Abstract
The fungal genus Nigrospora is known to be a plant pathogen, endophyte, and saprobe, and it is usually isolated from various substrates like soil and air. During the surveys of soil fungi in Hebei Province of China, two isolates of Nigrospora were obtained. A multi-locus phylogeny of combined loci of the 5.8S nuclear ribosomal gene with the two flanking transcribed spacers (ITS), part of the translation elongation factor 1-alpha (tef1), and the beta-tubulin (tub2) loci, in conjunction with morphological characters were used to identify the newly collected isolates. Nigrospora humicola sp. Nov. is described and proposed herein, which differs from its phylogenetically close species N. chinensis and N. globosa by the sequences of ITS, tef1, and tub2.
1. Introduction
Nigrospora was proposed based on N. panici, collected from dead leaves of Panicum amphibium in Indonesia, which had spherical to subspherical conidiogenous cells and black and globose to subglobose conidia [1]. Members of Nigrospora were traditionally distinguished by comparing the morphological features, especially the conidial dimensions [2]. However, a recent study showed that the conidial dimensions frequently overlapped among phylogenetically distinct species [3]. Nigrospora was currently classified in the family Apiosporaceae within Amphisphaeriales evidenced by the phylogeny of molecular data [4,5]. Subsequently, several novel species of Nigrospora were revealed on the basis of the molecular and morphological evidence [6,7,8,9,10,11,12,13].
Nigrospora is a cosmopolitan genus on various substrates, including multifarious plants, soil, and air [2,3,7]. In addition, some Nigrospora species were considered to be important plant pathogens [14,15]. For example, N. sphaerica causes Camellia sinensis leaf blight diseases in China [16], N. lacticolonia and N. sphaerica are associated with the reddish brown spot disease of Hylocereus polyrhizus [17], and N. oryzae results in the leaf spot of Hibiscus mutabilis [18]. In addition, species of Nigrospora are also commonly discovered in an indoor environment and sometimes from the soil [3,9].
Nigrospora taxa are considered as a source of natural products due to their industrial applications [19,20,21]. For example, N. sacchari produces metabolites that have remarkable herbicidal activity in greenhouse-grown plants [22]; N. spherical can produce phomalactone against mosquitoes [23]. Hence, species of this genus are worth studying to develop related natural products. In the present study, new isolates were obtained from the forest soil and identified using a combined method of morphology and phylogeny.
2. Materials and Methods
2.1. Isolation
Strains of Nigrospora in the present study were isolated from forest soils in the Hebei Province of China in July 2021. Soil samples were divided into 1 g per portion and spread on 15 cm petri dishes containing potato dextrose agar medium (PDA; 200 g potato, 20 g glucose, 16 g agar per liter) with streptomycin sulfate and ampicillin 100 mg/mL in each dish. Plates were incubated at 25 °C for 2 d, the colonies were obtained, and then, they were transferred to the new PDA plates. The cultures were deposited in the China Forestry Culture Collection Center and the specimens in the herbarium of the Chinese Academy of Forestry.
2.2. Morphology
Isolates obtained in the present study were observed and described in terms of the colony color and appearance, based on the colonies grown on PDA medium. Plates were incubated for a week in the dark at 25 °C. Micro-morphological features were observed and recorded by a Nikon Eclipse 80i compound microscope equipped with a Nikon digital sight DS-Ri2 high-definition color camera. A total of 50 conidiogenous cells and conidia were randomly selected, observed, and measured.
2.3. DNA Extraction, PCR Amplification, and Phylogenetic Analyses
The fungal DNA was extracted from cultures grown on PDA plates overlaid with cellophane using a CTAB method [24]. The primer pair ITS1/ITS4 was used to amplify the internal transcribed spacer region and intervening 5.8S nrRNA gene (ITS) [25]. The primer pair EF-688F/EF2 was used to amplify part of the translation elongation factor 1-alpha (tef1) [26]. Bt2a/Bt2b was used to amplify part of the Beta-tubulin gene (tub2) [27]. The polymerase chain reaction (PCR) conditions were as performed. The resulting PCR products were visualized on a 1.4% agarose gel with ethidium bromide under UV light, and then the PCR positive products were sent to the Shanghai Invitrogen Biological Technology Company Limited (Beijing, China) for sequencing reactions using an ABI PRISM® 3730XL DNA Analyzer with BigDye® Terminater Kit v.3.1 (Invitrogen, Beijing, China).
Reference sequences were retrieved from the National Center for Biotechnology Information (NCBI) based on recent publications on the genus Nigrospora [3,6,7,8,9,10,11,12,13], and sequences from the present study were deposited in GenBank (Table 1). Sequences were aligned using MAFFT v. 7 [28] and manually edited using MEGA7 [29]. The phylogenetic analyses of the combined ITS, tef1, and tub2 loci were conducted using both Maximum Likelihood (ML) and Bayesian Inference (BI) methods. ML was implemented on the website of CIPRES Science Gateway using RAxML-HPC BlackBox 8.2.10 [30], employing a GTRGAMMA substitution model with 1000 bootstrap replicates, while BI was performed by a Markov Chain Monte Carlo (MCMC) algorithm using MrBayes v. 3.0 [31]. Two MCMC chains, started from random trees for 1,000,000 generations and trees, were sampled every 100th generation, resulting in a total of 10,000 trees. The first 25% of trees were discarded as the burn-in of each analysis. Branches with significant Bayesian Posterior Probabilities (BPP) were estimated in the remaining 7500 trees. Phylogenetic trees were viewed and edited in FigTree v.1.3.1 and Adobe Illustrator CS5.

Table 1.
Isolates and GenBank accession numbers used in the phylogenetic analyses of Nigrospora.
3. Results
3.1. Phylogeny
The resulting phylogram based on a combined analysis of ITS, tef1, and tub2 loci was used to reveal the species relationship of the newly collected isolates within Nigrospora. The dataset consisted of 45 sequences including two outgroup taxa, namely Apiospora qinglingensis (CFCC 52303) and A. vietnamensis (IMI 99670). The dataset comprised 1511 characters after alignment including the gaps (547 for ITS, 535 for tef1, and 429 for tub2), which were included in the phylogenetic analysis. Of these, 876 characters were constant, 166 variable characters were parsimony uninformative, and 467 characters were parsimony informative. The topologies resulting from the ML and BI analyses of the concatenated dataset were congruent (Figure 1). Two isolates from the present study clustered into a distinct clade from the other species of this genus, which represents an undescribed Nigrospora species.
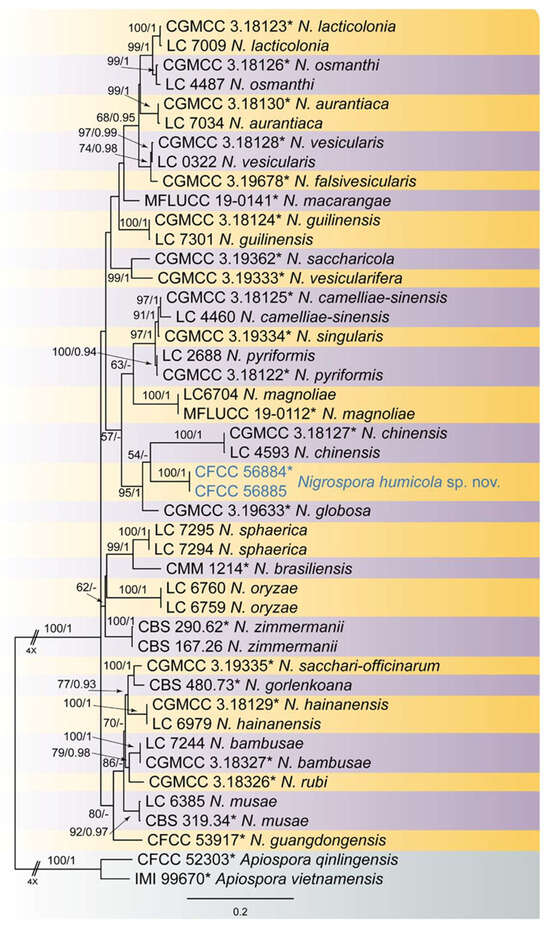
Figure 1.
Phylogenetical tree of Nigrospora of ML analysis on basis of combined ITS, tef1, and tub2 loci. Numbers above the branches indicate ML bootstraps (left, ML BS ≥ 50%) and Bayesian Posterior Probabilities (right, BPP ≥ 0.90). The tree is rooted with Apiospora qinlingensis (CFCC 52303) and A. vietnamensis (IMI 99670). New species from the present study are marked in blue, and ex-type strains are marked with *.
3.2. Taxonomy
Nigrospora humicola Q. Yang & Ning Jiang, sp. nov.
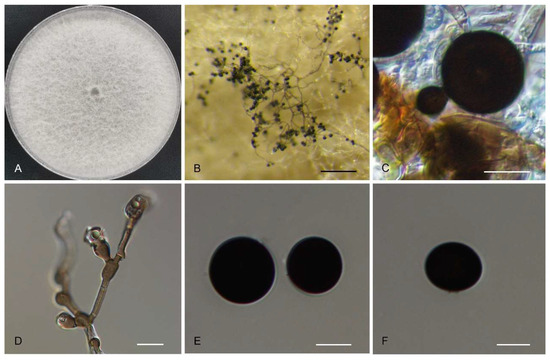
Figure 2.
Morphology of Nigrospora humicola (CFCC 56884). (A) Colony on PDA. (B) Conidiomata formed in culture. (C,D) Conidiogenous cells giving rise to conidia. (E,F) Conidia. Scale bars: (B) = 200 µm; (C–F) = 10 µm.
MycoBank no: 844103
Etymology: Referring to the substrate soil, where the type of strain originated.
Sexual morph undetermined. Asexual morph on PDA: Hyphae 2.5–6 μm diam., smooth, hyaline to brown, branched, septate. Conidiophores smooth, hyaline to brown, branched, septate, sometimes reduced to conidiogenous cells. Conidiogenous cells 4.5–15.5 × 2.5–12 μm, aggregated in clusters on hyphae, pale brown, subglobose to ampulliform. Conidia 12.5–23.5 × 9.5–16 μm (av. = 16.3 ± 3.4 × 13.2 ± 2.7 μm) solitary, globose to subglobose, black, shiny, smooth, aseptate.
Cultural characteristics: Colonies on PDA at 25 °C floccose, edge entire, initially white, becoming grey to brown with age, reaching 9 cm diam in 10 d, reverse smoke-grey with black patches.
Material examined: CHINA, Hebei Province, Chengde City, Tongshan Garden, from soil, Q. Yang, 5 July 2021 (holotype CAF 800052; ex-type culture CFCC 56884); ibid. (culture CFCC 56885).
Notes: Two saprophytic isolates in the soil obtained in this study clustered into a well-supported clade distinguished from the other known species (Figure 1). Nigrospora humicola is phylogenetically close to N. chinensis and N. globosa. Morphologically, these three species share similar conidial morphology and size (12.5–23.5 × 9.5–16 μm in N. humicola vs. 10–14.5 × 7.5–11 μm in N. chinensis vs. 11–14.5 × 9–13 μm in N. globosa). However, N. humicola differs from N. chinensis (ITS: 28/518; tef1: 110/481; tub2: 40/392) and N. globosa (ITS: 19/486; tub2: 38/392) by sequence data [3,9].
4. Discussion
Nigrospora is a recently redefined monophyletic genus, and the species were well distinguished based on the combined loci of ITS, tef1, and tub2 sequence data [3,7]. Currently, the type species of Nigrospora, N. panici from Panicum amphibium in Indonesia, is not available in molecular data, and the holotype has been lost [1]. Hence, new collections from the original region and host P. amphibium are necessary to improve the genus concept.
Nigrospora species are common during fungal investigations; however, the sexual morph is rarely observed. From the asexual morph, all members have spherical to subspherical conidiogenous cells and black and globose to subglobose conidia [3]. In recent publications, species were distinguished mainly by conidial sizes [6,7,8,9,10,11,12,13,32]. However, the new species from the present study, N. humicola, is difficult to distinguish from its related species N. chinensis and N. globosa. Hence, molecular data (ITS, tef1 and tub2) are necessary for the species identification and delimitation of Nigrospora.
Two other phylogenetically distinct genera within Apiosporaceae, Arthrinium and Apiospora, are morphologically similar to Nigrospora in producing deeply pigmented conidia [33,34,35]. The distinction between these three genera is obscure, but the most characteristic difference is the production of a single conidium produced on each conidiogenous cell in Nigrospora, while conidia are usually produced in clusters in Arthrinum and Apiospora [33,34,35].
A new Arthrinum-like genus named Neoarthrinium was recently proposed in Amphisphaeriales based on Neo. lithocarpicola, Neo. moseri (syn. Wardomyces moseri), Neo. trachycarpi (syn. A. trachycarpi), and Neo. urticae (syn. A. urticae) [36]. This paper further confirmed the classification of Arthrinum, Apiospora, Neoarthrinium, and Nigrospora in the order Amphisphaeriales [36]. More strains of Nigrospora are needed from different ecosystems to improve the phylogram of this genus and related genera in the future.
Author Contributions
Conceptualization, Y.-Y.Z. and T.Z.; methodology, N.J.; software, Q.Y.; validation, H.-Y.L., R.Z. and J.R.; formal analysis, Q.Y.; investigation, Q.Y.; resources, Y.-Y.Z.; writing—original draft preparation, N.J.; writing—review and editing, Y.-Y.Z.; visualization, N.J.; supervision, Y.-Y.Z.; project administration, H.-Y.L. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by Key R&D Program Projects in Hebei Province, grant number 21327306D, and Chengde Science and Technology Plan Basic Research Project 202205B064.
Institutional Review Board Statement
Not applicable.
Data Availability Statement
New sequences from the present study are listed in Table 1.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Zimmerman, A. Ueber einige an tropischen Kulturpflanzen beobachtete Pilze III. Zentralblatt Für Bakteriol. Parasitenkd. 1902, 8, 216–221. [Google Scholar]
- Mason, E.W. On species of the genus Nigrospora zimmermann recorded on monocotyledons. Trans. Br. Mycol. Soc. 1927, 12, 152-IN6. [Google Scholar] [CrossRef]
- Wang, M.; Liu, F.; Crous, P.W.; Cai, L. Phylogenetic reassessment of Nigrospora: Ubiquitous endophytes, plant and human pathogens. Persoonia 2017, 39, 118–142. [Google Scholar] [CrossRef]
- Hyde, K.D.; Norphanphoun, C.; Maharachchikumbura, S.S.N.; Bhat, D.J.; Jones, E.B.G.; Bundhun, D.; Chen, Y.J.; Bao, D.F.; Boonmee, S.; Calabon, M.S.; et al. Refined families of Sordariomycetes. Mycosphere 2020, 11, 305–1059. [Google Scholar] [CrossRef]
- Wijayawardene, N.N.; Hyde, K.D.; Al-Ani, L.K.; Tedersoo, L.; Haelewaters, D.; Rajeshkumar, K.C.; Zhao, R.L.; Aptroot, A.; Leontyev, D.V.; Saxena, R.K.; et al. Outline of Fungi and fungus-like taxa. Mycosphere 2020, 11, 1060–1456. [Google Scholar] [CrossRef]
- Crous, P.W.; Carnegie, A.J.; Wingfield, M.J.; Sharma, R.; Mughini, G.; Noordeloos, M.E.; Santini, A.; Shouche, Y.S.; Bezerra, J.D.; Dima, B.; et al. Fungal Planet description sheets: 868–950. Persoonia 2019, 42, 291–473. [Google Scholar] [CrossRef]
- Raza, M.; Zhang, Z.F.; Hyde, K.D.; Diao, Y.Z.; Cai, L. Culturable plant pathogenic fungi associated with sugarcane in southern China. Fungal Divers 2019, 99, 1–104. [Google Scholar] [CrossRef]
- Tian, L.Y.; Zhang, Y.F.; Liao, T.; Qin, C.S.; Xu, J.Z. Nigrospora guangdongensis sp. nov. from the needle of Cunninghamia lanceolata in China. Phytotaxa 2020, 449, 181–187. [Google Scholar] [CrossRef]
- Zhang, Z.F.; Zhou, S.Y.; Eurwilaichitr, L.; Ingsriswang, S.; Raza, M.; Chen, Q.; Zhao, P.; Liu, F.; Cai, L. Culturable mycobiota from Karst caves in China II, with descriptions of 33 new species. Fungal Divers 2020, 106, 29–136. [Google Scholar] [CrossRef]
- De Silva, N.I.; Maharachchikumbura, S.S.; Thambugala, K.M.; Bhat, D.J.; Karunarathna, S.C.; Tennakoon, D.S.; Phookamsak, R.; Jayawardena, R.S.; Lumyong, S.; Hyde, K.D. Morpho-molecular taxonomic studies reveal a high number of endophytic fungi from Magnolia candolli and M. garrettii in China and Thailand. Mycosphere 2021, 12, 163–237. [Google Scholar] [CrossRef]
- Tennakoon, D.S.; Kuo, C.H.; Maharachchikumbura, S.S.; Thambugala, K.M.; Gentekaki, E.; Phillips, A.J.; Bhat, D.J.; Wanasinghe, D.N.; de Silva, N.I.; Promputtha, I.; et al. Taxonomic and phylogenetic contributions to Celtis formosana, Ficus ampelas, F. septica, Macaranga tanarius and Morus australis leaf litter inhabiting microfungi. Fungal Divers 2021, 108, 1–215. [Google Scholar] [CrossRef]
- Tan, Y.P.; Bishop-Hurley, S.L.; Shivas, R.G.; Cowan, D.A.; Maggs-Kölling, G.; Maharachchikumbura, S.S.; Pinruan, U.; Bransgrove, K.L.; De la Peña-Lastra, S.; Larsson, E.; et al. Fungal Planet description sheets: 1436–1477. Persoonia 2022, 49, 261–350. [Google Scholar] [CrossRef] [PubMed]
- Chen, Q.; Bakhshi, M.; Balci, Y.; Broders, K.D.; Cheewangkoon, R.; Chen, S.F.; Fan, X.L.; Gramaje, D.; Halleen, F.; Jung, M.H.; et al. Genera of phytopathogenic fungi: GOPHY 4. Stud. Mycol. 2022, 101, 417–564. [Google Scholar] [CrossRef] [PubMed]
- Palmateer, A.J.; Mclean, K.S.; Van Santen, E.; Morgan-Jones, G. Occurrence of Nigrospora lint rot caused by Nigrospora oryzae on Cotton in Alabama. Plant Dis. 2003, 87, 873. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.M.; Liu, Y.J. First report of Nigrospora sphaerica causing leaf blight on Cunninghamia lanceolata in China. Plant Dis. 2017, 101, 389. [Google Scholar] [CrossRef]
- Liu, Y.J.; Tang, Q.; Fang, L. First report of Nigrospora sphaerica causing leaf blight on Camellia sinensis in China. Plant Dis. 2016, 100, 221. [Google Scholar] [CrossRef]
- Kee, Y.J.; Hafifi, A.B.; Huda-Shakirah, A.R.; Wong, K.L.; Jin, X.L.; Nordahliawate, M.S.; Zakaria, L.; Mohd, M.H. First report of reddish brown spot disease of red-fleshed dragon fruit (Hylocereus polyrhizus) caused by Nigrospora lacticolonia and Nigrospora sphaerica in Malaysia. Crop Prot. 2019, 122, 165–170. [Google Scholar] [CrossRef]
- Han, S.; Yu, S.; Zhu, T.; Li, S.; Qiao, T.; Liu, Y.; Lin, T.; Yang, C. Nigrospora oryzae causing black leaf spot disease of Hibiscus mutabilis in China. Plant Dis. 2021, 105, 2255. [Google Scholar] [CrossRef] [PubMed]
- Metwaly, A.M.; Kadry, H.A.; El-Hela, A.A.; Mohammad, A.I.; Ma, G.; Cutler, S.J.; Ross, S.A. Nigrosphaerin A a new isochromene derivative from the endophytic fungus Nigrospora sphaerica. Phytochem. Lett. 2014, 7, 1–5. [Google Scholar] [CrossRef]
- Ibrahim, D.; Chong, C.L.; Tong, W.Y.; Zakaria, L.; Sheh-Hong, L. Effect of the extract of endophytic fungus, Nigrospora sphaerica CL-OP 30, against the growth of methicillin-resistant Staphylococcus aureus (MRSA) and Klebsiella pneumonia cells. Trop. J. Pharm. Res. 2015, 14, 2091–2097. [Google Scholar] [CrossRef]
- Zhong, J.; Zhao, S.Q.; Li, G.F.; Pang, X.D.; Deng, X.J.; Zhu, H.J.; Da Gao, B.; Zhou, Q. A novel fusarivirus isolated from the phytopathogenic fungus Nigrospora oryzae. Virus Genes 2016, 52, 891–895. [Google Scholar] [CrossRef]
- Fukushima, T.; Tanaka, M.; Gohbara, M.; Fujimori, T. Phytotoxicity of three lactones from Nigrospora sacchari. Phytochemistry 1998, 48, 625–630. [Google Scholar] [CrossRef]
- Meepagala, K.M.; Becnel, J.J.; Estep, A.S. Phomalactone as the active constituent against mosquitoes from Nigrospora spherica. Agric. Sci. 2015, 6, 1195–1201. [Google Scholar] [CrossRef]
- Doyle, J.J.; Doyle, J.L. Isolation of plant DNA from fresh tissue. Focus 1990, 12, 39–40. [Google Scholar]
- White, T.J.; Bruns, T.; Lee, S.; Taylor, J. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. PCR Protoc. Guide Methods Appl. 1990, 18, 315–322. [Google Scholar]
- Carbone, I.; Kohn, L.M. A method for designing primer sets for speciation studies in filamentous ascomycetes. Mycologia 1999, 3, 553–556. [Google Scholar] [CrossRef]
- Glass, N.L.; Donaldson, G.C. Development of primer sets designed for use with the PCR to amplify conserved genes from filamentous ascomycetes. Appl. Environ. Microbiol. 1995, 61, 1323–1330. [Google Scholar] [CrossRef] [PubMed]
- Katoh, K.; Rozewicki, J.; Yamada, K.D. MAFFT online service: Multiple sequence alignment, interactive sequence choice and visualization. Brief. Bioinform. 2019, 20, 1160–1166. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [PubMed]
- Stamatakis, A. RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 2014, 30, 1312–1313. [Google Scholar] [CrossRef]
- Ronquist, F.; Huelsenbeck, J.P. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 2003, 19, 1572–1574. [Google Scholar] [CrossRef] [PubMed]
- Lee, W.; Kim, D.; Perera, R.H.; Kim, J.S.; Cho, Y.; Lee, J.W.; Seo, C.W.; Lim, Y.W. Diversity of Nigrospora (Xylariales, Apiosporaceae) species identified in Korean macroalgae including five unrecorded species. Mycobiology 2023, 51, 401–409. [Google Scholar] [CrossRef] [PubMed]
- Jiang, N.; Liang, Y.M.; Tian, C.M. A novel bambusicolous fungus from China, Arthrinium chinense (Xylariales). Sydowia 2020, 72, 77–83. [Google Scholar] [CrossRef]
- Pintos, A.; Alvarado, P.; Planas, J.; Jarling, R. Six new species of Arthrinium from Europe and notes about A. caricicola and other species found in Carex spp. hosts. MycoKeys 2019, 49, 15–48. [Google Scholar] [CrossRef]
- Pintos, A.; Alvarado, P. Phylogenetic delimitation of Apiospora and Arthrinium. Fungal Syst. Evol. 2021, 7, 197–221. [Google Scholar] [CrossRef]
- Jiang, N.; Voglmayr, H.; Ma, C.Y.; Xue, H.; Piao, C.G.; Li, Y. A new Arthrinium-like genus of Amphisphaeriales in China. MycoKeys 2022, 92, 27–43. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).