Abstract
Lichen-feeding termites occupy a distinctive ecological niche. This feeding behavior underscores a complex interplay between the termites’ digestive abilities and the biochemical properties of lichens, known for their resilience and production of secondary metabolites. Understanding the dietary preferences and digestive mechanisms of these termites offers insights into their ecological roles and the evolutionary adaptations that enable them to exploit such a specialized food source. We conducted experiments with Constrictotermes cyphergaster, feeding it with different combinations of its natural food sources: wood bark and lichen from host trees. Gut microbial communities were analyzed through 16S rRNA sequencing and shotgun metagenomics. Our results revealed that a diet containing lichens induces a shift in microbiota composition and increases the abundance of genes encoding an AA3 enzyme with a role in lignin digestion. This study emphasizes the potential role of lichens in enhancing the digestive capabilities of termites, highlighting the intricate relationships between diet, gut microbiota, and enzymatic activity in Termitidae.
1. Introduction
The diversification of Termitidae (encompassing over 80% of known termite species) [1] is attributed to the loss of unicellular gut protists and the acquisition of lignocellulolytic bacterial lineages. These adaptations enable termites to exploit a broader range of food sources, including wood, leaf litter, live or dead grasses, soil with high organic material content undergoing humification, animal excrement, the mycelium of certain fungi, and even microepiphytes [2,3,4,5,6].
Lichens result from the symbiotic association between fungi and green algae or cyanobacteria and are considered to have low palatability due to their high content of toxic compounds in some species [7]. For this reason, few organisms are specialized lichen-feeders [8]. Nevertheless, some termites are known to feed on lichens, with documented cases in species from the genera Hospitalitermes, Longipeditermes, Grallatotermes, and Constrictotermes [9,10,11,12]. Constrictotermes (Termitidae: Nasutitermitinae) is a Neotropical genus with two lichen-feeding species, C. cavifrons (Holmgren, 1910), and C. cyphergaster (Silvestri, 1901), [9]. Constrictotermes cyphergaster is an arboreal termite found in the savannas and dry forests extending from Brazil to Argentina [13,14] (Figure 1a). The workers of this species engage in nocturnal foraging along exposed trails on trees, where they consume not only the bark of wood but also various species of lichens present on tree trunks (Figure 1b) [15,16].

Figure 1.
(a) Nest of Constrictotermes cyphergaster at the Caatinga (dry-forest vegetation) in São João do Cariri Experimental Station, Brazil. (b) Detailed view of workers feeding on an unidentified lichen species.
The consumption of lichens could be a potential nitrogen source for these insects [11,17] or aid in the hydrolysis of certain lignocellulosic components [18]. However, aspects of lignocellulose digestion and the composition of the symbiotic microbiota in the digestive tract of termites that include lichens in their diet remain unknown.
The presence of lignocellulolytic enzymes has been reported in some lichen species [18,19]. Therefore, the incorporation of lichens into the diet could provide termites with an additional source of enzymes, potentially aiding in the more efficient digestion of lignocellulosic components in their food. This hypothesis will be investigated by evaluating: (i) the effect of lichen on bark wood consumption, (ii) the composition of gut microbiota in termites fed with or without lichen, (iii) the difference in lignocellulolytic activity between food diets, and (iv) the community functions based on metagenomic analyses in termites subjected to diets containing bark wood and/or lichen.
2. Materials and Methods
2.1. Termites and Study Site
Twelve colonies of Constrictotermes cyphergaster were collected in areas of dry-forest vegetation (Caatinga) at the Experimental Station of São João do Cariri (7°20′ S, 36°31′ W), Paraíba State, Brazil. (Figure S1). The climate of the region is semi-arid with an average temperature of 30 °C and annual rainfall of 400 mm, concentrated from February to July [20]. Sampling was authorized by the Brazilian Institute of Environment and Renewable Natural Resources (IBAMA), a Brazilian Ministry of the Environment’s enforcement agency (SISBIO n° 33269). No endangered species were collected in this study.
2.2. Lichen and Bark Wood Consumption
We conducted feeding experiments combining the availability of natural food sources of C. cyphergaster: wood bark and lichen from host trees. One hundred workers and five soldiers were carefully removed from each of four colonies and placed in 500 mL glass flasks containing sterilized, moistened sand and one of the following food treatments: (1) wood bark from the host tree Aspidosperma piryfolium (Apocynaceae); (2) a crustose lichen belonging to a single species within the genus Polymeridium. (Trypetheliaceae); and (3) a combination of wood bark and lichen simultaneously (Figure S2). The lichen sample was collected from one tree that contained one of the nests. The identification of lichen was conducted as described by Cáceres (2007) [21]. Each treatment was replicated ten times for each colony, providing sufficient material for subsequent metagenomic and enzymatic analyses. The experiment was conducted for 7 days in a climate-controlled room maintained at a constant temperature (25 °C) and relative humidity (70%). Dead termites were removed daily. Food consumption was calculated as the difference between initial and final fresh weight corrected by humidity loss in control flasks with food without termites. After 7 days, all surviving termites were removed from flasks and their digestive tracts were extracted and frozen at −22 °C following previously established methodologies [22]. An in situ sample (100 workers) from each colony was kept without any experimental treatment.
Generalized Mixed Models (GLMM) with Gaussian error distribution were conducted to assess differences in food consumption among treatments using the glmmTMB package [23]. Pairwise comparisons were performed using estimated marginal means with the package emmeans [24]. We assessed residual distribution using the DHARMa package [25].
2.3. DNA Extraction
The whole guts of 50 workers from the 4 colonies subjected to each food treatment and from the in situ sample were placed in 2 mL tubes containing 1 mL of lysis buffer (500 mM NaCl, 50 mM Tris-HCl, pH 8.0, 50 mM EDTA, and 4% sodium dodecyl sulfate. DNA extraction was carried out using a bead-beating protocol as previously described [26]. DNA integrity was assessed by agarose gel electrophoresis (1.0 w/v) and quantification was performed using a NanoDrop spectrophotometer by measuring the absorbance at 260 nm.
2.4. 16S rRNA Library Preparation, Sequencing, and Taxonomic Assignment
Gut microbial communities were accessed in termites from four colonies following the methodology of Menezes et al. (2018) [22]. The V4 region of the 16S rRNA gene was targeted using primers 515F (5′ GTGCCAGCMGCCGCGGTAA 3′) and 806R (5′ GGACTACHVGGGTWTCTAAT 3′) [26]. Libraries were prepared using two PCR steps. The first PCR step was performed using specific primers for each library using Phusion Polymerase (Thermo Scientific, Waltham, MA, USA) and 40 ng of template DNA from each sample. For 16S rRNA amplification, the thermocycler program was set at initial denaturing at 98 °C for 2 min, followed by 30 cycles of 98 °C (30 s), 60.1 °C (30 s) and 72 °C (40 s), ending with a final extension of 72 °C (5 min). The second PCR step was necessary to add Illumina sequencing adapters and dual index barcodes (Nextera Index Kit, Illumina, San Diego, CA, USA) to the amplified libraries using Phusion polymerase (Thermo Scientific) with 100 ng of purified PCR products from the previous step as template and indexing primers from Illumina. Amplification conditions included an initial denaturation at 98 °C for 3 min, followed by 5 cycles of 98 °C (30 s), 55 °C (30 s), and 72 °C (30 s), with a final extension of 72 °C for 5 min. Each sample was amplified in triplicates. Pooled samples were purified using Agencourt AMPure Magnetic Beads (Beckman Coulter, Brea, CA, USA) and quantified using Qubit Fluorometer 2.0 (Invitrogen). Sequencing was performed on the Illumina Miseq Platform (available at the Brazilian Bioethanol Science and Technology Laboratory—CTBE/CNPEM), using a Miseq Reagent Kits V3 (600 cycles).
The dataset was processed using the UPARSE pipeline [27]. Briefly, paired-end reads were first merged using fastq_mergepairs. Only reads with a minimum overlap of 250 bp and a maximum expected error of 0.5 were used for downstream analysis. The quality-filtered reads were thus grouped into zero-radius operational taxonomic units (zOTUs, also known as Amplicon Sequence Variants (ASVs), 97% similarity threshold) using the UPARSE-ASV algorithm. The identified ASVs were further compared to the Gold database as a reference to filter chimera sequences using the chimera UCHIME algorithm [28], which is implemented in the USEARCH package. ASVs with no more than one read in at least 10% of the samples were removed. Reads were clustered at 97% sequence similarity. Taxonomic assignment was performed using the RDP classifier implemented in MOTHUR and the sintax command in the SILVA database [29]. Relative abundances were calculated as the number of reads per taxon. The analysis was performed on the ASV table, which was rarefied to the smallest library size.
2.5. Metagenomic Sequencing, Assembly, and Gene Annotation
A library was constructed, using the Nextera library preparation kit (Illumina), according to the manufacturer’s instructions. The prepared library was validated using the Agilent bioanalyzer 2100 system with a 12,000 DNA assay kit (Agilent, Santa Clara, CA, USA) and quantified using Kapa Biosystems next-generation sequencing library qPCR kit (Kapa Biosystems, Wilmington, MA, USA), respectively following the methodology of Moreira et al. (2018) [6]. Sequencing was performed at the LNBR NGS facility (CNPEM—Campinas, São Paulo, Brazil) using an Illumina HiSeq 2500 platform and applying the paired-end protocol (2 × 100-bp paired ends). Metagenome (MG) raw reads were quality-checked using FastQC https://www.bioinformatics.babraham.ac.uk/projects/fastqc/ (accessed on 3 January 2024) and quality-filtered using Trimmomatic v.0.36 [30]. Next, the whole set of MG trimmed reads were de novo co-assembled using Megahit [31]. Reads were taxonomically assigned using MMseqs [32]. Gene prediction and annotation were conducted utilizing Prokka version 1.11 [33], while annotation of Carbohydrate-active enzymes (CAZy) genes was carried out using the dbCAN3 database [34]. Only bacteria genes were used to run a search against dbCAN3. The normalized abundance of protein-coding genes was calculated by dividing the number of reads from the metagenome mapped to a single gene (×106) by its length, and subsequently dividing this result by the sum of the number of reads from the metagenome mapped to all genes, which was also divided by their respective lengths.
2.6. Microbial Diversity and Community Structure Analyses
We used R version 3.5.0 [35] to conduct analyses using different packages. Downstream analysis, including α- and β-diversity analysis (see below), was calculated using the microeco v.1.9.0 and MicrobiotaProcess v.3.19 packages [36,37]. The ASVs table was merged with relevant metadata into a microeco object. Generalized linear mixed models (GLMM) were performed to check for overall significant differences in α-diversity estimates and the total number of ASVs among samples. We fitted multivariate generalized linear models (mvabund package) [38] to test the effects of the diet on the microbial relative abundance. Models were fitted with a negative binomial distribution using 999 bootstrap iterations. The manyglm function was used to carry out the analysis. Ordination analyses were used to evaluate microbial composition at the ASV level. We used ALDEx2 analysis (ANOVA-Like Differential Expression tool for compositional data) to identify ASVs that define the differences between termite microbiomes [39,40]. Comparison with other termite feeding guilds was conducted using Raw Illumina sequences deposited in ENA with accession no. PRJEB17080. The sequences include the litter-feeding species, Cornitermes cumulans (Kollar, 1832), Syntermes dirus (Burmeister, 1839) (Termitidae: Syntermitinae), Velocitermes heteropterus (Silvestri, 1901) (Termitidae: Nasutitermitinae), and Ruptitermes sp. (Termitidae: Apicotermitinae), and the intermediate feeding species Silvestritermes euamignathus (Silvestri, 1901) and Procornitermes araujoi Emerson, 1952 (Termitidae: Syntermitinae).
2.7. Assays of Cellulolytic and Hemicellulolytic Activities of Worker Guts
The activity against cellulose and hemicellulose substrates was assayed using crude enzyme extracts from the gut of workers from four colonies subjected to different food diets, following the methodology of Menezes et al. (2018) [22]. In situ samples from eight colonies were also analyzed. The assays were conducted to assess the activity of the soluble fraction of protein extracts against natural polysaccharides and synthetic oligosaccharides with varying monomer compositions and branching. Crude protein extractions were carried out from the guts of 50 workers from each colony. The samples were homogenized in 2 mL of 100 mM sodium acetate buffer at pH 5.5. Subsequently, the crude extract was centrifuged at 20,100× g for 30 min at 4 °C, the supernatant was collected, and 1 µL of Protease Inhibitor Cocktail (Anresco) per mL of crude extract was added. The protein concentration in each crude extract was determined using the Bradford method [41].
The enzymatic activity essay was conducted following the methodology previously described [42], consisting of 10 µL of crude protein extract incubated at 37 °C for 40 min with 40 µL of 50 mM sodium acetate buffer pH 5.5 and 50 µL of 0.5% specific carbohydrate (in water), in triplicate. Enzymatic reactions were stopped by adding 100 µL of dinitrosalicilic acid (DNSA) and heating at 99 °C for 5 min. The color change was analyzed at 412 nm using a TECAN M2000 plate reader. Results were expressed in mmol of glucose equivalents produced per mg of protein. One unit of enzymatic activity was defined as the amount of enzyme that released 1 mmol of reducing sugar. Blank reactions were performed as described above with 100 µL of DNSA added to the reaction prior to incubation with the protein.
For the nitrophenyl-monosaccharide (pNP-G), 10 µL of crude extracts was incubated with 50 µL of 5 mM pNP-G and 40 µL of 50 mM sodium acetate buffer, pH 5.5. The reactions were stopped after the addition of 100 µL of 1 M Sodium Carbonate (Na2CO3). The activity of pNP-G was expressed in terms of the mM of p-nitrophenyl released. Blank reactions were performed as described above with 100 µL of 1 M Sodium Carbonate (Na2CO3) added to the reaction before the incubation. The color change was analyzed at 412 nm using a TECAN M2000 plate reader. Glucose and p-nitrophenyl were used for standard curve construction.
Carbohydrates included CMC (carboxymethyl cellulose low viscosity) (β-1,4-carboxymethylglucan); β-glucan from Barley (low viscosity) (β-1,4-glucan), Xylan from Oat Spelt (β-1,4-xylan), Rye Arabinoxylan (α-2,3-arabinose-β-1,4-xylan) from Citrus, Lichenin from moss, and pNP-G (4-nitrophenyl β-1,4-D-glucopyranoside). The effect of diet on the enzymatic activity of termite workers was analyzed using Wilks’ Lambda type nonparametric multivariate inference with the package npmv [43].
3. Results
3.1. The Gut Microbiota of a Lichen-Feeding Termite and Its Relation with Other Feeding Guilds
We identified a total of 1834 bacterial ASVs (319,593 reads) and 14 archaeal ASVs (19,771 reads) associated with the gut of C. cyphergaster workers. In situ samples revealed the presence of 19 bacterial phyla and 122 genera, along with 3 archaeal phyla and 4 genera (Table S1). Rarefaction curves indicated adequate sampling (Figure S3). The most abundant phyla (Firmicutes, Bacteroidota, and Spirochaetota) accounted for 74.2% of sequence reads. Firmicutes dominated the microbial community, with an average abundance of 51.1%, whereas Bacteroidota and Spirochaetota accounted for 12.5 and 10.6%, respectively. Treponema (Spirochaetota) and two Ruminococcaceae and Lachnospiraceae unknown organisms were the most abundant taxa in the gut of C. cyphergaster workers (Figure 2).
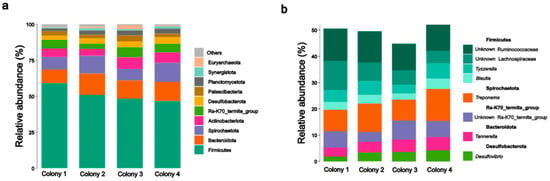
Figure 2.
Taxonomic composition of the microbial communities associated with the gut of Constrictotermes cyphergaster workers from four colonies: (a) Relative abundances of the ten most prevalent phyla. (b) Relative abundances of the eight most prevalent genera or the lowest taxonomic level.
The proportions of the most abundant members of the gut microbiota of C. cyphergaster were similar when compared with other feeding guilds, showing a predominance of the phyla Firmicutes, Spirochaetota, and Bacteroidota. Firmicutes predominated in the gut of C. cyphergaster, Silvestritermes euamignathus, and Ruptitermes sp. In contrast, Spirochaetota was the most abundant phylum in litter-feeder species, except for S. dirus (Figure 3a). However, C. cyphergaster displayed a distinct gut microbiota composition compared with intermediate feeders (manyglm; Dev = 4957; df = 2; p = 0.015) and litter feeders (Dev = 4197; df = 2; p = 0.015) termites (Figure 3b).
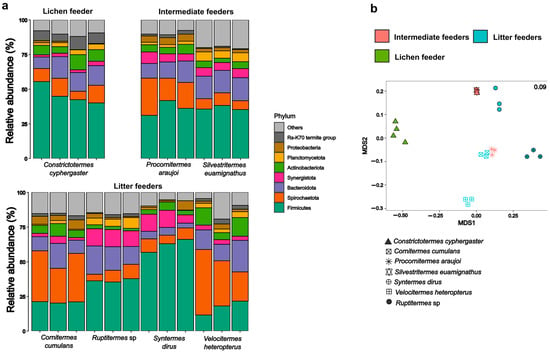
Figure 3.
Comparison between gut microbial communities of lichen-feeder Constrictotermes cyphergaster and other feeding guilds: (a) Relative abundance of microbial phyla among termite species for each feeding guild. (b) NMDS plot showing gut microbial communities of different termite feeding guilds. Each stacked bar represents a microbial community from one colony.
3.2. Glucose Polymers Enzymatic Activity and Encoding Genes in the Gut of Constrictotermes Cyphergaster
The soluble fraction of protein extracts from worker guts showed high activity against all the polysaccharides assessed in this study. Significant variations were observed, with the highest values recorded for all the glucose polymers evaluated in this study: β-glucan (β = 558.24; p < 0.001), lichenin (β = 112.23; p < 0.001) pNP-G (β = 149.81; p < 0.001), and CMC (β = 112.23; p < 0.001) (Table S2, Figure 4a).
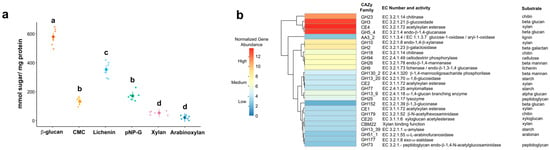
Figure 4.
(a) Glucose and xylose-polymer activities of the soluble fraction of gut protein extracts of Constrictotermes cyphergaster worker guts. Substrates abbreviations: CMC (carboxymethyl cellulose) (β-1,4-carboxymethylglucan); β-glucan (β-1,4-glucan), Xylan (β-1,4-xylan), Arabinoxylan (α-2,3-arabinose-β-1,4-xylan), Liquenin, and pNP-G (4-nitrophenyl-β-1,4 -D-glucopyranoside). In the plot, the modeled means are represented by large circles, bars are the modeled 95% confidence intervals, and the model-adjusted individual response values are represented by the small dots. Different letters indicate significance at p < 0.05 (comparisons of least square means). (b) Genes encoding carbohydrate active enzymes (CAZy) in the gut of C. cyphergaster workers. The clustering heatmap shows normalized abundance of protein-coding genes of the most abundant CAZy families. The dendrogram was produced by hierarchical clustering representing Pearson correlation between rows (CAZy genes).
Metagenomic analyses revealed abundance of the main CAZy gene families in the gut microbiota of C. cyphergaster, which might be involved in the hydrolysis of these glucose polymers were GH3 (beta-glucosidase; EC 3.2.1.21), GH5_4 (beta-glucanase; EC 3.2.1.4), and GH9 (lichenase; EC 3.2.1.79) (Table S3 and Figure 4b).
Other abundant CAZy gene families include GH23 (chitinase, EC 3.2.1.14), GH10 (endo-1,4-beta-xlanase, EC 3.2.1.8), CE4 (acetylxylan esterase, EC 3.1.1.72), GH2 (beta-galactosidase, EC 3.2.1.23), and GH26 (endo-beta-1,4-mannanase, EC 3.2.1.78), and lysozyme (GH25) (Figure 4b).
3.3. Lichen Availability Influences Gut Microbiota and CAZy Gene Abundance
Workers of C. cyphergaster exhibited consistent consumption patterns of lichen and wood bark, irrespective of whether these food sources were presented individually (t = −1.855, p = 0.072) or in combination (t = −0.835, p = 0.4098). The average consumption quantities for termites were 40.70 ± 9.58 mg of lichen and 62.50 ± 8.53 mg of wood bark when provided separately, and 43.90 ± 10.52 mg of lichen and 55.10 ± 9.98 mg of wood bark when offered together (Table S4, Figure S4).
The inclusion of lichen in the diet significantly influenced the composition of the gut microbiota in workers of C. cyphergaster (Table S5; Figure 5a). A diet-related shift in the microbial community was observed when termites were fed with a combination of lichen and wood, as indicated by the manyglm analysis (Deviance = 3.793; p = 0.017) (Figure 5b). Furthermore, a significant association with Treponema (Spirochaetes) was identified through Aldex2 analyses (p = 0.042; Wilcoxon signed-rank test) when compared to termites that were exclusively fed with lichen or wood (Figure 5c).
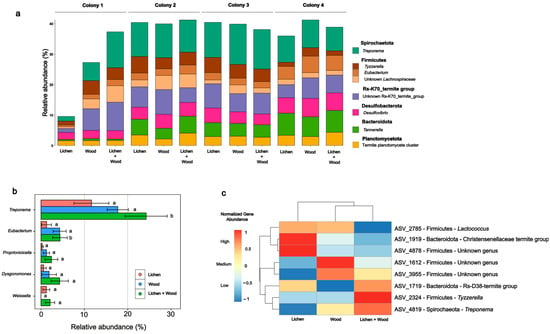
Figure 5.
Effect of diet on gut microbial communities of Constrictotermes cyphergaster: (a) Relative abundances of the eight most prevalent Amplicon Sequence Variants (ASVs) at the genus or the lowest taxonomic level. (b) Aldex2 differential analysis of genus relative abundances among diet treatments. Results are shown for five genera with the highest contribution to differences across diet treatments. Different letters indicate significance at p < 0.05 (Wilcoxon signed-rank test). (c) Taxa responsible for differences in gut microbiota composition between diet treatments (manyglm; Dev = 3793; p = 0.017). The clustering heatmap shows normalized abundance of ASVs among diet treatments. The dendrograms were produced by hierarchical clustering representing Pearson correlation between rows (ASVs).
No significant variation was observed in the enzymatic activity of gut extracts in response to glucose and xylose polymers across the different feeding treatments (Wilks’ Lambda = 0.332; df = 12, 20; p = 0.987) (Table S6). However, a shift was detected in the microbial gene encoding the enzyme AA3_2 following the addition of lichen to the diet, as well as in the genes GH23 and GH152 when wood was included in the feeding treatments (Tables S3 and S7, and Figure 6).

Figure 6.
Genes encoding carbohydrate-active enzymes (CAZy) and their substrates in workers of C. cyphergaster under different diets. The clustering heatmap shows normalized abundance of protein-coding genes of the twelve most abundant CAZy families. The dendrograms were produced by hierarchical clustering representing Pearson correlation between rows (CAZy genes).
4. Discussion
4.1. The Gut Microbiome of the Lichen-Feeder Termite Constrictotermes Cyphergaster
While termites are primarily known for consuming lignocellulose-based materials like wood and litter, there have been instances of termites consuming microepiphytes. Although uncommon among termites, this behavior may occur under specific conditions or in certain species [11].
Constrictotermes cyphergaster exhibits dietary flexibility by consuming wood and several species of crustose lichens [17]. Lichens might serve as a nutrient source, providing carbohydrates and nitrogen that may be advantageous for termites, especially in environments with limited nutrients like the dry forests inhabited by C. cyphergaster. However, most termites that feed on lichens are found in nutrient-rich habitats such as rainforests [9,11,44]. Furthermore, lichens produce a diverse array of secondary metabolites, some of which inhibit enzyme activity or exhibit antimicrobial effects, thereby acting against lichen-eating invertebrates and pathogens [45,46,47]. Consequently, consuming lichens may affect various aspects of termite digestion beyond the nutritional composition.
The gut microbiota of C. cyphergaster is characterized by a high abundance of Ruminococcaceae and Lachnospiraceae (Firmicutes), families that include numerous species with notable xylanolytic and cellulolytic capabilities [48,49]. The Treponema genus, also abundant in the gut community of C. cyphergaster serves as a significant source of enzymes that aid in the breakdown of wood polysaccharides [50], which is confirmed by the enzymatic activities of its crude protein extract. These characteristics are typical of wood-feeding termites and may elucidate C. cyphergaster’s ability to utilize wood and wood bark in its habitats. However, its gut microbiota composition differs extensively from other termite-feeding guilds, suggesting that a lichen-based diet has an impact on the microbial communities [5,51,52].
Our study revealed high enzymatic activity on glucose polymers and the presence of microbial genes encoding cellulases and hemicellulases in the gut of C. cyphergaster. These findings are likely associated with the high abundance of Treponema sp., as well as from the families Lachnospiraceae and Ruminococcaceae. These bacterial taxa are essential for providing cellulases and hemicellulases in termites, facilitating the degradation of wood polysaccharides [50,53,54]. The wood from the Caatinga dry forest is known for its high levels of cellulose, lignin, and hemicellulose [55]. Furthermore, our analysis revealed high proportions of genes encoding enzymes such as beta-glucosidase, beta-glucanase, xylanases, galactosidase, mannanase, and cellulase, which is consistent with the wood consumption by this termite species.
Other CAZy genes detected in the gut microbiome of C. cyphergaster include licheninase, chitinase, and lysozyme-encoding genes. The primary cell wall polysaccharides identified in lichen mycobiont comprise β-1,3 glucans and chitin. Licheninase is responsible for degrading the β-1,3;1,4-glucans present in lichen cell walls [56]. Chitin, a fundamental constituent of fungal cell walls, is structured with β-1,4 linked N-acetylglucosamine [57]. Chitin can be a source of nitrogen for C. cyphergaster, and chitinase facilitates the nonspecific cleavage of internal bonds in chitin, generating smaller oligosaccharides, from which N-acetylglucosamine units are liberated through the activity of beta-N-acetylglucosaminidases [58]. Thus, the presence of licheninases and chitinases in the gut of C. cyphergaster is consistent with the incorporation of lichen in the diet of this species.
4.2. Does a Lichen Diet Affect Lignocellulose Digestion in Constrictotermes Cyphergaster?
The results of our study revealed that C. cyphergaster workers consume equivalent amounts of lichen and wood bark in controlled laboratory conditions, regardless of whether they are presented individually or in combination. We did not detect changes in the gut enzymatic activity of C. cyphergaster workers against glucose and xylose polymers when lichen was added to the diet. However, when workers consumed a combination of lichen and wood bark there was an increase in Treponema.
We observed shifts in the abundance of microbial genes associated with the AA3_2 subfamily when only lichen was accessible to the termites. The AA3 family contains enzymes that play a role in lignin degradation [59]. Lignin is a recalcitrant component of plant cell walls that contains a complex network of aromatic compounds, forming an insoluble barrier that restricts access to the cellulose and hemicellulose [60]. The process of lignocellulose digestion requires the presence of effective cellulases and a variety of other glycoside hydrolases to break down the components of the cell wall. Nevertheless, it is crucial to first overcome the barrier posed by the lignin matrix [61].
The subfamily AA3_2 is composed of oxidoreductases, which are reported in the digestive systems of various termite species and certain fungi. This subfamily comprises enzymes that target phenolic aromatic aldehydes and acids originating from lignin, while catalyzing the conversion of O2 to H2O2. The produced H2O2 can be utilized by peroxidases to degrade lignin [62,63,64,65,66]. Genes categorized as AA3_2 have also been detected in metagenomic analyses of the gut microbiomes of other termite species, potentially aiding in the decomposition or alteration of ingested lignin structures [54,65,66]. Notably, Treponema genomes found in termite gut microbiomes are recognized for encoding AA3 enzymes [67]. In addition, AA3_2 enzymes are capable of oxidizing some toxic compounds derived from lignin degradation or from microorganisms [68].
We also observed a shift in microbial genes encoding chitinase (GH23) and β-1,3-glucanase (GH152) when wood was provided to the termites. Both enzymes are responsible for hydrolyzing chitin and fungal polysaccharides present in the cell walls of fungi [69,70]. They are linked to a mycolytic gut bacterial community in certain termite species, particularly those that feed on wood, where wood-degrading fungi may form a significant part of the termite diet [71], or serve as a defense mechanism against pathogenic fungi [72]. Further research is required to understand the reasons behind the increased abundance of chitinase microbial genes following wood ingestion.
In this context, the consumption of lichen by C. cyphergaster workers could aid in the degradation of the lignin barrier present in wood, thereby acting as a preliminary stage in reaching the carbohydrate content and transforming it into an energy reservoir. This holds importance due to the close chemical bonding of lignin with cellulose and hemicellulose in the cell walls of land plants.
5. Conclusions
Termites, recognized for their lignocellulose-based diet, demonstrate dietary adaptability by consuming microepiphytes. Previous research has linked this behavior to their nutrient-deficient diets, with lichens serving as an alternative nutritional source offering supplementary carbohydrates and nitrogen. Nevertheless, our findings indicate that the ingestion of lichens promotes lignocellulose digestion over their nutritional content.
Our study demonstrates that lichen ingestion by Constrictotermes cyphergaster increased the abundance of gut carbohydrate-active enzymes (CAZy) genes that encode an oxidoreductase enzyme with a role in lignin digestion and xenobiotic degradation. Consequently, consuming lichens may help to break the recalcitrant lignin barrier of their main food, the hardwood of dry forest trees in Brazil. From the perspective of termite evolution, this study advances the knowledge regarding the association between gut symbionts and termites and their role in diet diversification of Termitidae. Future research should focus on species-specific behaviors, microbial analyses of lichens, and the influence of environmental seasonal factors of the dry forest on termite dietary choices. Understanding the conditions under which termites consume lichens, as well as identifying the specific species involved, might offer valuable insights into their foraging behavior and ecological adaptability.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/d16100623/s1, Figure S1: Map of study site where Constrictotermes cyphergaster were collected in the microregion do Cariri Oriental located at the Municipality of São João de Cariri, northeastern Brazil, in the state of Paraíba; Figure S2: Detail of the bioassays elaborated for consumption of lichen and wood by Constrictotermes cyphergaster; Figure S3: Rarefaction curves showing the number of Amplicon Sequence Variants (ASVs) in the gut microbiome of Constrictotermes cyphergaster; Figure S4: Lichen and wood consumption by Constrictotermes cyphergaster workers when exposed to the two types of food individually and simultaneously. In the plot, the modelled means are represented by large circles, bars are the modelled 95% confidence intervals, and the model-adjusted individual response values are represented by the small dots. p values < 0.05 indicate significance (lsmeans pairwise analysis). Table S1: Taxonomical assignment of bacteria and archaea from the gut of Constrictotermes cyphergaster based on 16S rRNA amplicon sequencing; Table S2: Generalized linear mixed model explaining enzymatic activity of workers guts on different polysaccharides nested under colony number; Table S3: Metagenome sequencing, assembly and annotation summary; Table S4: Generalized linear mixed model explaining food consumption by workers between lichen and wood diets (individually or combined) nested under colony number; Table S5: Taxonomical assignment of bacteria and archaea from the gut of Constrictotermes cyphergaster subjected to different diet treatments based on 16S rRNA amplicon sequencing; Table S6: Non parametric MANOVA model explaining enzymatic activity of workers guts on polysaccharides from termites submitted to different diet treatments; Table S7: Abundance of Carbohydrate Active Enzymes (CAZy) genes associated to gut of Constrictotermes cyphergaster under different diet treatments. The abundance of protein-coding genes was calculated by dividing the number of reads from the metagenome mapped to a single gene (×106) by its length, and subsequently dividing this result by the sum of the number of reads from the metagenome mapped to all genes, which was also divided by their respective lengths.
Author Contributions
The authors A.A., M.A.B.-G., H.C., D.S.-D. and A.M.C.-L. designed the study. L.R.d.M., J.P.L.F.C., M.S.L.R. and M.H.d.O. performed the experiments. A.A., L.R.d.M., R.A.C.d.S. and L.C.P. analyzed data. A.A., J.P.L.F.C. and L.C.P. wrote the paper. All authors have read and agreed to the published version of the manuscript.
Funding
This study was supported by the São Paulo Research Foundation (FAPESP), grants #2015/21497-6 and #2018/22839-6.
Institutional Review Board Statement
Not applicable.
Data Availability Statement
The original data presented in the study are openly available at NCBI (https://www.ncbi.nlm.nih.gov), Bioproject number PRJNA912562.
Acknowledgments
We would like to thank the Brazilian Biorenewables National Laboratory (LNBR/CNPEM) NGS Sequencing Facility for generating the metagenomics sequencing data described here.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Krishna, K.; Grimaldi, D.A.; Krishna, V.; Engel, M.S. Treatise on the Isoptera of the World. Bull. Am. Mus. Nat. Hist. 2013, 377, 2433–2705. [Google Scholar] [CrossRef]
- Donovan, S.E.; Eggleton, P.; Bignell, D.E. Gut Content Analysis and a New Feeding Group Classification of Termites. Ecol. Entomol. 2001, 26, 356–366. [Google Scholar] [CrossRef]
- Eggleton, P.; Tayasu, I. Feeding Groups, Lifetypes and the Global Ecology of Termites. Ecol. Res. 2001, 16, 941–960. [Google Scholar] [CrossRef]
- Barbosa-Silva, A.M.; Farias, M.A.A.; de Mello, A.P.; de Souza, A.E.F.; Garcia, H.H.M.; Bezerra-Gusmão, M.A. Lignocellulosic Fungi in Nests and Food Content of Constrictotermes cyphergaster and Inquilinitermes fur (Isoptera, Termitidae) from the Semiarid Region of Brazil. Fungal Ecol. 2016, 20, 75–78. [Google Scholar] [CrossRef]
- Mikaelyan, A.; Dietrich, C.; Köhler, T.; Poulsen, M.; Sillam-Dussès, D.; Brune, A. Diet Is the Primary Determinant of Bacterial Community Structure in the Guts of Higher Termites. Mol. Ecol. 2015, 24, 5284–5295. [Google Scholar] [CrossRef] [PubMed]
- Moreira, E.A.; Alvarez, T.M.; Persinoti, G.F.; Paixão, D.A.A.; Menezes, L.R.; Cairo, J.P.F.; Squina, F.M.; Costa-Leonardo, A.M.; Carrijo, T.; Arab, A. Microbial Communities of the Gut and Nest of the Humus- and Litter-Feeding Termite Procornitermes araujoi (Syntermitinae). Curr. Microbiol. 2018, 75, 1609–1618. [Google Scholar] [CrossRef]
- Hawksworth, D.L.; Hill, D.J. The Lichen-Forming Fungi, 1st ed.; Springer US: Boston, MA, USA, 1984; ISBN 978-0-216-91634-0. [Google Scholar]
- Pöykkö, H.; Bačkor, M.; Bencúrová, E.; Molcanová, V.; Bačkorová, M.; Hyvärinen, M. Host Use of a Specialist Lichen-Feeder: Dealing with Lichen Secondary Metabolites. Oecologia 2010, 164, 423–430. [Google Scholar] [CrossRef]
- Martius, C.; Amelung, W.; Garcia, M.V.B. The Amazonian Forest Termite (Isoptera: Termitidae) (Constrictotermes cavifrons) Feeds on Microepiphytes. Sociobiology 2000, 35, 379–383. [Google Scholar]
- Roisin, Y.; Pasteels, J.M. The Nasute Termites (Isoptera: Nasutitermitinae) of Papua New Guinea. Invertebr. Syst. 1996, 10, 507–616. [Google Scholar] [CrossRef]
- Miura, T.; Matsumoto, T. Diet and Nest Material of the Processional Termite Hospitalitermes, and Cohabitation of Termes (Isoptera, Termitidae) on Borneo Island. Insectes Soc. 1997, 44, 267–275. [Google Scholar] [CrossRef]
- Gay, F.J. Notes on Grallatotermes grallator (Desneux) and the Taxonomic Status of the Genus Grallatotermes (Isoptera: Termitidae: Nasutitermitinae). Pac. Insects 1971, 13, 41–48. [Google Scholar]
- Melo, A.C.S.; Bandeira, A.G. A Qualitative and Quantitative Survey of Termites (Isoptera) in an Open Shrubby Caatinga in Northeast Brazil. Sociobiology 2004, 44, 707–716. [Google Scholar]
- Torales, G.J.; Laffont, E.R.; Godoy, M.C.; Coronel, J.M.; Arbino, M.O. Update on Taxonomy and Distribution of Isoptera from Argentina. Sociobiology 2005, 45, 853–886. [Google Scholar]
- Barbosa-Silva, A.M. Líquens Associados à Alimentação de Constrictotermes cyphergaster (Silvestre, 1901) (Isoptera: Termitidae) No Semiárido Brasileiro. Master’s Thesis, Universidade Estadual da Paraiba, Campina Grande, Brasil, 2014. [Google Scholar]
- Barbosa-Silva, A.M.; Silva, A.C.; Pereira, E.C.G.; Buril, M.L.L.; Silva, N.H.; Cáceres, M.E.S.; Aptroot, A.; Bezerra- Gusmão, M.A. Richness of Lichens Consumed by Constrictotermes cyphergaster in the Semi-Arid Region of Brazil. Sociobiology 2019, 66, 154–160. [Google Scholar] [CrossRef]
- Barbosa-Silva, A.M.; Vasconcellos, A. Consumption Rate of Lichens by Constrictotermes cyphergaster (Isoptera): Effects of C, N, and P Contents and Ratios. Insects 2019, 10, 23. [Google Scholar] [CrossRef]
- Beckett, R.P.; Zavarzina, A.G.; Liers, C. Oxidoreductases and Cellulases in Lichens: Possible Roles in Lichen Biology and Soil Organic Matter Turnover. Fungal Biol. 2013, 117, 431–438. [Google Scholar] [CrossRef]
- Bates, S.T.; Cropsey, G.W.G.; Caporaso, J.G.; Knight, R.; Fierer, N. Bacterial Communities Associated with the Lichen Symbiosis. Appl. Environ. Microbiol. 2011, 77, 1309–1314. [Google Scholar] [CrossRef]
- Prado, D.E. As Caatingas Da América Do Sul. In Ecologia e Conservação da Caatinga; Leal, I.R., Tabarelli, M., Cardoso, J.M.S., Eds.; Universidade Federal de Pernambuco: Recife, Brasil, 2003; pp. 3–73. ISBN 978-8573152159. [Google Scholar]
- Caceres, M.E.S. Corticolous Crustose and Microfoliose Lichens of Northeastern Brazil; IHW Verlag: Eching, Germany, 2007; ISBN 9783930167685. [Google Scholar]
- Menezes, L.; Alvarez, T.M.; Persinoti, G.F.; Franco, J.P.; Squina, F.; Moreira, E.A.; Alvaredo Paixão, D.A.; Costa-Leonardo, A.M.; da Silva, V.X.; Clerici, M.T.P.S.; et al. Food Storage by the Savanna Termite Cornitermes cumulans (Syntermitinae): A Strategy to Improve Hemicellulose Digestibility? Microb. Ecol. 2018, 76, 492–505. [Google Scholar] [CrossRef]
- Brooks, M.E.; Kristensen, K.; Benthem, K.J.V.; Magnusson, A.; Berg, C.W.; Nielsen, A.; Skaug, H.J.; Mächler, M.; Bolker, B.M. GlmmTMB Balances Speed and Flexibility Among Packages for Zero-Inflated Generalized Linear Mixed Modeling. R J. 2017, 9, 378–400. [Google Scholar] [CrossRef]
- Lenth, R.V. Least-Squares Means: The R Package Lsmeans. J. Stat. Softw. 2016, 69, 1–33. [Google Scholar] [CrossRef]
- Hartig, F. DHARMa: Residual Diagnostics for Hierarchical (Multi-Level/Mixed) Regression Models. R Package 2022. Available online: https://cir.nii.ac.jp/crid/1370580229833186830 (accessed on 29 September 2024).
- Caporaso, J.G.; Lauber, C.L.; Walters, W.A.; Berg-Lyons, D.; Lozupone, C.A.; Turnbaugh, P.J.; Fierer, N.; Knight, R. Global Patterns of 16S RRNA Diversity at a Depth of Millions of Sequences per Sample. Proc. Natl. Acad. Sci. USA 2011, 108, 4516–4522. [Google Scholar] [CrossRef] [PubMed]
- Edgar, R.C. UPARSE: Highly Accurate OTU Sequences from Microbial Amplicon Reads. Nat. Methods 2013, 10, 996–998. [Google Scholar] [CrossRef] [PubMed]
- Edgar, R.C.; Haas, B.J.; Clemente, J.C.; Quince, C.; Knight, R. UCHIME Improves Sensitivity and Speed of Chimera Detection. Bioinformatics 2011, 27, 2194–2200. [Google Scholar] [CrossRef]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glöckner, F.O. The SILVA Ribosomal RNA Gene Database Project: Improved Data Processing and Web-Based Tools. Nucleic Acids Res. 2013, 41, D590–D596. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A Flexible Trimmer for Illumina Sequence Data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef]
- Li, D.; Liu, C.M.; Luo, R.; Sadakane, K.; Lam, T.W. MEGAHIT: An Ultra-Fast Single-Node Solution for Large and Complex Metagenomics Assembly via Succinct de Bruijn Graph. Bioinformatics 2015, 31, 1674–1676. [Google Scholar] [CrossRef] [PubMed]
- Hauser, M.; Steinegger, M.; Söding, J. MMseqs Software Suite for Fast and Deep Clustering and Searching of Large Protein Sequence Sets. Bioinformatics 2016, 32, 1323–1330. [Google Scholar] [CrossRef] [PubMed]
- Seemann, T. Prokka: Rapid Prokaryotic Genome Annotation. Bioinformatics 2014, 30, 2068–2069. [Google Scholar] [CrossRef]
- Zhang, H.; Yohe, T.; Huang, L.; Entwistle, S.; Wu, P.; Yang, Z.; Busk, P.K.; Xu, Y.; Yin, Y. DbCAN2: A Meta Server for Automated Carbohydrate-Active Enzyme Annotation. Nucleic Acids Res. 2018, 46, W95–W101. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing; R. Foundation for Statistical Computing: Vienna, Austria, 2021; Available online: http://www.R-project.org (accessed on 29 September 2024).
- Liu, C.; Cui, Y.; Li, X.; Yao, M. Microeco: An R Package for Data Mining in Microbial Community Ecology. FEMS Microbiol. Ecol. 2021, 97, fiaa255. [Google Scholar] [CrossRef]
- Xu, S.; Zhan, L.; Tang, W.; Wang, Q.; Dai, Z.; Zhou, L.; Feng, T.; Chen, M.; Wu, T.; Hu, E.; et al. MicrobiotaProcess: A Comprehensive R Package for Deep Mining Microbiome. Innovation 2023, 4, 100388. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Naumann, U.; Wright, S.T.; Warton, D.I. Mvabund- an R Package for Model-Based Analysis of Multivariate Abundance Data. Methods Ecol. Evol. 2012, 3, 471–474. [Google Scholar] [CrossRef]
- Nearing, J.T.; Douglas, G.M.; Hayes, M.G.; MacDonald, J.; Desai, D.K.; Allward, N.; Jones, C.M.A.; Wright, R.J.; Dhanani, A.S.; Comeau, A.M.; et al. Microbiome Differential Abundance Methods Produce Different Results across 38 Datasets. Nat. Commun. 2022, 13, 342. [Google Scholar] [CrossRef]
- Fernandes, A.D.; Reid, J.N.S.; Macklaim, J.M.; McMurrough, T.A.; Edgell, D.R.; Gloor, G.B. Unifying the Analysis of High-Throughput Sequencing Datasets: Characterizing RNA-Seq, 16S RRNA Gene Sequencing and Selective Growth Experiments by Compositional Data Analysis. Microbiome 2014, 2, 15. [Google Scholar] [CrossRef]
- Bradford, M.M. A Rapid and Sensitive Method for the Quantitation of Microgram Quantities of Protein Utilizing the Principle of Protein-Dye Binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Franco Cairo, J.P.L.; Leonardo, F.C.; Alvarez, T.M.; Ribeiro, D.A.; Büchli, F.; Costa-Leonardo, A.M.; Carazzolle, M.F.; Costa, F.F.; Paes Leme, A.F.; Pereira, G.A.; et al. Functional Characterization and Target Discovery of Glycoside Hydrolases from the Digestome of the Lower Termite Coptotermes gestroi. Biotechnol. Biofuels 2011, 4, 50. [Google Scholar] [CrossRef]
- Ellis, A.R.; Burchett, W.W.; Harrar, S.W.; Bathke, A.C. Nonparametric Inference for Multivariate Data: The R Package Npmv. J. Stat. Softw. 2017, 76, 1–18. [Google Scholar] [CrossRef]
- Collins, N.M. Observations on the Foraging Activity of Hospitalitermes umbrinus (Haviland), (Isoptera, Termitidae) in the Gunong-Mulu-National-Park, Sarawak. Ecol. Entomol. 1979, 4, 231–238. [Google Scholar] [CrossRef]
- Asplund, J.; Wardle, D.A. The Impact of Secondary Compounds and Functional Characteristics on Lichen Palatability and Decomposition. J. Ecol. 2013, 101, 689–700. [Google Scholar] [CrossRef]
- Pöykkö, H.; Hyvärinen, M.; Bačkor, M. Removal of Lichen Secondary Metabolites Affects Food Choice and Survival of Lichenivorous Moth Larvae. Ecology 2005, 86, 2623–2632. [Google Scholar] [CrossRef]
- de Oliveira, M.H.; Lacerda-Rolim, M.D.S.; Barbosa-Silva, A.M.; Abad, A.C.A.; Mota, R.A.; Pereira, E.C.; Martins, M.C.B.; de Lima, L.M.; Bezerra-Gusmão, M.A. Inhibitory Effect of Usnic Acid on the Gut Microbiota of the Termite Constrictotermes cyphergaster. Symbiosis 2023, 89, 329–335. [Google Scholar] [CrossRef]
- Auer, L.; Lazuka, A.; Sillam-Dussès, D.; Miambi, E.; O’Donohue, M.; Hernandez-Raquet, G. Uncovering the Potential of Termite Gut Microbiome for Lignocellulose Bioconversion in Anaerobic Batch Bioreactors. Front. Microbiol. 2017, 8, 2623. [Google Scholar] [CrossRef] [PubMed]
- Stackebrandt, E. The Family Lachnospiraceae. In The Prokaryotes; Rosenberg, E., DeLong, E.F., Lory, S., Stackebrandt, E., Thompson, F., Eds.; Springer: Berlin/Heidelberg, Germany, 2014; pp. 197–201. ISBN 978-3-642-30120-9. [Google Scholar]
- Warnecke, F.; Luginbühl, P.; Ivanova, N.; Ghassemian, M.; Richardson, T.H.; Stege, J.T.; Cayouette, M.; McHardy, A.C.; Djordjevic, G.; Aboushadi, N.; et al. Metagenomic and Functional Analysis of Hindgut Microbiota of a Wood-Feeding Higher Termite. Nature 2007, 450, 560–565. [Google Scholar] [CrossRef] [PubMed]
- Dietrich, C.; Köhler, T.; Brune, A. The Cockroach Origin of the Termite Gut Microbiota: Patterns in Bacterial Community Structure Reflect Major Evolutionary Events. Appl. Environ. Microbiol. 2014, 80, 2261–2269. [Google Scholar] [CrossRef] [PubMed]
- Shukla, S.P.; Sanders, J.G.; Byrne, M.J.; Pierce, N.E. Gut Microbiota of Dung Beetles Correspond to Dietary Specializations of Adults and Larvae. Mol. Ecol. 2016, 25, 6092–6106. [Google Scholar] [CrossRef]
- Romero Victorica, M.; Soria, M.A.; Batista-García, R.A.; Ceja-Navarro, J.A.; Vikram, S.; Ortiz, M.; Ontañon, O.; Ghio, S.; Martínez-Ávila, L.; Quintero García, O.J.; et al. Neotropical Termite Microbiomes as Sources of Novel Plant Cell Wall Degrading Enzymes. Sci. Rep. 2020, 10, 3864. [Google Scholar] [CrossRef]
- Arora, J.; Kinjo, Y.; Šobotník, J.; Buček, A.; Clitheroe, C.; Stiblik, P.; Roisin, Y.; Žifčáková, L.; Park, Y.C.; Kim, K.Y.; et al. The Functional Evolution of Termite Gut Microbiota. Microbiome 2022, 10, 78. [Google Scholar] [CrossRef]
- Almeida, M.C.P.d.S.; Silva, J.E.d.; Batista, W.G.d.S.; Alves, J.L.F.; Melo, D.M.d.A.; Pimenta, A.S.; Braga, R.M. Valorization of Wood Residues from Vegetation Suppression during Wind Energy Plant Implementation and Its Potential for Renewable Phenolic Compounds through Flash Pyrolysis: A Case Study in Northeast Brazil’s Semi-Arid Region. Forests 2024, 15, 621. [Google Scholar] [CrossRef]
- Perrot, T.; Pauly, M.; Ramírez, V. Emerging Roles of β-Glucanases in Plant Development and Adaptative Responses. Plants 2022, 11, 1119. [Google Scholar] [CrossRef]
- Lenardon, M.D.; Munro, C.A.; Gow, N.A.R. Chitin Synthesis and Fungal Pathogenesis. Curr. Opin. Microbiol. 2010, 13, 416–423. [Google Scholar] [CrossRef]
- Arakane, Y.; Muthukrishnan, S. Insect Chitinase and Chitinase-like Proteins. Cell. Mol. Life Sci. 2010, 67, 201–216. [Google Scholar] [CrossRef] [PubMed]
- Sützl, L.; Foley, G.; Gillam, E.M.J.; Bodén, M.; Haltrich, D. The GMC Superfamily of Oxidoreductases Revisited: Analysis and Evolution of Fungal GMC Oxidoreductases. Biotechnol. Biofuels 2019, 12, 118. [Google Scholar] [CrossRef]
- Ruiz-Dueñas, F.J.; Martínez, Á.T. Microbial Degradation of Lignin: How a Bulky Recalcitrant Polymer Is Efficiently Recycled in Nature and How We Can Take Advantage of This. Microb. Biotechnol. 2009, 2, 164–177. [Google Scholar] [CrossRef] [PubMed]
- Brune, A. Symbiotic Digestion of Lignocellulose in Termite Guts. Nat. Rev. Microbiol. 2014, 12, 168–180. [Google Scholar] [CrossRef] [PubMed]
- Mathieu, Y.; Piumi, F.; Valli, R.; Aramburu, J.C.; Ferreira, P.; Faulds, C.B.; Record, E. Activities of Secreted Aryl Alcohol Quinone Oxidoreductases from Pycnoporus cinnabarinus Provide Insights into Fungal Degradation of Plant Biomass. Appl. Environ. Microbiol. 2016, 82, 2411–2423. [Google Scholar] [CrossRef] [PubMed]
- Levasseur, A.; Drula, E.; Lombard, V.; Coutinho, P.M.; Henrissat, B. Expansion of the Enzymatic Repertoire of the CAZy Database to Integrate Auxiliary Redox Enzymes. Biotechnol. Biofuels 2013, 6, 41. [Google Scholar] [CrossRef]
- Daniel, G.; Volc, J.; Filonova, L.; Plíhal, O.; Kubátová, E.; Halada, P. Characteristics of Gloeophyllum trabeum Alcohol Oxidase, an Extracellular Source of H2O2 in Brown Rot Decay of Wood. Appl. Environ. Microbiol. 2007, 73, 6241–6253. [Google Scholar] [CrossRef]
- Franco Cairo, J.P.L.; Carazzolle, M.F.; Leonardo, F.C.; Mofatto, L.S.; Brenelli, L.B.; Gonçalves, T.A.; Uchima, C.A.; Domingues, R.R.; Alvarez, T.M.; Tramontina, R.; et al. Expanding the Knowledge on Lignocellulolytic and Redox Enzymes of Worker and Soldier Castes from the Lower Termite Coptotermes gestroi. Front. Microbiol. 2016, 7, 1518. [Google Scholar] [CrossRef]
- Geng, A.; Cheng, Y.; Wang, Y.; Zhu, D.; Le, Y.; Wu, J.; Xie, R.; Yuan, J.S.; Sun, J. Transcriptome Analysis of the Digestive System of a Wood-Feeding Termite (Coptotermes formosanus) Revealed a Unique Mechanism for Effective Biomass Degradation. Biotechnol. Biofuels 2018, 11, 24. [Google Scholar] [CrossRef]
- Calusinska, M.; Marynowska, M.; Bertucci, M.; Untereiner, B.; Klimek, D.; Goux, X.; Sillam-Dussès, D.; Gawron, P.; Halder, R.; Wilmes, P.; et al. Integrative Omics Analysis of the Termite Gut System Adaptation to Miscanthus Diet Identifies Lignocellulose Degradation Enzymes. Commun. Biol. 2020, 3, 275. [Google Scholar] [CrossRef]
- Tramontina, R.; Brenelli, L.B.; Sodré, V.; Franco Cairo, J.P.; Travália, B.M.; Egawa, V.Y.; Goldbeck, R.; Squina, F.M. Enzymatic Removal of Inhibitory Compounds from Lignocellulosic Hydrolysates for Biomass to Bioproducts Applications. World J. Microbiol. Biotechnol. 2020, 36, 166. [Google Scholar] [CrossRef] [PubMed]
- Kfoury, B.; Rodrigues, W.F.C.; Kim, S.J.; Brandizzi, F.; Del-Bem, L.E. Multiple Horizontal Gene Transfer Events Have Shaped Plant Glycosyl Hydrolase Diversity and Function. New Phytol. 2024, 242, 809–824. [Google Scholar] [CrossRef] [PubMed]
- Merzendorfer, H.; Zimoch, L. Chitin Metabolism in Insects: Structure, Function and Regulation of Chitin Synthases and Chitinases. J. Exp. Biol. 2003, 206, 4393–4412. [Google Scholar] [CrossRef] [PubMed]
- Hu, H.; da Costa, R.R.; Pilgaard, B.; Schiøtt, M.; Lange, L.; Poulsen, M. Fungiculture in Termites Is Associated with a Mycolytic Gut Bacterial Community. mSphere 2019, 4. [Google Scholar] [CrossRef]
- Rosengaus, R.B.; Schultheis, K.F.; Yalonetskaya, A.; Bulmer, M.S.; DuComb, W.S.; Benson, R.W.; Thottam, J.P.; Godoy-Carter, V. Symbiont-Derived β-1,3-Glucanases in a Social Insect: Mutualism beyond Nutrition. Front. Microbiol. 2014, 5, 607. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).