Two New Lyophyllum Species from Yunnan, China
Abstract
1. Introduction
2. Materials and Methods
2.1. Morphological Studies
2.2. DNA Extraction, PCR Amplification and Sequencing
2.3. Sequence Alignment and Phylogenetic Analyses
2.4. ITS Genetic Distances Calculation
3. Result
3.1. Phylogenetic Analyses
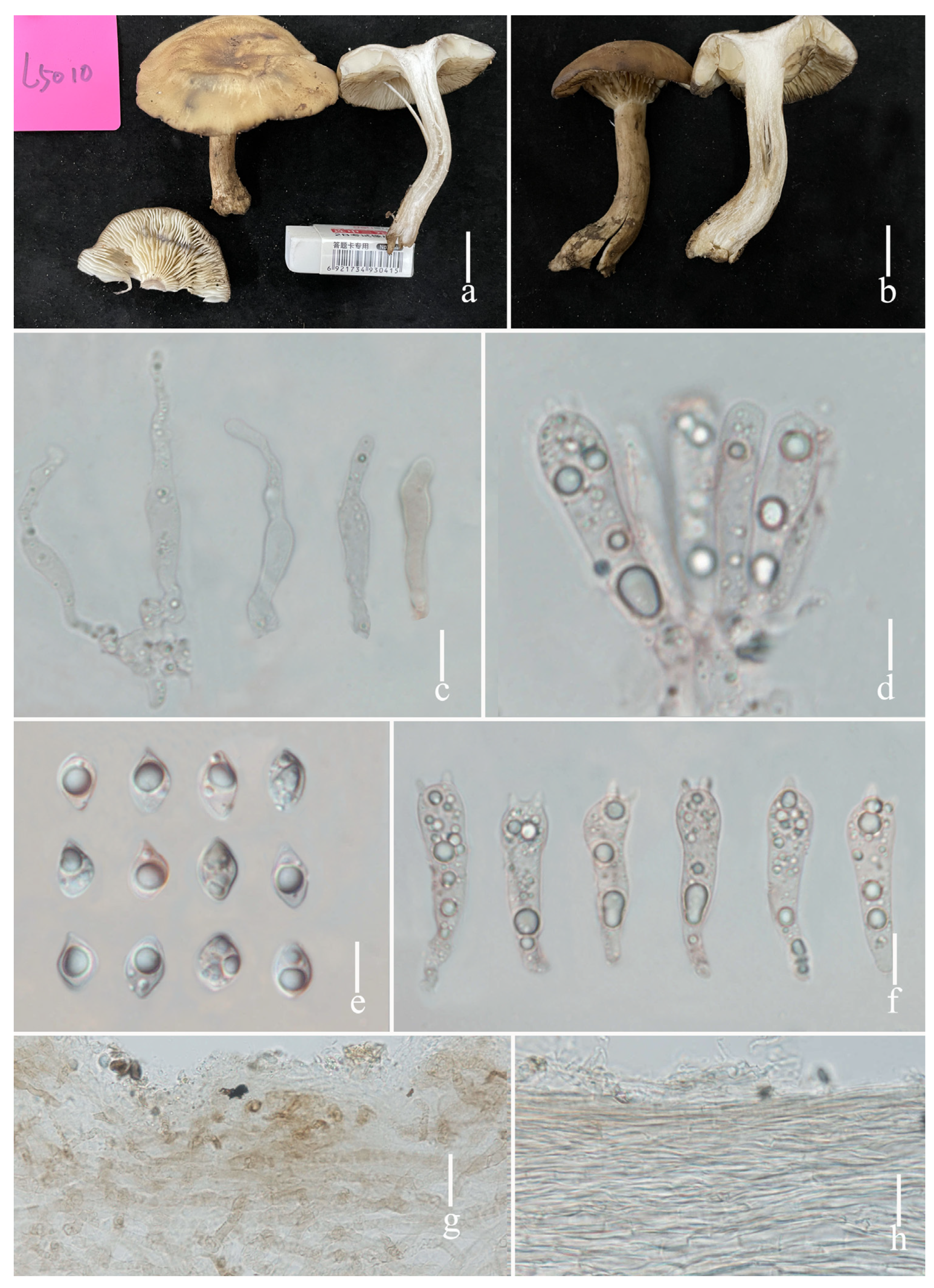
3.2. Taxonomy
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Karsten, P.A. Hymenomycetes Fennici enumerati. Acta Soc. Fauna Flora Fenn. 1881, 2, 4. [Google Scholar]
- Dähncke, R.M.; Contu, M.; Vizzini, A. Two new species of Lyophyllum s.l. (Basidiomycota, Agaricomycetes) from La Palma (Canary Islands, Spain). Mycotaxon 2011, 115, 65–71. [Google Scholar] [CrossRef]
- Cooper, J.A. New species and combinations of some New Zealand agarics belonging to Clitopilus, Lyophyllum, Gerhardtia, Clitocybe, Hydnangium, Mycena, Rhodocollybia and Gerronema. Mycosphere 2014, 5, 263–288. [Google Scholar] [CrossRef]
- Bellanger, J.M.; Moreau, P.A.; Corriol, G.; Bidaud, A.; Chalange, R.; Dudova, Z.; Richard, F. Plunging hands into the mushroom jar: A phylogenetic framework for Lyophyllaceae (Agaricales, Basidiomycota). Genetica 2015, 143, 169–194. [Google Scholar] [CrossRef]
- Lomberg, M.L.; Renker, C.; Buchalo, A.S.; Solomko, E.F.; Kirchhoff, B. Micromorphological and Molecular Biological Study of Culinary-Medicinal Mushroom Hypsizygus marmoreus (Peck) Bigel. (Agaricomycetideae). Int. J. Med. Mushrooms 2003, 5, 307–312. [Google Scholar] [CrossRef]
- Dai, F. A Summary of Chinese Fungi; Science Press: Beijing, China, 1979. [Google Scholar]
- Kühner, R. Utilisation du carmin acétique dans la classification des agarics leucosporés. Publ. Société Linnéenne Lyon 1938, 7, 204–211. [Google Scholar] [CrossRef]
- Gminder, A. Nomenclatural novelties. Index Fungorum 2016, 302, 1. [Google Scholar]
- Li, X.; Li, Y. Research progress on fungus of the genus Lyophyllum in China. J. Edible Fungi 2009, 16, 75–79. [Google Scholar]
- Larsson, E.; Sundberg, H. Lyophyllum shimeji, a species associated with lichen pine forest in northern Fennoscandia. Mycoscience 2011, 52, 289–295. [Google Scholar] [CrossRef]
- Li, S.H.; Tang, S.M.; He, J.; Zhou, D. Two new edible Lyophyllum species from Tibetan areas, China. Diversity 2023, 15, 1027. [Google Scholar] [CrossRef]
- Ma, Y.H.; Liu, P.; Zhao, Z.Y.; Chen, W.M.; Zhao, Y.C. Lyophyllum deqinense (Lyophyllaceae, Agaricales), a new species from southwestern China. Phytotaxa 2023, 598, 219–228. [Google Scholar] [CrossRef]
- Wei, S.W.; Lu, B.Y.; Wang, Y.; Dou, W.J.; Wang, Q.; Li, Y. Morphology and phylogeny of Lyophylloid mushrooms in china with description of four new species. J. Fungi 2023, 9, 77. [Google Scholar] [CrossRef] [PubMed]
- Sesli, E.; Vizzini, A.; Contu, M. Lyophyllum turcicum (Agaricomycetes: Lyophyllaceae), a new species from Turkey. Turk. J. Bot. 2015, 39, 512–519. [Google Scholar] [CrossRef]
- Boa, E. Los Hongos Silvestres Comestibles: Perspectiva Global de Su USO E Importancia Para la Población; FAO: Roma, Italy, 2005; No. 17. [Google Scholar]
- Pokhrel, C.P.; Sumikawa, S.; Iida, S.; Ohga, S. Growth and productivity of Lyophyllum decastes on compost enriched with various supplements. Micol. Apl. Int. 2006, 18, 21–28. [Google Scholar]
- Yamanaka, K. Commercial cultivation of Lyophyllum shimeji. Mushroom news. In Proceedings of the 6th International Conference on Mushroom Biology and Mushroom Products, Bonn, Germany, 29 September–3 October 2008. [Google Scholar]
- Ohta, A. Some cultural characteristics of mycelia of a mycorrhizal fungus, Lyophyllum shimeji. Mycoscience 1994, 35, 83–87. [Google Scholar] [CrossRef]
- Visnovsky, S.B.; Cummings, N.; Guerin-Laguette, A.; Wang, Y.; Yamada, A.; Kobayashi, H.; Kawai, M.; Pitman, A.R. Detection of the edible ectomycorrhizal fungus Lyophyllum shimeji colonising seedlings of cultivated conifer species in New Zealand. Mycorrhiza 2014, 24, 453–463. [Google Scholar] [CrossRef] [PubMed]
- Kornerup, A.; Wanscher, J.H. Methuen Handbook of Colour, 3rd ed.; Eyre Methuen: London, UK, 1978. [Google Scholar]
- Vellinga, E.C. Glossary. In Flora Agaricina Neerlandica; Bas, C., Kuijper, T.W., Noordeloos, M.E., Vellinga, E.C., Eds.; CRC Press: Boca Raton, FL, USA, 1998; Volume 1, pp. 54–64. [Google Scholar]
- White, T.J.; Bruns, T.D.; Lee, S.; Taylor, J.W. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics science direct. PCR Protoc. 1990, 18, 315–322. [Google Scholar]
- Katoh, K.; Standley, D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef] [PubMed]
- Hall, T. BioEdit v7. 2007. Available online: http://www.mbio.ncsu.edu/BioEdit/BioEdit.html (accessed on 10 September 2023).
- Stamatakis, A. RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 2014, 30, 1312–1313. [Google Scholar] [CrossRef] [PubMed]
- Miller, M.A.; Pfeiffer, W.; Schwartz, T. Creating the CIPRES Science Gateway for inference of large phylogenetic trees. In Proceedings of the Gateway Computing Environments Workshop (GCE), New Orleans, LA, USA, 14 November 2010. [Google Scholar]
- Nylander, J.A.A. MrModeltest, version 2.2.; Program distributed by the author; Uppsala University, Department of Systematic Zoology: Uppsala, Sweden, 2004. [Google Scholar]
- Rambaut, A.; Drummond, A.J.; Xie, D.; Baele, G.; Suchard, M.A. Posterior summarization in Bayesian phylogenetics using Tracer 1.7. Syst. Biol. 2018, 67, 901–904. [Google Scholar] [CrossRef]
- Hofstetter, V.; Clémençon, H.; Vilgalys, R.; Moncalvo, J.M. Phylogenetic analyses of the Lyophylleae (Agaricales, Basidiomycota) based on nuclear and mitochondrial rDNA sequences. Mycol. Res. 2002, 106, 1043–1059. [Google Scholar] [CrossRef]
- Ma, Y.H.; Liu, P.; Zhao, Z.Y.; Chen, W.M.; Zhao, Y.C. Lyophyllum pallidofumosum sp. nov. (Lyophyllaceae, Agaricales), from southwestern China. Phytotaxa 2022, 576, 173–183. [Google Scholar] [CrossRef]
- Tudor, D. Fungal Pigment Formation in Wood Substrate. Ph.D. Thesis, University of Toronto, Toronto, ON, Canada, 2013. [Google Scholar]
- Wang, X.Q.; Zhou, D.Q.; Zhao, Y.C.; Zhang, X.L.; Li, L.; Li, S.H. Lyophyllum rhombisporum sp. nov. from China. Mycotaxon 2013, 123, 473–477. [Google Scholar] [CrossRef]
- Hongo, T.; Clémençon, H. A new species of Lyophyllum from Japan. Mycol. Helv. 1983, 1, 43–46. [Google Scholar]
- Miura, T.; Kubo, M.; Itoh, Y.; Iwamoto, N.; Kato, M.; Park, S.R.; Yuuichi, U.; Yukio, K.; Ikukatsu, S.; Suzuki, I. Antidiabetic activity of Lyophyllum decastes in genetically type 2 diabetic mice. Biol. Pharm. Bull. 2002, 25, 1234–1237. [Google Scholar] [CrossRef] [PubMed]



| Taxon | Specimen Number | Country | ITS | Reference |
|---|---|---|---|---|
| Calocybe gambosa | HC 78/64 | Switzerland | AF357027 | [29] |
| C. carnea | CBS 552.50 | Switzerland | AF357028 | [29] |
| C. persicolor | HC 80/99 | Switzerland | AF357026 | [29] |
| L. ambustum | CBS 452.87 | Switzerland | AF357057 | [29] |
| L. anthracophilum | HC 79/132 | Switzerland | AF357055 | [29] |
| L. atratum | CBS 709.87 | Switzerland | AF357053 | [29] |
| L. atrofuscum | HMJAU 63461 | China | OP605493 | [13] |
| L. atrofuscum | HMJAU 63456 * | China | OP605494 | [13] |
| L. bulborhizum | L5083 * | China | PP406873 | This study |
| L. bulborhizum | L5092 | China | PP406874 | This study |
| L. bulborhizum | L5093 | China | PP406875 | This study |
| L. caerulescens | HC 80-140 | Switzerland | AF357052 | [29] |
| L. caerulescens | V.15759 | USA | JF908339 | [14] |
| L. crassifolium | V.5077 | Italy | JF908331 | [14] |
| L. decastes | Dd 08054 | China | FJ810160 | [14] |
| L. decastes | Ld 418 | China | HM119485 | [14] |
| L. deqinense | YAAS M6949 * | China | OQ418117 | [12] |
| L. deqinense | YAAS M6948 | China | OQ418116 | [12] |
| L. deliberatum | V.15032 | Slovenia | JF908338 | [14] |
| L. favrei | BSI94cp2 | Switzerland | AF357035 | [29] |
| L. favrei | V.6334 | Italy | JF908333 | [14] |
| L. fumosum | SJ02/006 | Sweden | HM572539 | [11] |
| L. fumosum | LAS00/144 | Sweden | HM572541 | [11] |
| L. fumosum | V.16077 | Italy | JF908340 | [14] |
| L. fumosum | LfumNlf24 | Japan | JN983977 | [14] |
| L. fumosum | L2010512371 | China | JX966310 | [14] |
| L. fumosum | YAAS M6135 | China | ON681708 | [30] |
| L. fumosum | YAAS M6340 | China | ON681709 | [30] |
| L. gangraenosum | V.12332 | Italy | JF908335 | [31] |
| L. heimogu | L3026 * | China | KY434100 | [11] |
| L. heimogu | L3033 | China | KY434101 | [11] |
| L. heimogu | L3035 | China | KY434102 | [11] |
| L. infumatum | V.10152 | Italy | JF908334 | [14] |
| L. leucophaeatum | Hae251.97 | Switzerland | AF357032 | [29] |
| L. littoralis | CA 20091130 | Italy | JX280410 | [14] |
| L. lixivium | HKAS 129929 | China | OR506463 | Unpublished |
| L. loricatum | V.13175 | USA | JF908336 | [14] |
| L. loricatum | CA 20090202.03 | Italy | JX280406 | [14] |
| L. loricatum | 01.12.09 | Italy | JX280407 | [14] |
| L. moncalvoanum | PDD 96328 * | New Zealand | NR_137615 | [10] |
| L. moncalvoanum | PDD 102581 | New Zealand | KJ461912 | [10] |
| L. nigrum | L5091 * | China | PP406876 | This study |
| L. nigrum | L5186 | China | PP406877 | This study |
| L. nigrum | L5187 | China | PP406878 | This study |
| L. ochraceum | BSI94.cp1 | Switzerland | AF357033 | [29] |
| L. ochraceum | V.537 | Italy | JF908329 | [14] |
| L. rhombisporum | L1762 * | China | JX966307 | [32] |
| L. rhombisporum | L2082 | China | JX966308 | [32] |
| L. rhombisporum | L5010 | China | PP406879 | This study |
| L. rhombisporum | L5084 | China | PP406880 | This study |
| L. semitale | HC 85/13 | Switzerland | AF357049 | [29] |
| L. semitale | EL 187-09 | Sweden | HM572552 | [24] |
| L. shimeji | Olsen 813006 | Sweden | HM572530 | [24] |
| L. shimeji | NZ4Q 88 | New Zealand | JN983985 | [14] |
| L. shimeji | L 2010512377 | China | JX966311 | [14] |
| L. sykosporum | IFO 30978 | Japan | AF357050 | [29] |
| L. subalpinarum | HMJAU 63447 * | China | OP605490 | [13] |
| L. subalpinarum | HMJAU 63453 | China | OP605491 | [13] |
| L. subdecastes | HMJAU 63470 | China | OP605488 | [13] |
| L. subdecastes | HMJAU 63467 * | China | OP605489 | [13] |
| L. turcicum | KATO-2971 * | Turkey | KJ158159 | [14] |
| L. yiqunyang | L4206 | China | KY434104 | [11] |
| L. yiqunyang | L2989 * | China | KY434103 | [11] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, S.; Tang, S.; He, J.; Zhou, D. Two New Lyophyllum Species from Yunnan, China. Diversity 2024, 16, 210. https://doi.org/10.3390/d16040210
Li S, Tang S, He J, Zhou D. Two New Lyophyllum Species from Yunnan, China. Diversity. 2024; 16(4):210. https://doi.org/10.3390/d16040210
Chicago/Turabian StyleLi, Shuhong, Songming Tang, Jun He, and Dequn Zhou. 2024. "Two New Lyophyllum Species from Yunnan, China" Diversity 16, no. 4: 210. https://doi.org/10.3390/d16040210
APA StyleLi, S., Tang, S., He, J., & Zhou, D. (2024). Two New Lyophyllum Species from Yunnan, China. Diversity, 16(4), 210. https://doi.org/10.3390/d16040210





