Abstract
Biofloc technology (BFT) systems heavily rely on microbiota to mitigate ammonia toxicity and manage essential nutrient cycling. Understanding the diversity and functional role of microbiota within BFT-applied aquaculture systems is crucial for ensuring sustainable operations. Though some studies exist on BFT microbiota, research on microbial differences in Japanese eel aquaculture is still limited, hindering the wider application of BFT systems. In this study, we analyzed the characteristics of water quality factors and microbiota in Japanese eel (Anguilla japonica) breeding water, applying the BFT system. Using a metabarcoding approach, the diversity and community structure of aquatic microbiota were compared between BFT and continuous flow (CF) systems. The pH was significantly higher in CF water, while total ammonia nitrogen (TAN) and nitrite (NO2−-N) was higher in BFT water. Alpha diversity was significantly higher in BFT compared to CF systems, and it was correlated significantly with pH and TAN. In both BFT and CF water, the phyla Proteobacteria and Bacteroidota were found to be the most abundant. In the BFT water, a diverse array of bacterial taxa, including BFT-specific clades, were consistently present, while the microbiota in CF water was more variable and contained fewer specific taxa. In addition, bacterial functions related to nitrate reduction, sulfur compound oxidation, and chitinolysis were significantly more abundant in BFT than in CF systems. These findings highlight differences in water quality and microbiota between aquaculture systems, which can inform future research on the use of BFT for sustainable fish farming.
1. Introduction
The rising global population continues to fuel the demand for aquaculture food, necessitating the expansion and intensification of aquaculture production [1,2]. To meet the seafood needs of the projected world population in 25 years, aquaculture production must increase by nearly 60% over 2018 levels [3]. Aquaculture poses significant environmental concerns, including feed waste, water pollution, disease outbreaks, carcass accumulation, and excessive use of marine pharmaceuticals [4]. Efficiently managing waste, like nitrogen and phosphorus compounds, is crucial in aquaculture. It is important to address parameters like total ammonia nitrogen (TAN), nitrite (NO2−-N) levels, and biochemical oxygen demand (BOD) to maintain water quality [5]. In response to growing concerns, there has been a rising interest in sustainable aquaculture technologies like recirculating aquaculture systems (RAS) and biofloc technology (BFT)-based farming offer promising solutions to enhance aquaculture productivity and reduce the negative impact on the environment by minimizing water exchange and waste discharge [6,7].
Bioflocs are biological aggregates of microorganisms and other kinds of organic matter such as feces and uneaten feed [8]. BFT is an aquaculture technique that effectively removes ammoniacal nitrogen, which originates from feed residues and the waste produced by aquaculture organisms [9]. This is achieved by assimilating ammonia nitrogen into bioflocs through precise control of the carbon-to-nitrogen (C:N) ratio, utilizing beneficial microorganisms present in the feed water during aquaculture, all without the need for water exchange [10]. By minimizing the exchange of feed water, BFT aquaculture systems can mitigate environmental impacts stemming from excess nutrients or pathogens [11]. The primary mechanism behind biofloc formation is the proliferation of heterotrophic bacteria on provided carbohydrates [12]. Since the bioflocs formed in the feed water can be utilized as food for aquaculture organisms, it has been reported to contribute to the growth promotion of aquaculture organisms by increasing feed efficiency and immunity against pathogens [13], and it is known to be economical compared to conventional aquaculture systems [14,15].
The various interactions of microbial communities have a direct impact on productivity and water quality factors in aquaculture systems [16,17]. Microbiota may also provide solutions to these problems, as previous studies suggest that manipulating the microbial communities associated with fish can improve water quality, as well as reduce the number of fish pathogenic bacteria and improve larval fecundity [18,19]. It has also been reported that greater BFT-associated microbial diversity improves waste mineralization processes [20], protein utilization [21], and pathogen control [22]. Identifying BFT-associated microbiota can help fine-tune BFT systems, reduce environmental degradation, and improve production of the aquaculture industry [23,24,25]. A thorough understanding of microbial diversity and their ecological functions is deemed essential for understanding the mechanism of BFT systems and the effective management of BFT systems, which enables the maintenance of consistency and predictability in aquaculture production. The establishment of next-generation sequencing (NGS) technologies has enabled the analysis of large amounts of metagenomic data due to the faster speed, lower cost, and higher data throughput of metagenomic methods compared to traditional methods [26]. While previous studies investigating microbiota using NGS technology have revealed diverse bacterial communities and their functions in the aquaculture systems for common fish (e.g., carp) and shrimp, research on Japanese eel is limited [12,27,28,29].
Japanese eels (Anguilla japonica) are a catadromous species found across Korea, Japan, Taiwan, China, and the northern Philippines [30]. It holds significant commercial importance as a high-valued fish and is a popular inland aquaculture species [31] that is widely recognized for its high-quality protein, collagen, vitamins, and other health benefits [32]. Studies have been conducted to compare the growth of Japanese eels using BFT and conventional aquaculture systems [33,34]. However, there is a lack of research analyzing the microbiota of Japanese eels by employing metabarcoding techniques on breeding water in BFT systems. In this study, we investigated the characteristics of microbiota and water quality factors in the breeding water of Japanese eels, applying the BFT system. Water quality factors in BFT systems were compared to those in a conventional continuous flow (CF) systems. Using a metabarcoding approach, the alpha diversity, community structures, and the predicted function of microbiota were analyzed from the BFT and CF water. Through this research, we offer fundamental data for eel cultivation in BFT aquaculture systems.
2. Materials and Methods
2.1. Experimental Design of BFT and CF Systems
The protocol for animal experiments was reviewed and approved by the Institutional Animal Care and Use Committee of the National Institute of Fisheries Science, Republic of Korea (No. 2024-NIFS-IACUC-23). The aquaculture experiments were conducted in a 0.5 ton (∅ 50 cm × H 50 cm) polypropylene (PP) circular tank. The Japanese eels used in the experiment were sourced from a general aquaculture store during the glass eel season and randomly selected from individuals housed at the Advanced Aquaculture Demonstration Center (Jinhae, Republic of Korea). Eels, averaging 170.5 ± 9.53 g in weight, were allocated into each tank with a total weight of 4.99 ± 0.07 kg and a density of 9.98 ± 0.15 kg/m3 per tank. The experiment was conducted after a 1-week in-tank acclimation period to ensure fish acclimation. Throughout the experiment, the fish were fed a commercial feed named “Gold-Eel” (crude protein 52%, crude fat 12%, Ca 1.0%, moisture 12%, P 2.7%, crude fiber 6.0%, and crude ash 6.0%) manufactured by Cargill Agri Purina (Seongnam, Republic of Korea). The feeding rate was 1% of the total weight per day (c.a. 50 g/day).
Two rearing conditions were tested: a BFT system and a CF system, with three triplicates for each. The CF tanks were supplied with water at a rate of 100 mL per minute to maintain a turnover rate of 53.3% per day. The BFT tanks were prepared by inoculating them with 10 ppm of BFT-specific bacteria (Bacillus amyloliquefaciens, Bacillus licheniformis, Bacillus subtilis, Cellulomonas denitrificans, Cellulomonas sp., Nitrobacter winogradskyi, Nitrosomonas europaea, Pseudomonas stutzeri, and Rhodopseudomonas palustris) in the form of lyophilized power (BFT-ST, EgeeTech, Ltd., Irvine, CA, USA). The carbon-to-nitrogen (C:N) ratio of BFT water followed the calculation method of a previous study [8,35]. To make an input C:N ratio of 15:1, 70 g of molasses was added when 50 g of feed was given every day. The feed, which contains 50% carbon and 8.06% nitrogen, alone had a C:N ratio of approximately 6.2:1. The addition of 70 g of molasses, which has 50% carbon, was calculated to balance the carbon-to-nitrogen ratio to the desired 15:1. One quarter of the water in the BFT tank was replaced all at once every week, and 1 g of fishery environmental enhancer (Atapon-aqua, Handong, Republic of Korea) was added at that time.
The water temperature was maintained at 25 ± 1 °C using a 2 kW heater (OKE-HE-100, Sewon OKE, Busan, Republic of Korea), and dissolved oxygen (DO) levels were regulated above 7 mg/L while maintaining the suspension of bioflocs by aerating each tank with two air stones. Additionally, in the BFT tank, DO was maintained by installing an additional high-pressure diffuser. Water temperature, DO, and pH levels were measured five times weekly before the morning feeding session using a multi-parameter water quality meter (YSI-650; Yellow Springs Instruments, Yellow Springs, OH, USA). Total ammonia nitrogen (TAN) and nitrite nitrogen (NO2−-N) levels were measured three times, weekly, prior to feeding and analyzed using a colorimetric method with a Spectroquant® Ammonium Test kit and Nitrite Test kit (Merck KGaA, Darmstadt, Germany), respectively, using an UV/VIS spectrophotometer Prove 600 plus (Merck KGaA, Darmstadt, Germany). Water quality data collected on the fourth week of cultivation were used for statistical analyses (e.g., t-test) in R v.4.1.2 [36].
2.2. Molecular Experiments
After the four-week breeding period, a total of 1 L of water sample was collected from each tank and individually filtered through a 0.22 µm membrane filter (Hyundai Micro, Anseong, Republic of Korea) using a Vacuum Membrane Filter Holder (AccuResearch Korea Inc., Seoul, Republic of Korea). Genomic DNA was then extracted from the membrane filter employing a DNeasy PowerSoil Pro Kit (Qiagen, Hilden, Germany), adhering to the manufacturer’s instructions. The amplification of bacterial nuclear 16S ribosomal RNA (rRNA) regions was performed using primers 515F and 806R [37,38] with Illumina adaptors. PCR amplifications were conducted using a SimpliAmp™ Thermal Cycler (Applied Biosystems, Waltham, MA, USA) and AccuPower PCR premix (Bioneer, Daejeon, Republic of Korea) with the following cycling conditions: initial denaturation at 94 °C for 5 min, followed by 30 cycles at 94 °C for 30 s, 55 °C for 30 s, 72 °C for 40 s, and a final extension at 72 °C for 10 min. Each PCR reaction comprised a total volume of 20 μL, containing 1 μL of each primer (10 pmol) and 1 μL of genomic DNA. PCR products were assessed via gel electrophoresis on a 1% agarose gel and subsequently purified using the Expin PCR Purification Kit (GeneAll Biotechnology, Seoul, Republic of Korea). To minimize stochastic PCR bias, each sample underwent triplicate amplification, and resulting amplicons were combined into a single sample. DNA sequencing was conducted utilizing an Illumina MiSeq platform (Macrogen, Seoul, Republic of Korea). All sequences generated during this study were archived in the NCBI Sequence Read Archive (SRA) under project number PRJNA1114445.
The abundance of bacterial 16S rRNA gene copies was measured using quantitative real-time polymerase chain reaction (qPCR). The PCR conditions were similar to the conventional PCR described above, with modifications for the reagent kit and instrument: Luna Universal qPCR Master Mix kit (New England Biolabs, Frankfurt, Germany) and QuantStudio 1 Real-Time PCR system (ThermoFisher Scientific, Waltham, MA, USA) at the Gyeongnam Bio and Anti-aging Core Facility Center (Changwon National University, Republic of Korea). The relative abundance of 16S rRNA gene copies was determined using the ΔCt method, calculated as log2 ΔCt = −(Ct value of the sample − mean Ct value of CF samples). All samples were analyzed in triplicate.
2.3. Bioinformatics and Statistical Analysis
The raw sequence data underwent processing using QIIME 2 [39]. Paired sequences were denoised and merged through the DADA2 pipeline [40] to filter out low-quality sequences. Amplicon sequence variants (ASVs) were then generated from denoised sequences and classified using a Naïve Bayesian classifier [41] against the Silva database v. 138 [42]. Subsequently, a phylogenetic tree of ASVs was constructed using FastTree [43] after aligning the sequences with MAFFT [44]. To mitigate technical bias arising from differences in sequence numbers, all samples were normalized to a minimum of 32,000 reads before further analysis. Alpha diversity and community analyses were performed using the pyloseq [45] and vegan [46] packages in R. Diversity indices such as Chao1 richness, Shannon’s diversity and evenness, and Faith’s phylogenetic diversity were calculated in QIIME 2 and compared using statistical tests including t-tests or ANOVA in R. Community structures were visualized through non-metric multidimensional scaling (NMDS) analysis based on weighted UniFrac distance. The significance of community differences was tested using PERMANOVA with 999 permutations, employing adonis2 in the vegan package. The influence of water quality factors on the microbiota was analyzed using the envfit function in the vegan package. Linear discriminant analysis effect size (LEfSe) analysis was performed using the microeco package [47] for detecting distinctive phylogenetic signals according to the aquaculture system. The function of bacterial communities was predicted based on the Functional Annotation of Prokaryotic Taxa (FAPROTAX) database [48] and visualized with the ggplot2 package [49].
3. Results
3.1. Water Quality Factors and Alpha Diversity of the Microbiota
Water quality factors were measured over four weeks (Figure S1), and the data from the fourth week were used for analysis. Among the water quality factors of both the BFT and CF rearing systems, significant differences were observed in pH and TAN levels (Table 1). The pH of the BFT water averaged 7.69, with TAN levels at 6.06, while in the CF system, the pH was significantly higher at 8.01, with significantly lower TAN levels at 2.94. Although it was not significant, the NO2−-N level was higher in the BFT system compared to that in the CF system. As expected, the operationally controlled water quality factors (temperature and DO) did not differ between the two systems.

Table 1.
Water quality factors of Japanese eel cultivating water in BFT and CF systems. The values of water quality factors are presented as the mean ± standard error (SE). The p-values from t-tests are provided to indicate the statistical significance of differences between the BFT and CF systems for each factor. Significant differences are indicated by * for p < 0.05 and ** for p < 0.01.
All indices of alpha diversity, calculated from the microbiota in the breeding water of the BFT system, exhibited greater diversity than in the CF water (Table 2). Diversity and evenness were significantly higher in BFT water than in CF water (p < 0.05). Further, the relative abundance of 16S rRNA gene copy numbers was about two times higher in BFT water than in CF water (p = 0.008). Among water quality factors, Shannon–Weaver’s diversity in aquaculture systems showed a significant positive correlation with TAN (r = 0.87, p = 0.025) and a significant negative correlation with pH (r = −0.82, p < 0.046) (Figure 1).

Table 2.
Alpha diversity indices of microbiota in cultivating water from BFT and CF systems with Japanese eel. The alpha diversity indices include observed ASV number (richness), Shannon–Weaver diver-sity index (diversity), Pielou’s evenness (evenness), and Faith’s phylogenetic diversity (PD). Alpha diversity and the relative abundance of 16S rRNA gene copy (ΔCt 16S rRNA) are presented as the mean ± standard error (SE). The p-values from t-tests are provided to indicate the statistical sig-nificance of differences between the BFT and CF systems for each factor. Significant differences are indicated by * for p < 0.05 and ** for p < 0.01.
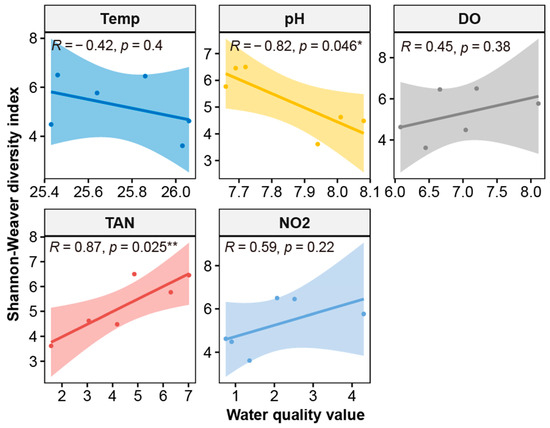
Figure 1.
Correlation of water quality factors and Shannon–Weaver diversity index of microbiota in Japanese eel cultivating water. Correlation coefficients (R values) and significance levels (p-values) are calculated using Pearson’s correlation method. Asterisks denote statistically significant correlations (*: p < 0.05 and **: p < 0.01).
3.2. Community Structure and Predicted Functions of Microbiota
For beta diversity of microbiota, NMDS plots and perMANOVA showed significant differences between BFT and CF systems (R2 = 0.446, p = 0.049) (Figure 2). Further analysis for the relationship of water quality factors and microbiota indicated that pH values significantly influenced bacterial community (r = 0.86, p = 0.03) (Figure 2). Regarding the most abundant phyla, Proteobacteria (29.2–47.2%) and Bacteroidota (21.0–47.3%) dominated both BFT and CF systems. The major phyla dominated in the BFT system were Actinobacteriota (9.9–14.7%), Patescibacteria (2.7–5.0%), and Verrucomicrobiota (4.7–6.0%), while Deinococcota (0.6–2.5%) was the major phylum in the CF water (Figure 3A). At the class level, Bacteroidia (21.0–47.2%) and Gammaproteobacteria (25.0–36.0%) were the most abundant in both BFT and CF water (Figure S2A). The major class in all samples of BFT water was Alphaproteobacteria (5.9–10.4%), Actinobacteria (9.7–14.5%), Gracilibacteria (2.0–4.8%), and Verrucomicrobiae (4.7–5.9%), while Deinococci (0.6–2.5%) was specific in all samples of CF water. At the order level, Burkholderiales (23.1–30.4%) appeared to be the most common order in both BFT and CF water (Figure S2B). Some orders (e.g., Flavobacteriales, Micrococcales, Cytophagales, Rhodobacterales, Sphingobacteriales, Absconditabacteriales, Chitinophagales, Rhizobiales, and Verrucomicrobiales) were represented in all three samples in BFT tanks, whereas in CF water, there was no specific order distributed across all samples, showing the large variation between samples. At the family level, Comamonadaceae (8.1–9.5%), Rhodobacteraceae (2.2–3.2%), unclassified Absconditabacteriales (SR1) (2.0–4.7%), and Chitinophagaceae (3.8–4.7%) were common in all BFT water (Figure 3B). In the case of CF water, there was a large variation between samples, except for Alcaligenaceae (4.4–11.4%). The heatmap for the major bacterial genera showed a high abundance of Flavobacterium (3.2–16.2%) in both BFT and CF water (Figure 4). Polynucleobacter (19.3–19.5%), unclassified Absconditabacteriales (SR1) (2.0–4.7%), Luteolibacter (2.3–3.7%), and Pedobacter (2.6–3.1%) were common among the three samples of BFT water (Figure 4). In CF water, Deinococcus (0.6–2.5%), Flavobacterium (3.2–16.1%), Flectobacillus (0.8–18.9%), and Undibacterium (0.8–11.6%) were abundant in all three samples, though they showed high variation between samples. LEfSe analysis was used to identify a phylogenetic clade showing specific occurrence patterns according to the aquaculture systems: Different farming systems resulted in differences in phylogenetic signals between the aquaculture systems (Figure 5). Two distinctive clades (Gammaproteobacteria and Verrucomicrobiae) were identified in the BFT system, while the Deinococci clade was specifically found in the CF water. Polynucleobacter (Burkholderiaceae, Burkholderiales) and Luteolibacter (Rubritaleaceae, Verrucomicrobiales) were characteristic of the BFT water. In contrast, Deinococcus (Deinococcaceae, Deinococcales) was characteristic in CF water. Additionally, the number of specific phylogenetic clades was higher in BFT compared to the CF system.
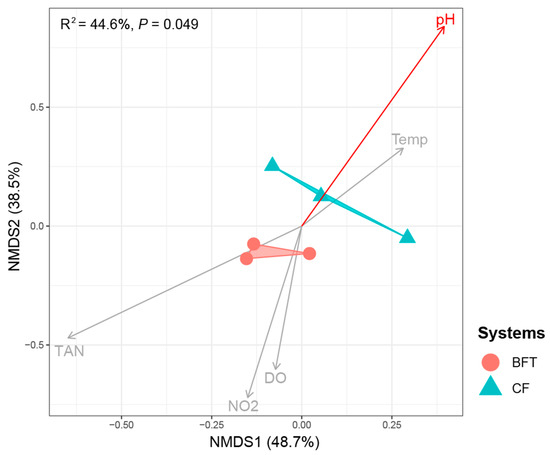
Figure 2.
Non-metric multi-dimensional scaling (NMDS) plot for the microbiota of Japanese eel cultivating water based on weighted UniFrac distance. The significance of the difference in the bacterial communities is compared between BFT and CF systems using the Adonis test, and the explanatory power (R2) and statistical significance levels (p-values) are indicated on the plot. The length and color of the arrows represent the strength and significance of the correlation between each water quality factor and the ordination space, respectively. Longer arrows indicate stronger correlations, and the red color indicates statistical significance (p < 0.05).
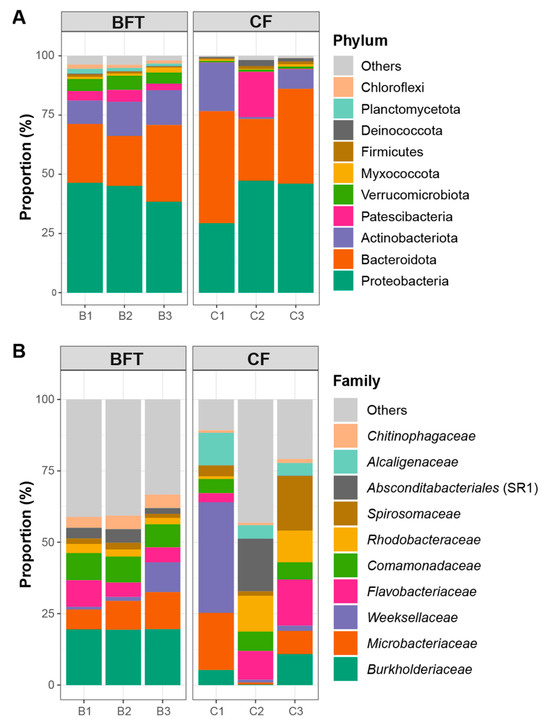
Figure 3.
Major taxonomic composition (top 10) of the microbiota in Japanese eel breeding water for (A) phylum and (B) family levels.
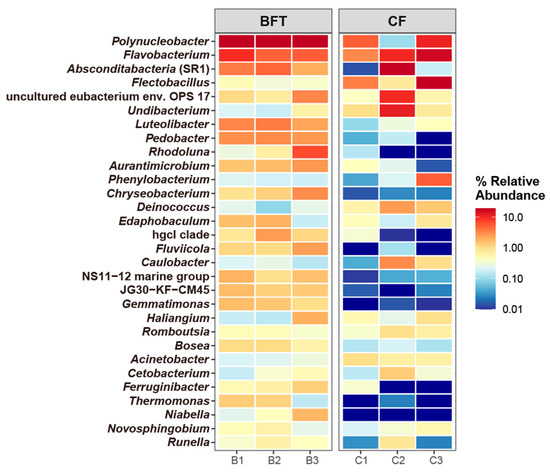
Figure 4.
Heatmap of major genera (top 30) of the microbiota in Japanese eel breeding water.
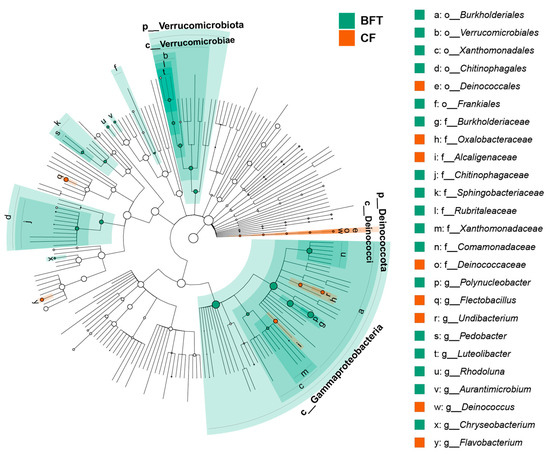
Figure 5.
Cladogram based on LEfSe analysis showing the distinctive taxa according to aquaculture systems. The color of the node indicates differentially abundant taxa between BFT and CF systems.
When predicting the function of microbiota based on the FARPROTAX database, bacterial functions related to nitrate reduction, sulfur compound oxidation, and chitinolysis were significantly more abundant in the BFT system compared to the CF system (p < 0.05) (Figure 6). Additionally, bacteria with chemoheterotrophy and ureolysis functions were more prevalent in BFT water, although not statistically different. Conversely, bacterial function associated with aromatic compound degradation was significantly more abundant in the CF system (p < 0.05).
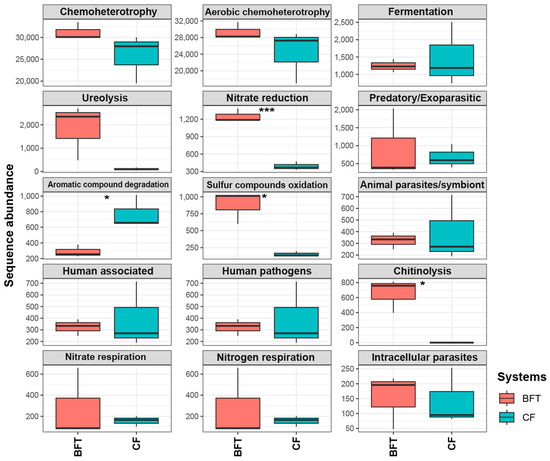
Figure 6.
Predicted functions (top 15) of the microbiota in Japanese eel cultivating water. Asterisks denote statistically significant differences from t-tests (*: p < 0.05 and ***: p < 0.001).
4. Discussions
In this study, we analyzed the characteristics of water quality and microbiota during eel cultivation in BFT and CF aquaculture. BFT systems apply the bacteria to recycle carbon and nitrogen from external carbon sources and fish feces [10]. The maintenance of BFT systems depends mainly on water quality factors, especially DO, TAN, NO2−-N, alkalinity, and pH [50,51]. In this study, the pH level was observed to be lower in BFT systems compared to CF systems, consistent with findings from earlier research. Previous studies have indicated that the decreases in pH in BFT systems are due to reduced dissolved inorganic carbon [19] and the nitrification process [52]. However, the small difference in pH observed in this study (average difference: 0.32) may not adequately reflect the full impact of these processes on the overall system. In the BFT system, TAN levels were significantly higher than those in the CF water. Although not statistically significant, NO2−-N also exhibited higher values compared to CF. Previous studies comparing BFT to a conventional CF system with similar turnover rates to this experiments showed lower TAN and NO2−-N levels in the CF water [19]. Because ammonia did not accumulate due to the exchange of feed water in the CF system, the lower levels of TAN and NO2−-N in CF water are an expected result. TAN is the sum of the concentration of un-ionized ammonia (NH3) and ionized ammonia (NH4+), while the level of un-ionized ammonia, which varies depending on pH and temperature, is critical to aquaculture [53]. High concentrations of NH3-N are toxic to fish, slowing growth, causing physiological imbalances, and leading to death [54,55]. We observed a slightly elevated TAN level (6.06 mg/L) in our BFT system. However, the NH3-N levels, approximately 0.18 mg/L (3% of TAN under these conditions) [56], remained within the acceptable range for freshwater aquaculture (<0.2 mg/L) [57]. It is important to note that this system was operated with limited water exchange, a condition that may have contributed to elevated TAN and NO2-N levels. Despite this, caution is warranted when interpreting our results, as the elevated levels of TAN and NO2-N suggest that our BFT system may not have been fully established during the experimental period. Further studies are necessary to confirm this possibility and to determine whether longer acclimation periods or adjustments to water exchange rates could facilitate the development of a fully functional biofloc, potentially yielding lower nitrogen concentrations.
Alpha diversity and relative abundance of the 16S rRNA gene were significantly higher in BFT water compared to CF water, indicating a greater diversity of bacterial groups in BFT systems, likely due to a stable and nutrient-rich aquaculture environment compared to the CF system [58]. Furthermore, the alpha diversity exhibited a positive correlation with TAN levels and a negative correlation with pH. These findings agree with previous research indicating a significant positive correlation between inorganic nitrogen (e.g., NH4+-N and NO2−-N) and bacterial diversity in shrimp-crab polyculture aquaculture systems [59]. Elevated levels of TAN can disrupt the balance of nitrifying microbial populations, thereby impacting alpha diversity [51,60]. pH also plays a crucial role in nitrification and microbial community structure, potentially influencing changes in bacterial diversity, and it can decrease due to biofloc formation processes and nitrification [50].
Using a metabarcoding analysis, we found that diverse bacterial taxa exist in the Japanese eel aquaculture system. The dominant phyla observed in both BFT and CF systems were Proteobacteria (Gammaproteobacteria and Alphaproteobacteria) and Bacteroidota (Bacteroidia). Proteobacteria have consistently been identified as a dominant phylum in BFT water and in various aquatic environments, including natural and aquaculture environments [19,61]. As they are known for their significant role in nutrient cycling and organic compound mineralization, Proteobacteria contribute substantially to ecosystem functioning [62]. Proteobacteria are generally recognized for their active involvement in nitrogen conversion processes such as nitrification, with chemically autotrophic microorganisms classified into the classes Betaproteobacteria and Gammaproteobacteria [63,64]. For example, Nitrosococcus, known as nitrifying bacteria, belongs to the class Gammaproteobacteria [65]. In addition, Bacteroidota are specialized in degrading complex organic matter in the biosphere, particularly polysaccharides and hydrocarbons [62].
The notable phyla in BFT system include Actinobacteriota and Verrucomicrobiota. Previous studies have also reported an enrichment of Actinobacteriota in aquaculture environments employing the BFT system [61], while Verrucomicrobiota distribution was found to be higher in BFT water compared to RAS or CF (50% water exchange/day) systems [19]. Verrucomicrobiota are heterotrophic bacteria that use monosaccharides and polysaccharides for growth and are commonly found in water and soil [66]. They are widely distributed in various environments, including drinking water, freshwater lakes, and marine sediments [61]. Actinobacteriota is a major bacterial phylum commonly found in aquaculture water as well as in the gut of farmed organisms [67]. At the genus level, several genera were notably abundant in BFT water, including Polynucleobacter, unclassified Absconditabacteriales (SR1), Luteolibacter, and Pedobacter. Among these, Polynucleobacter is known for producing adhesive polymers that facilitate the formation of microbial biofilms [68], playing a crucial role in biofloc formation in BFT systems. Additionally, Luteolibacter has been frequently detected in the fish gut [69] and is more abundant in larger fish compared to smaller individuals [70], suggesting its potential for promoting growth. In the healthy gut of Atlantic salmon (Salmo salar), Pedobacter was more abundant compared to unhealthy fish [71]. Therefore, it seems that the predominant bacteria in BFT water contribute to the formation of bioflocs, essential for fostering a favorable environment for fish farming or to support fish health.
In CF water, most phyla, except for Proteobacteria and Bacteroidota, were highly variable between samples; however, in contrast to BFT, Deinococcota was distinctly present. Deinococcota are known to be highly resistant to environmental stress and play an important role in denitrification processes [72,73]. A distinctive pattern of difference between BFT and CF systems is evident at the order level. In the BFT system, all three samples exhibited similar proportions of orders such as Flavobacteriales, Verrucomicrobiales, Cytophagales, Rhodobacterales, and Micrococcales, while substantial differences between samples were observed in the CF system. At the genus level, Deinococcus, Flavobacterium, Flectobacillus, and Undibacterium were distinctive taxa in CF water. Some Flectobacillus species are known as fish pathogens, causing flectobacillosis [74]. Additionally, a study on microbial diversity in rainbow trout (Oncorhynchus mykiss) aquaculture impacted by red mark syndrome (RMS) revealed that Flavobacterium and Undibacterium were more abundant in RMS-affected water, even though Flavobacterium is present in both RMS-free and RMS-affected conditions [75]. Therefore, the distinctive taxa in CF water may suggest a higher susceptibility to disease.
When considering the more abundant functions in BFT systems, it becomes apparent that there are significantly more bacteria with the functions for nitrate reduction, sulfur compound oxidation, and chitinolysis. In BFT aquaculture systems, there are two main pathways for ammonia removal: chemoautotrophic bacterial nitrification (CBN) and heterotrophic bacterial assimilation (HBA) [76]. Heterotrophic bacteria form ammonium ions through dissimilatory nitrate reduction to ammonium (DNRA), and the resulting ammonia is removed as protein in the form of biomass [63]. The ability to perform DNRA is widespread across both bacterial and archaeal domains, with certain bacteria in the phylum Proteobacteria known to exhibit this capability. DNRA involves the reduction of NO3− and NO2− alongside the oxidation of organic compounds or hydrogen sulfide [77,78]. Xanthomonadaceae was identified as characteristic in BFT breeding water, and this taxon contains many chitinolytic and biocontrol agent bacteria such as Lysobacter [79,80]. The higher abundance of aromatic compound degradation in CF water may be associated with the nutrient availability in water and the reproductive strategy of bacteria. In the BFT system, the higher input of carbon causes a change in the bacterial composition from autotrophic to heterotrophic by adding high carbon for the rapid growth of heterotrophic bacteria [12]. Thus, the nutrient environment in BFT water is more nutrient-rich, which is favored by r-strategy bacteria, while the CF water is more oligotrophic, which is favored by K-strategy bacteria [81,82]. Among bacteria, K-strategists often showed higher performance in the degradation of aromatic hydrocarbons [83], likely due to adaptation to low nutrient availability in surrounding environments.
In conclusion, this study showed how the microbial structure and diversity in stock water varied between BFT and CF systems when culturing Japanese eels. Microbial diversity was high in the BFT system, and specific phyla were present in a constant proportion in the BFT water. In addition, some bacterial functions possibly associated with the improvement of water quality were significantly prevalent in BFT compared to the CF system. These findings underscore the potential of BFT systems for water purification and enhancing aquaculture productivity. Moreover, reducing antibiotic use in aquaculture could further support the stability and effectiveness of such systems.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/d16100601/s1, Figure S1: Water quality factors over 4 weeks in Japanese eel aquaculture; Figure S2: Major taxonomic composition (top 10) of microbiota in Japanese eel cultivating water for (A) class and (B) order level.
Author Contributions
H.C.: Investigation, Writing—Original Draft Preparation, Writing—Review and Editing; J.S.P.: Conceptualization, Investigation, Project Administration; J.-A.H.: Investigation, Project Administration; S.-K.K.: Investigation; Y.C.: Investigation; S.-Y.O.: Formal Analysis, Supervision, Writing—Original Draft Preparation, Writing—Review and Editing. All authors have read and agreed to the published version of the manuscript.
Funding
This study was supported by the National Institute of Fisheries Science, Ministry of Oceans and Fisheries, Korea (grant number: R2024024) and Korea Basic Science Institute (National Research Facilities and Equipment Center) grant funded by the Ministry of Education (Grant No. 2023R1A6C101B022).
Institutional Review Board Statement
The animal study protocol was approved by the Institutional Animal Care and Use Committee of the National Institute of Fisheries Science, South Korea (No. 2024-NIFS-IACUC-23).
Data Availability Statement
The original data presented in the study are openly available in NCBI Sequence Read Archive (SRA) at PRJNA1114445.
Conflicts of Interest
The authors declare that there are no conflicts of interest.
References
- Davidson, K.; Pan, M.; Hu, W.; Poerwanto, D. Consumers’ willingness to pay for aquaculture fish products vs. wild-caught seafood-A case study in Hawaii. Aquac. Econ. Manag. 2012, 16, 136–154. [Google Scholar] [CrossRef]
- Tacon, A.G.J.; Metian, M. Feed matters: Satisfying the feed demand of aquaculture. Rev. Fish. Sci. Aquac. 2015, 23, 1–10. [Google Scholar] [CrossRef]
- Boyd, C.E.; McNevin, A.A.; Davis, R.P. The contribution of fisheries and aquaculture to the global protein supply. Food Secur. 2022, 14, 805–827. [Google Scholar] [CrossRef] [PubMed]
- Buhmann, A.K.; Waller, U.; Wecker, B.; Papenbrock, J. Optimization of culturing conditions and selection of species for the use of halophytes as biofilter for nutrient-rich saline water. Agric. Water Manag. 2015, 149, 102–114. [Google Scholar] [CrossRef]
- John, E.M.; Krishnapriya, K.; Sankar, T.V. Treatment of ammonia and nitrite in aquaculture wastewater by an assembled bacterial consortium. Aquaculture 2020, 526, 735390. [Google Scholar] [CrossRef]
- Laktuka, K.; Kalnbalkite, A.; Sniega, L.; Logins, K.; Lauka, D. Towards the sustainable intensification of aquaculture: Exploring possible ways forward. Sustainability 2023, 15, 16952. [Google Scholar] [CrossRef]
- Zhang, R.; Chen, T.; Wang, Y.; Short, M. Systems approaches for sustainable fisheries: A comprehensive review and future perspectives. Sustain. Prod. Consum. 2023, 41, 242–252. [Google Scholar] [CrossRef]
- Hargreaves, J.A. Biofloc production systems for aquaculture. In Biofloc Technology: A Practical Guide Book, 3rd ed.; Avnimelech, Y., De Schryver, P., Emerenciano, M., Kuhn, D., Ray, A., Taw, N., Eds.; World Aquaculture Society: Baton Rouge, LA, USA, 2013; pp. 1–11. [Google Scholar]
- Crab, R.; Defoirdt, T.; Bossier, P.; Verstraete, W. Biofloc technology in aquaculture: Beneficial effects and future challenges. Aquaculture 2012, 356–357, 351–356. [Google Scholar] [CrossRef]
- Avnimelech, Y. Carbon/nitrogen ratio as a control element in aquaculture systems. Aquaculture 1999, 176, 227–235. [Google Scholar] [CrossRef]
- Emerenciano, M.; Ballester, E.L.C.; Cavalli, R.O.; Wasielesky, W. Biofloc technology application as a food source in a limited water exchange nursery system for pink shrimp Farfantepenaeus brasiliensis (Latreille, 1817). Aquac. Res. 2012, 43, 447–457. [Google Scholar] [CrossRef]
- Liu, H.; Li, H.; Wei, H.; Zhu, X.; Han, D.; Jin, J.; Xie, S. Biofloc formation improves water quality and fish yield in a freshwater pond aquaculture system. Aquaculture 2019, 506, 256–269. [Google Scholar] [CrossRef]
- Van Rijn, J. Waste treatment in recirculating aquaculture systems. Aquac. Eng. 2013, 53, 49–56. [Google Scholar] [CrossRef]
- De Schryver, P.; Crab, R.; Defoirdt, T.; Boon, N.; Verstraete, W. The basics of bio-flocs technology: The added value for aquaculture. Aquaculture 2008, 277, 125–137. [Google Scholar] [CrossRef]
- Kim, S.-K.; Pang, Z.; Seo, H.-C.; Cho, Y.-R.; Samocha, T.; Jang, I.-K. Effect of bioflocs on growth and immune activity of Pacific white shrimp, Litopenaeus vannamei postlarvae. Aquac. Res. 2014, 45, 362–371. [Google Scholar] [CrossRef]
- Moriarty, D.J.W. The role of microorganisms in aquaculture ponds. Aquaculture 1997, 151, 333–349. [Google Scholar] [CrossRef]
- López, A.L.; Zaballos, M. Diversidad y actividad procariótica en ecosistemas marinos. Ecosistemas 2005, 14, 30–40. [Google Scholar]
- Bentzon-Tilia, M.; Sonnenschein, E.C.; Gram, L. Monitoring and managing microbes in aquaculture–Towards a sustainable industry. Microb. Biotechnol. 2016, 9, 576–584. [Google Scholar] [CrossRef]
- Tabarrok, M.; Seyfabadi, J.; Salehi Jouzani, G.; Younesi, H. Comparison between recirculating aquaculture and biofloc systems for rearing juvenile common carp (Cyprinus carpio): Growth performance, haemato-immunological indices, water quality and microbial communities. Aquac. Res. 2020, 51, 4881–4892. [Google Scholar] [CrossRef]
- Martínez-Córdova, L.R.; Emerenciano, M.; Miranda-Baeza, A.; Martínez-Porchas, M. Microbial-based systems for aquaculture of fish and shrimp: An updated review. Rev. Aquac. 2015, 7, 131–148. [Google Scholar] [CrossRef]
- Avnimelech, Y. Feeding with microbial flocs by tilapia in minimal discharge bio-flocs technology ponds. Aquaculture 2007, 264, 140–147. [Google Scholar] [CrossRef]
- Emerenciano, M.; Gaxiola, G.; Cuzon, G.; Emerenciano, M.; Gaxiola, G.; Cuzon, G. Biofloc Technology (BFT): A Review for Aquaculture Application and Animal Food Industry, Biomass Now-Cultivation and Utilization; IntechOpen: London, UK, 2013. [Google Scholar]
- Emerenciano, M.G.; Martínez-Córdova, L.R.; Martínez-Porchas, M.; Miranda-Baeza, A. Biofloc technology (BFT): A tool for water quality management in aquaculture. In Water Quality; IntechOpen: London, UK, 2017; pp. 92–109. [Google Scholar]
- Kumar, V.; Roy, S.; Behera, B.K.; Swain, H.S.; Das, B.K. Biofloc microbiome with bioremediation and health benefits. Front. Microbiol. 2021, 12, 741164. [Google Scholar] [CrossRef] [PubMed]
- Abakari, G.; Wu, X.; He, X.; Fan, L.; Luo, G. Bacteria in biofloc technology aquaculture systems: Roles and mediating factors. Rev. Aquac. 2022, 14, 1260–1284. [Google Scholar] [CrossRef]
- Rashid, M.; Stingl, U. Contemporary molecular tools in microbial ecology and their application to advancing biotechnology. Biotechnol. Adv. 2015, 33, 1755–1773. [Google Scholar] [CrossRef] [PubMed]
- Tepaamorndech, S.; Nookaew, I.; Higdon, S.M.; Santiyanont, P.; Phromson, M.; Chantarasakha, K.; Visessanguan, W. Metagenomics in bioflocs and their effects on gut microbiome and immune responses in Pacific white shrimp. Fish Shellfish. Immunol. 2020, 106, 733–741. [Google Scholar] [CrossRef] [PubMed]
- Yun, H.-S.; Kim, D.-H.; Kim, J.-G.; Kim, Y.-S.; Yoon, H.-S. The microbial communities (bacteria, algae, zooplankton, and fungi) improved biofloc technology including the nitrogen-related material cycle in Litopenaeus vannamei farms. Front. Bioeng. Biotechnol. 2022, 10, 883522. [Google Scholar] [CrossRef] [PubMed]
- Hwang, J.-A.; Park, J.S.; Jeong, H.S.; Kim, H.; Oh, S.-Y. Productivity of fish and crop growth and characteristics of bacterial communities in the FLOCponics system. Fishes 2023, 8, 422. [Google Scholar] [CrossRef]
- Kuroki, M.; Aoyama, J.; Miller, M.J.; Yoshinaga, T.; Shinoda, A.; Hagihara, S.; Tsukamoto, K. Sympatric spawning of Anguilla marmorata and Anguilla japonica in the western North Pacific Ocean. J. Fish Biol. 2009, 74, 1853–1865. [Google Scholar] [CrossRef]
- Hamidoghli, A.; Bae, J.; Won, S.; Lee, S.; Kim, D.-J.; Bai, S.C. A review on Japanese eel (Anguilla japonica) aquaculture, with special emphasis on nutrition. Rev. Fish. Sci. Aquac. 2019, 27, 226–241. [Google Scholar] [CrossRef]
- Seo, J.-S.; Choi, J.-H.; Seo, J.-H.; Ahn, T.-H.; Chong, W.-S.; Kim, S.-H.; Ahn, J.C. Comparison of major nutrients in eels Anguilla japonica cultured with different formula feeds or at different farms. Fish. Aquat. Sci. 2013, 16, 85–92. [Google Scholar] [CrossRef]
- Hwang, J.-A.; Lee, J.-H.; Park, J.S.; Choe, J.R.; Lee, D.; Kim, H. Effect on eel Anguilla japonica and crop growth by the development of a biofloc technology (BFT) aquaponic system. Korean J. Fish. Aquat. Sci. 2021, 54, 418–425. [Google Scholar]
- Jeong, H.S.; Park, J.S.; Hwang, J.-A. A comparison study on the growth performance of eel Anguilla japnica and far eastern catfish Silurus asotus and caipira Lactuca sativa in biofloc technology and flocponics systems. J. Fish. Mar. Sci. Educ. 2024, 36, 236–244. [Google Scholar]
- Azim, M.E.; Little, D.C.; Bron, J.E. Microbial protein production in activated suspension tanks manipulating C:N ratio in feed and the implications for fish culture. Bioresour. Technol. 2008, 99, 3590–3599. [Google Scholar] [CrossRef] [PubMed]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2021. [Google Scholar]
- Apprill, A.; McNally, S.; Parsons, R.; Weber, L. Minor revision to V4 region SSU rRNA 806R gene primer greatly increases detection of SAR11 bacterioplankton. Aquat. Microb. Ecol. 2015, 75, 129–137. [Google Scholar] [CrossRef]
- Parada, A.E.; Needham, D.M.; Fuhrman, J.A. Every base matters: Assessing small subunit rRNA primers for marine microbiomes with mock communities, time series and global field samples. Environ. Microbiol. 2016, 18, 1403–1414. [Google Scholar] [CrossRef]
- Bolyen, E.; Rideout, J.R.; Dillon, M.R.; Bokulich, N.A.; Abnet, C.C.; Al-Ghalith, G.A.; Caporaso, J.G. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 2019, 37, 852–857. [Google Scholar] [CrossRef]
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.A.; Holmes, S.P. DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef]
- Bokulich, N.A.; Kaehler, B.D.; Rideout, J.R.; Dillon, M.; Bolyen, E.; Knight, R.; Gregory Caporaso, J. Optimizing taxonomic classification of marker-gene amplicon sequences with QIIME 2′s q2-feature-classifier plugin. Microbiome 2018, 6, 90. [Google Scholar] [CrossRef]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Glöckner, F.O. The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 2013, 41, D590–D596. [Google Scholar] [CrossRef]
- Price, M.N.; Dehal, P.S.; Arkin, A.P. FastTree: Computing Large Minimum Evolution Trees with Profiles instead of a Distance Matrix. Mol. Biol. Evol. 2009, 26, 1641–1650. [Google Scholar] [CrossRef]
- Katoh, K.; Standley, D.M. MAFFT Multiple Sequence Alignment Software Version 7: Improvements in Performance and Usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef]
- McMurdie P Holmes, S. phyloseq: An R Package for Reproducible Interactive Analysis and Graphics of Microbiome Census Data. PLoS ONE 2013, 8, e61217. [Google Scholar] [CrossRef] [PubMed]
- Oksanen, J.; Blanchet, F.G.; Friendly, M.; Kindt, R.; Legendre, P.; McGlinn, D.; Oksanen, M.J. Vegan: Community Ecology Package. Community Ecol. Package 2020, 2, 1–263. [Google Scholar]
- Liu, C.; Cui, Y.; Li, X.; Yao, M. microeco: An R package for data mining in microbial community ecology. FEMS Microbiol. Ecol. 2021, 97, fiaa255. [Google Scholar] [CrossRef] [PubMed]
- Louca, S.; Parfrey, L.W.; Doebeli, M. Decoupling function and taxonomy in the global ocean microbiome. Science 2016, 353, 1272–1277. [Google Scholar] [CrossRef]
- Wickham, H. Package ‘ggplot2’: Elegant Graphics for Data Analysis; Springer: New York, NY, USA, 2016. [Google Scholar]
- Zhang, K.; Pan, L.; Chen, W.; Wang, C. Effect of using sodium bicarbonate to adjust the pH to different levels on water quality, the growth and the immune response of shrimp Litopenaeus vannamei reared in zero-water exchange biofloc-based culture tanks. Aquac. Res. 2017, 48, 1194–1208. [Google Scholar] [CrossRef]
- Gullian Klanian, M.; Delgadillo Díaz, M.; Sánchez Solís, M.J.; Aranda, J.; Moreno Moral, P. Effect of the content of microbial proteins and the poly-β-hydroxybutyric acid in biofloc on the performance and health of Nile tilapia (Oreochromis niloticus) fingerlings fed on a protein-restricted diet. Aquaculture 2020, 519, 734872. [Google Scholar] [CrossRef]
- Boyd, C.E. General relationship between water quality and aquaculture performance in ponds. In Fish Diseases; Jeney, G., Ed.; Academic Press: New York, NY, USA, 2017; pp. 147–166. [Google Scholar]
- Edwards, T.M.; Puglis, H.J.; Kent, D.B.; Durán, J.L.; Bradshaw, L.M.; Farag, A.M. Ammonia and aquatic ecosystems-A review of global sources, biogeochemical cycling, and effects on fish. Sci. Total Environ. 2024, 907, 167911. [Google Scholar] [CrossRef]
- Zhang, W.; Xia, S.; Zhu, J.; Miao, L.; Ren, M.; Lin, Y.; Sun, S. Growth performance, physiological response and histology changes of juvenile blunt snout bream, Megalobrama amblycephala exposed to chronic ammonia. Aquaculture 2019, 506, 424–436. [Google Scholar] [CrossRef]
- Liu, M.-J.; Guo, H.-Y.; Zhu, K.-C.; Liu, B.-S.; Liu, B.; Guo, L.; Zhang, D.C. Effects of acute ammonia exposure and recovery on the antioxidant response and expression of genes in the Nrf2-Keap1 signaling pathway in the juvenile golden pompano (Trachinotus ovatus). Aquat. Toxicol. 2021, 240, 105969. [Google Scholar] [CrossRef]
- Emerson, K.; Russo, R.C.; Lund, R.E.; Thurston, R.V. Aqueous ammonia equilibrium calculations: Effect of ph and temperature. J. Fish. Res. Board Can. 1975, 32, 2379–2383. [Google Scholar] [CrossRef]
- Bhatnagar, A.; Singh, G. Culture fisheries in village ponds: A multi-location study in Haryana, India. Agric. Biol. J. N. Am. 2010, 1, 961–968. [Google Scholar] [CrossRef]
- Abakari, G.; Luo, G.; Kombat, E.O.; Alhassan, E.H. Supplemental carbon sources applied in biofloc technology aquaculture systems: Types, effects and future research. Rev. Aquac. 2021, 13, 1193–1222. [Google Scholar] [CrossRef]
- Sun, F.; Wang, C.; Yang, H. Physicochemical factors drive bacterial communities in an aquaculture environment. Front. Environ. Sci. 2021, 9, 709541. [Google Scholar] [CrossRef]
- Jiang, W.; Tian, X.; Li, L.; Dong, S.; Zhao, K.; Li, H.; Yong, C.Y. Temporal bacterial community succession during the start-up process of biofilters in a cold-freshwater recirculating aquaculture system. Bioresour. Technol. 2019, 287, 121441. [Google Scholar] [CrossRef]
- Chen, X.; He, Z.; Zhao, J.; Liao, M.; Xue, Y.; Zhou, J.; Sun, C. Metagenomic analysis of bacterial communities and antibiotic resistance genes in penaeus monodon biofloc-based aquaculture environments. Front. Mar. Sci. 2022, 8, 762345. [Google Scholar] [CrossRef]
- Yang, Y.; Xia, J.; Liu, Y.; Dong, J.; Xu, N.; Yang, Q.; Ai, X. Safety evaluation for the use of Bacillus amyloliquefaciens in freshwater fish cultures. Aquac. Rep. 2021, 21, 100822. [Google Scholar] [CrossRef]
- Abakari, G.; Luo, G.; Kombat, E.O. Dynamics of nitrogenous compounds and their control in biofloc technology (BFT) systems: A review. Aquac. Fish. 2021, 6, 441–447. [Google Scholar] [CrossRef]
- McCusker, S.; Warberg, M.B.; Davies, S.J.; Valente, C.d.S.; Johnson, M.P.; Cooney, R.; Wan, A.H. Biofloc technology as part of a sustainable aquaculture system: A review on the status and innovations for its expansion. Aquac. Fish Fish. 2023, 3, 331–352. [Google Scholar] [CrossRef]
- Flores-Valenzuela, E.; Miranda-Baeza, A.; Rivas-Vega, M.E.; Miranda-Arizmendi, V.; Beltrán-Ramírez, O.; Emerenciano, M.G.C. Water quality and productive response of Litopenaeus vannamei reared in biofloc with addition of commercial strains of nitrifying bacteria and Lactobacillus rhamnosus. Aquaculture 2021, 542, 736869. [Google Scholar] [CrossRef]
- Gou, J.; Hong, C.U.; Deng, M.; Chen, J.; Hou, J.; Li, D.; He, X. Effect of carbon to nitrogen ratio on water quality and community structure evolution in suspended growth bioreactors through biofloc technology. Water 2019, 11, 1640. [Google Scholar] [CrossRef]
- Abakari, G.; Luo, G.; Shao, L.; Abdullateef, Y.; Cobbina, S.J. Effects of biochar on microbial community in bioflocs and gut of Oreochromis niloticus reared in a biofloc system. Aquac. Int. 2021, 29, 1295–1315. [Google Scholar] [CrossRef]
- Cai, W.; Li, Y.; Niu, L.; Zhang, W.; Wang, C.; Wang, P.; Meng, F. New insights into the spatial variability of biofilm communities and potentially negative bacterial groups in hydraulic concrete structures. Water Res. 2017, 123, 495–504. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Xie, Y.; Zhang, G.; Pan, L. Microbial community structure and diversity in fish-flower (mint) symbiosis. AMB Express 2023, 13, 46. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Z.; Zhao, H.; Zhang, L.; Huang, Z.; Ke, H.; Liu, Y.; Li, Q. Integrated analysis of how gender and body weight affect the intestinal microbial diversity of Gymnocypris chilianensis. Sci. Rep. 2023, 13, 8811. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Sun, G.; Li, S.; Li, X.; Liu, Y. Intestinal microbiota of healthy and unhealthy Atlantic salmon Salmo salar L. in a recirculating aquaculture system. J. Oceanol. Limnol. 2018, 36, 414–426. [Google Scholar] [CrossRef]
- Griffiths, E.; Gupta, R.S. Identification of signature proteins that are distinctive of the Deinococcus-Thermus phylum. Int. Microbiol. 2007, 10, 201–208. [Google Scholar]
- Qu, J.; Yang, H.; Liu, Y.; Qi, H.; Wang, Y.; Zhang, Q. The study of natural biofilm formation and microbial community structure for recirculating aquaculture system. IOP Conf. Ser. Earth Environ. Sci. 2021, 742, 012018. [Google Scholar] [CrossRef]
- Adikesavalu, H.; Patra, A.; Banerjee, S.; Sarkar, A.; Abraham, T.J. Phenotypic and molecular characterization and pathology of Flectobacillus roseus causing flectobacillosis in captive held carp Labeo rohita (Ham.) fingerlings. Aquaculture 2015, 439, 60–65. [Google Scholar] [CrossRef]
- Bruno, A.; Cafiso, A.; Sandionigi, A.; Galimberti, A.; Magnani, D.; Manfrin, A.; Bazzocchi, C. Red mark syndrome: Is the aquaculture water microbiome a keystone for understanding the disease aetiology? Front. Microbiol. 2023, 14, 1059127. [Google Scholar] [CrossRef]
- Luo, G.; Xu, J.; Meng, H. Nitrate accumulation in biofloc aquaculture systems. Aquaculture 2020, 520, 734675. [Google Scholar] [CrossRef]
- Herrmann, M.; Taubert, M. Biogeochemical cycling of carbon and nitrogen in groundwater—Key processes and microbial drivers. In Encyclopedia of Inland Waters, 2nd ed.; Mehner, T., Tockner, K., Eds.; Elsevier: Oxford, UK, 2022; pp. 412–427. [Google Scholar]
- Voss, M.; Choisnard, N.; Bartoli, M.; Bonaglia, S.; Bourbonnais, A.; Frey, C.; Weston, K. Coastal nitrogen cycling–biogeochemical processes and the impacts of human activities and climate change. In Treatise on Estuarine and Coastal Science, 2nd ed.; Baird, D., Elliott, M., Eds.; Academic Press: Oxford, UK, 2024; pp. 225–250. [Google Scholar]
- Saraihom, S.; Kobayashi, D.Y.; Lotrakul Prasongsuk, S.; Eveleigh, D.E.; Punnapayak, H. First report of a tropical Lysobacter enzymogenes producing bifunctional endoglucanase activity towards carboxymethylcellulose and chitosan. Ann. Microbiol. 2016, 66, 907–919. [Google Scholar] [CrossRef]
- Drenker, C.; El Mazouar, D.; Bücker, G.; Weißhaupt, S.; Wienke, E.; Koch, E.; Linkies, A. Characterization of a disease-suppressive isolate of Lysobacter enzymogenes with broad antagonistic activity against bacterial, oomycetal and fungal pathogens in different crops. Plants 2023, 12, 682. [Google Scholar] [CrossRef] [PubMed]
- Schramm, A.; De Beer, D.; Gieseke, A.; Amann, R. Microenvironments and distribution of nitrifying bacteria in a membrane-bound biofilm. Environ. Microbiol. 2000, 2, 680–686. [Google Scholar] [CrossRef]
- Kim, D.-J.; Kim, S.-H. Effect of nitrite concentration on the distribution and competition of nitrite-oxidizing bacteria in nitratation reactor systems and their kinetic characteristics. Water Res. 2006, 40, 887–894. [Google Scholar] [CrossRef] [PubMed]
- Brzeszcz, J.; Steliga, T.; Kapusta, P.; Turkiewicz, A.; Kaszycki, P. r-strategist versus K-strategist for the application in bioremediation of hydrocarbon-contaminated soils. Int. Biodeterior. Biodegrad. 2016, 106, 41–52. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).