wiens, J.J.; Baselga, A. Climatic niche divergence drives patterns of diversification and richness among mammal families. Sci. Rep. 2018, 8, 8781. [Google Scholar] [CrossRef] [PubMed]
- McLellan, B.; Reiner, D.C. A review of bear evolution. Int. Conf. Bear Res. Manag. 1980, 9, 85–96. [Google Scholar] [CrossRef]
- Talbot, S.L.; Shields, G.F. A Phylogeny of the bears (Ursidae) inferred from complete sequences of three mitochondrial genes. Mol. Phylogenet. Evol. 1996, 5, 567–575. [Google Scholar] [CrossRef] [PubMed]
- Juárez-Casillas, L.A.; Varas, C. Genética evolutiva y molecular de la Familia Ursidae: Una revisión bibliográfica actualizada. Therya 2011, 2, 47–65. [Google Scholar] [CrossRef]
- Kutschera, V.E.; Bidon, T.; Hailer, F.; Rodi, J.L.; Fain, S.R.; Janke, A. Bears in a forest of gene trees: Phylogenetic inference is complicated by incomplete lineage sorting and gene flow. Mol. Biol. Evol. 2014, 31, 2004–2017. [Google Scholar] [CrossRef] [PubMed]
- Wooten, J.A.; Gibbs, H.L. Niche divergence and lineage diversification among closely related Sistrurus rattlesnakes. J. Evol. Biol. 2012, 25, 317–328. [Google Scholar] [CrossRef]
- Stuart-Smith, R.D.; Edgar, G.J.; Bates, A.E. Thermal limits to the geographic distributions of shallow-water marine species. Nat. Ecol. Evol. 2017, 1, 1846–1852. [Google Scholar] [CrossRef]
- Blonder, B. Hypervolume concepts in niche-and trait-based ecology. Ecography 2018, 41, 1441–1455. [Google Scholar] [CrossRef]
- Wiens, J.A.; Stralberg, D.; Jongsomjit, D.; Howell, C.A.; Snyder, M.A. Niches, models, and climate change: Assessing the assumptions and uncertainties. Proc. Natl. Acad. Sci. USA 2009, 106, 19729–19736. [Google Scholar] [CrossRef] [PubMed]
- Pérez-Consuegra, S.G.; Sánchez, L.; Rodríguez-Tapia, G.; Castañeda-Rico, S.; Vázquez-Domínguez, E. Late Pleistocene altitudinal segregation and demography define future climate change distribution of the Peromyscus mexicanus species group: Conservation implications. Animals 2023, 13, 1753. [Google Scholar] [CrossRef]
- IUCN. The IUCN Red List of Threatened Species. Version 2023-1. Available online: https://www.iucnredlist.org (accessed on 14 February 2023).
- Aiello-Lammens, M.E.; Boria, R.A.; Radosavljevic, A.; Vilela, B.; Anderson, R.P. SpThin: An R package for spatial thinning of species occurrence records for use in ecological niche models. Ecography 2015, 38, 541–545. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing, Vienna. 2021; Available online: https://www.R-project.org (accessed on 14 February 2023).
- Hijmans, R.J.; Cameron, S.E.; Parra, J.L.; Jones, P.G.; Jarvis, A. Very high resolution interpolated climate surfaces for global land areas. Intl. J. Climat. 2005, 25, 1965–1978. [Google Scholar] [CrossRef]
- Osorio-Olvera, L.; Barve, V.; Soberón, J. Nichetoolbox: From Getting Biodiversity Data to Evaluating Species Distribution Models in a Friendly GUI Environment. R Package Version 0.2.0.0. 2017. Available online: https://github.com/luismurao/ntbox (accessed on 14 February 2023).
- Segurado, P.A.G.E.; Araújo, M.B.; Kunin, W.E. Consequences of spatial autocorrelation for niche-based models. J. Appl. Ecol. 2006, 43, 433–444. [Google Scholar] [CrossRef]
- Hawkins, B.A.; Field, R.; Cornell, H.V.; Currie, D.J.; Guégan, J.F.; Kaufman, D.M.; Kerr, J.T.; Mittelbach, G.G.; Oberdorff, T.; O’Brien, E.M.; et al. Energy, water, and broad-scale geographic patterns of species richness. Ecology 2003, 84, 3105–3117. [Google Scholar] [CrossRef]
- Lovejoy, T.E.; Hannah, L. Climate Change and Biodiversity; Yale University Press: New Haven, CY, USA, 2005. [Google Scholar]
- Swanson, H.K.; Lysy, M.; Power, M.; Stasko, A.D.; Johnson, J.D.; Reist, J.D. A new probabilistic method for quantifying n-dimensional ecological niches and niche overlap. Ecology 2015, 96, 318–324. [Google Scholar] [CrossRef]
- Boria, R.A.; Blois, J.L. The effect of large sample sizes on ecological niche models: Analysis using a North American rodent, Peromyscus maniculatus. Ecol. Model. 2018, 386, 83–88. [Google Scholar] [CrossRef]
- Shcheglovitova, M.; Anderson, R.P. Estimating optimal complexity for ecological niche models: A jackknife approach for species with small sample sizes. Ecol. Model. 2013, 269, 9–17. [Google Scholar] [CrossRef]
- Wickham, H.; François, R.; Henry, L.; Müller, K.; Vaughan, D. dplyr: A Grammar of Data Manipulation. R Package Version 1.1.4. 2023. Available online: https://dplyr.tidyverse.org (accessed on 14 February 2023).
- Yu, L.; Li, Y.W.; Ryder, O.A.; Zhang, Y.P. Analysis of complete mitochondrial genome sequences increases phylogenetic resolution of bears (Ursidae), a mammalian family that experienced rapid speciation. BMC Evol. Biol. 2007, 7, 198. [Google Scholar] [CrossRef]
- Kearse, M.; Moir, R.; Wilson, A.; Stones-Havas, S.; Cheung, M.; Sturrock, S.; Buxton, S.; Cooper, A.; Markowitz, S.; Duran, C.; et al. Geneious Basic: An integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 2012, 28, 1647–1649. [Google Scholar] [CrossRef] [PubMed]
- Darriba, D.; Taboada, G.L.; Doallo, R.; Posada, D. JModelTest 2: More models, new heuristics and parallel computing. Nat. Methods 2012, 9, 772. [Google Scholar] [CrossRef] [PubMed]
- Guindon, S.; Dufayard, J.F.; Lefort, V.; Anisimova, M.; Hordijk, W.; Gascuel, O. New algorithms and methods to estimate maximum-likelihood phylogenies: Assessing the performance of PhyML 3.0. Syst. Biol. 2010, 59, 307–321. [Google Scholar] [CrossRef] [PubMed]
- Lefort, V.; Longueville, J.-E.; Gascuel, O. SMS: Smart model selection in phyml. Mol. Biol. Evol. 2017, 34, 2422–2424. [Google Scholar] [CrossRef] [PubMed]
- Arnason, U.; Gullberg, A.; Janke, A.; Kullberg, M. Mitogenomic analyses of caniform relationships. Mol. Phyl. Evol. 2007, 45, 863–874. [Google Scholar] [CrossRef] [PubMed]
- Nyakatura, K.; Bininda-Emonds, O.R. Updating the evolutionary history of Carnivora (Mammalia): A new species-level supertree complete with divergence time estimates. BMC Biol. 2012, 10, 12. [Google Scholar] [CrossRef] [PubMed]
- Rambaut, A.; Drummond, A.J.; Xie, D.; Baele, G.; Suchard, M.A. Posterior summarisation in Bayesian phylogenetics using Tracer 1.7. Syst. Biol. 2018, 67, 901–904. [Google Scholar] [CrossRef] [PubMed]
- Miller, E.T.; Zanne, A.E.; Ricklefs, R.E. Niche conservatism constrains Australian honeyeater assemblages in stressful environments. Ecol. Lett. 2013, 16, 1186–1194. [Google Scholar] [CrossRef] [PubMed]
- Revell, L.J. phytools: An R package for phylogenetic comparative biology (and other things). Methods Ecol. Evol. 2012, 3, 217–223. Available online: http://www.respond2articles.com/MEE/ (accessed on 14 February 2023). [CrossRef]
- Khaliq, I.; Fritz, S.A.; Prinzinger, R.; Pfenninger, M.; Böhning-Gaese, K.; Hof, C. Global variation in thermal physiology of birds and mammals: Evidence for phylogenetic niche conservatism only in the tropics. J. Biogeog. 2015, 42, 2187–2196. [Google Scholar] [CrossRef]
- Schluter, D.; Price, T.; Mooers, A.Ø.; Ludwig, D. Likelihood of ancestor states in adaptive radiation. Evolution 1997, 51, 1699–1711. [Google Scholar] [CrossRef] [PubMed]
- Krause, J.; Unger, T.; Noçon, A.; Malaspinas, A.S.; Kolokotronis, S.O.; Stiller, M.; Bray, S.C. Mitochondrial genomes reveal an explosive radiation of extinct and extant bears near the Miocene-Pliocene boundary. BMC Evol. Biol. 2008, 8, 220. [Google Scholar] [CrossRef] [PubMed]
- Pages, M.; Calvignac, S.; Klein, C.; Paris, M.; Hughes, S.; Hänni, C. Combined analysis of fourteen nuclear genes refines the Ursidae phylogeny. Mol. Phylogenet. Evol. 2008, 47, 73–83. [Google Scholar] [CrossRef] [PubMed]
- Rangel, T.F.; Edwards, N.R.; Holden, P.B.; Diniz-Filho, J.A.F.; Gosling, W.D.; Coelho, M.T.P.; Colwell, R.K. Modeling the ecology and evolution of biodiversity: Biogeographical cradles, museums, and graves. Science 2018, 361, eaar5452. [Google Scholar] [CrossRef] [PubMed]
- Saupe, E.E.; Barve, N.; Owens, H.L.; Cooper, J.C.; Hosner, P.A.; Peterson, A.T. Reconstructing ecological niche evolution when niches are incompletely characterized. Syst. Biol. 2017, 67, 428–438. [Google Scholar] [CrossRef] [PubMed]
- Pearman, P.B.; Guisan, A.; Broennimann, O.; Randin, C.F. Niche dynamics in space and time. Trends Ecol. Evol. 2008, 23, 149–158. [Google Scholar] [CrossRef] [PubMed]
- Luna-Aranguré, C.; Soberón, J.; Vázquez-Domínguez, E. A tale of four bears: Environmental signal on the phylogeographical patterns within the extant Ursus species. J. Biogeog. 2020, 47, 472–486. [Google Scholar] [CrossRef]
- Goursi, U.H.; Anwar, M.; Bosso, L.; Nawaz, M.A.; Kabir, M. Spatial distribution of the threatened Asiatic black bear in northern Pakistan. Ursus 2021, 2021, 1–5. [Google Scholar] [CrossRef]
- Ahmad, F.; Nawaz, M.A.; Salim, M.; Rehan, M.; Farhadinia, M.; Bosso, L.; Kabir, M. Patterns of spatial distribution, diel activity and human-bear conflict of Ursus thibetanus in the Hindu Kush mountains, Pakistan. Global Ecol. Conserv. 2022, 37, e02145. [Google Scholar] [CrossRef]
- Smith, B.T.; Bryson Jr, R.W.; Houston, D.D.; Klicka, J. An asymmetry in niche conservatism contributes to the latitudinal species diversity gradient in New World vertebrates. Ecol. Lett. 2012, 15, 1318–1325. [Google Scholar] [CrossRef]
- Obbard, M.E.; Cattet, M.R.; Howe, E.J.; Middel, K.R.; Newton, E.J.; Kolenosky, G.B.; Greenwood, C.J. Trends in body condition in polar bears (Ursus maritimus) from the Southern Hudson Bay subpopulation in relation to changes in sea ice. Arct. Sci. 2016, 2, 15–32. [Google Scholar] [CrossRef]
- Luna-Aranguré, C.; Vázquez-Domínguez, E. Of pandas, fossils and bamboo forests: Ecological niche modeling of the giant panda (Ailuropoda melanoleuca) during the Last Glacial Maximum. J. Mammal. 2021, 102, 718–730. [Google Scholar] [CrossRef]
- Schaller, G.B.; Hu, J.; Pan, W.; Zhu, J. The Giant Pandas of Wolong; University of Chicago Press: Chicago, IL, USA, 1985. [Google Scholar]
- Liu, J.; Viña, A. Pandas, plants, and people. Ann. Mo. Bot. Gard. 2014, 100, 108–125. [Google Scholar] [CrossRef]
- Sheng, G.L.; Barlow, A.; Cooper, A.; Hou, X.D.; Ji, X.P.; Jablonski, N.G.; Zhong, B.-J.; Liu, H.; Flynn, L.J.; Yuan, J.-X.; et al. Ancient DNA from giant panda (Ailuropoda melanoleuca) of south-western China reveals genetic diversity loss during the Holocene. Genes 2018, 9, 198. [Google Scholar] [CrossRef] [PubMed]
- Lindqvist, C.; Schuster, S.C.; Sun, Y.; Talbot, S.L.; Qi, J.; Ratan, A.; Tomsho, L.P.; Kasson, L.; Zeyl, E.; Aars, J.; et al. Complete mitochondrial genome of a Pleistocene jawbone unveils the origin of polar bear. Proc. Natl. Acad. Sci. USA 2010, 107, 5053–5057. [Google Scholar] [CrossRef]
- McLellan, B.N.; Proctor, M.F.; Huber, D.; Michel, S. Ursus arctos (amended version of 2017 assessment). IUCN Red List Threat. Species 2017, e.T41688A121229971. [Google Scholar] [CrossRef]
- Wong, T.S.; Servheen, C.; Ambu, L. Food habits of malayan sun bears in lowland tropical forests of Borneo. Ursus 2002, 13, 127–136. [Google Scholar]
- Laurie, A.; Seidensticker, J. Behavioural ecology of the sloth bear (Melursus ursinus). J. Zool. 1977, 182, 187–204. [Google Scholar] [CrossRef]
- Eggleton, P.; Tayasu, I. Feeding groups, lifetypes and the global ecology of termites. Ecology 2001, 82, 253–261. [Google Scholar] [CrossRef]
- Bignell, D.E.; Roisin, Y.; Lo, N. Biology of Termites: A Modern Synthesis; Springer Science & Business Media: Berlin, Germany, 2011. [Google Scholar]
- Steinmetz, R.; Garshelis, D.L.; Chutipong, W.; Seuaturien, N. The shared preference niche of sympatric asiatic black bears and sun bears in a tropical forest mosaic. PLoS ONE 2011, 6, e14509. [Google Scholar] [CrossRef]
- Steinmetz, R.; Garshelis, D.L.; Chutipong, W.; Seuaturien, N. Foraging ecology and coexistence of Asiatic black bears and sun bears in a seasonal tropical forest in Southeast Asia. J. Mammal. 2013, 94, 1–18. [Google Scholar] [CrossRef]

| A. mel | T. orn | M. urs | H. mal | U. thi | U. ame | U. arc | U. mar | |
|---|---|---|---|---|---|---|---|---|
| A. mel | 1.30 | 0.38 | 0 | 93.22 | 46.85 | 0 | 0 | |
| T. orn | 0 | 9.02 | 16.56 | 9.62 | 23.31 | 0 | 0 | |
| M. urs | 0 | 2.20 | 15.22 | 28.02 | 0.60 | 0 | 0 | |
| H. mal | 0 | 15.15 | 22.89 | 30.96 | 0.79 | 0 | 0 | |
| U. thi | 0.04 | 1.43 | 5.72 | 2.06 | 39.13 | 0.11 | 0.03 | |
| U. ame | 0.01 | 0.29 | 0.15 | 0.02 | 49.05 | 7.16 | 0.45 | |
| U. arc | 0 | 0.01 | 0 | 0 | 19.32 | 98.66 | 8.22 | |
| U. mar | 0 | 0.02 | 0 | 0 | 5.20 | 66.19 | 7.66 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Luna-Aranguré, C.; Vázquez-Domínguez, E. Bears into the Niche-Space: Phylogeography and Phyloclimatic Model of the Family Ursidae. Diversity 2024, 16, 223. https://doi.org/10.3390/d16040223
Luna-Aranguré C, Vázquez-Domínguez E. Bears into the Niche-Space: Phylogeography and Phyloclimatic Model of the Family Ursidae. Diversity. 2024; 16(4):223. https://doi.org/10.3390/d16040223
Chicago/Turabian StyleLuna-Aranguré, Carlos, and Ella Vázquez-Domínguez. 2024. "Bears into the Niche-Space: Phylogeography and Phyloclimatic Model of the Family Ursidae" Diversity 16, no. 4: 223. https://doi.org/10.3390/d16040223
APA StyleLuna-Aranguré, C., & Vázquez-Domínguez, E. (2024). Bears into the Niche-Space: Phylogeography and Phyloclimatic Model of the Family Ursidae. Diversity, 16(4), 223. https://doi.org/10.3390/d16040223






