An Update of Knowledge of the Bacterial Assemblages Associated with the Mexican Caribbean Corals Acropora palmata, Orbicella faveolata, and Porites porites
Abstract
1. Introduction
2. Materials and Methods
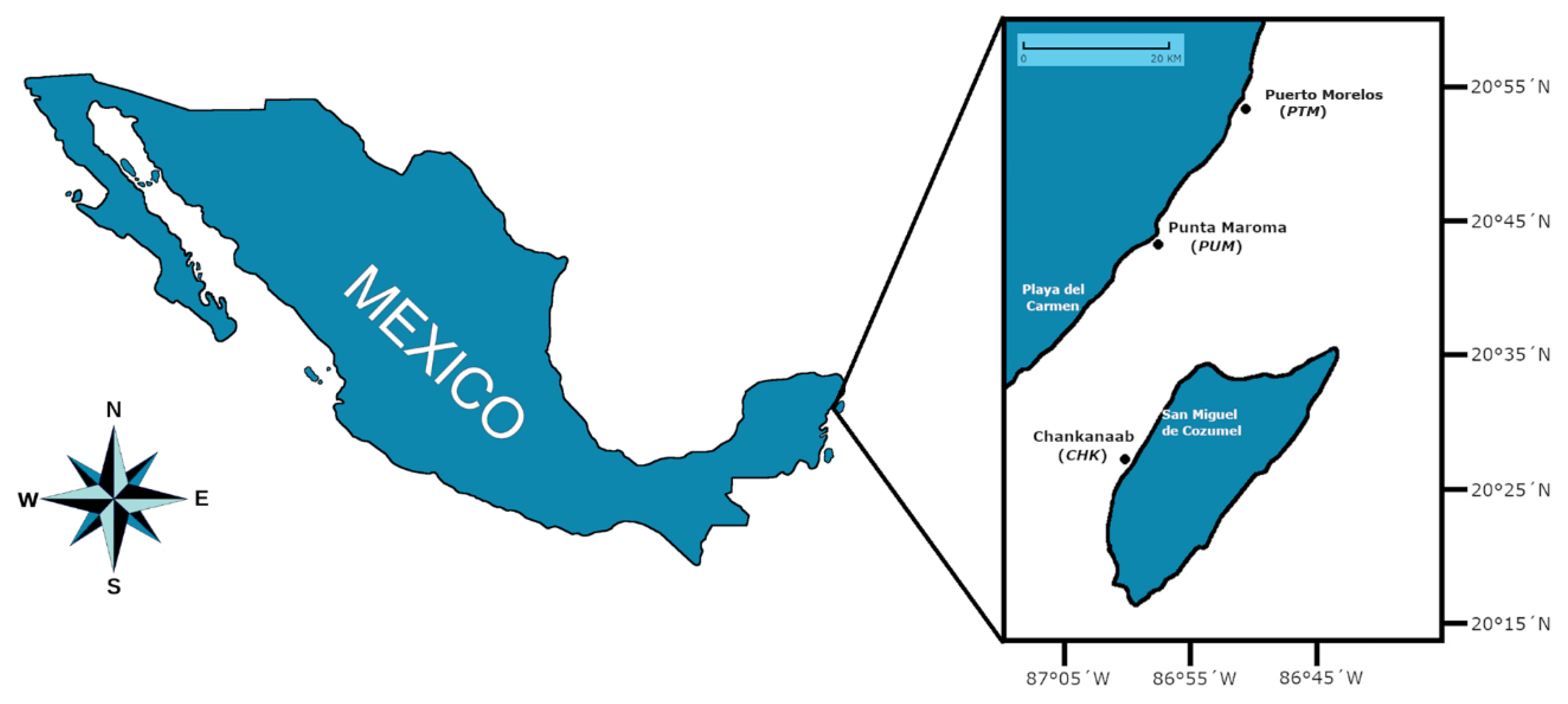
2.1. Area of Study and Fieldwork
2.2. Sample Collection
2.3. Sample Pretreatment and DNA Extraction
2.4. PCR Amplification of the 16S rRNA Gene
2.5. Sequencing of the 16S rRNA Gene
2.6. Sequence Analysis and Taxonomic Identification Using the SILVA Database
2.7. Data Analysis
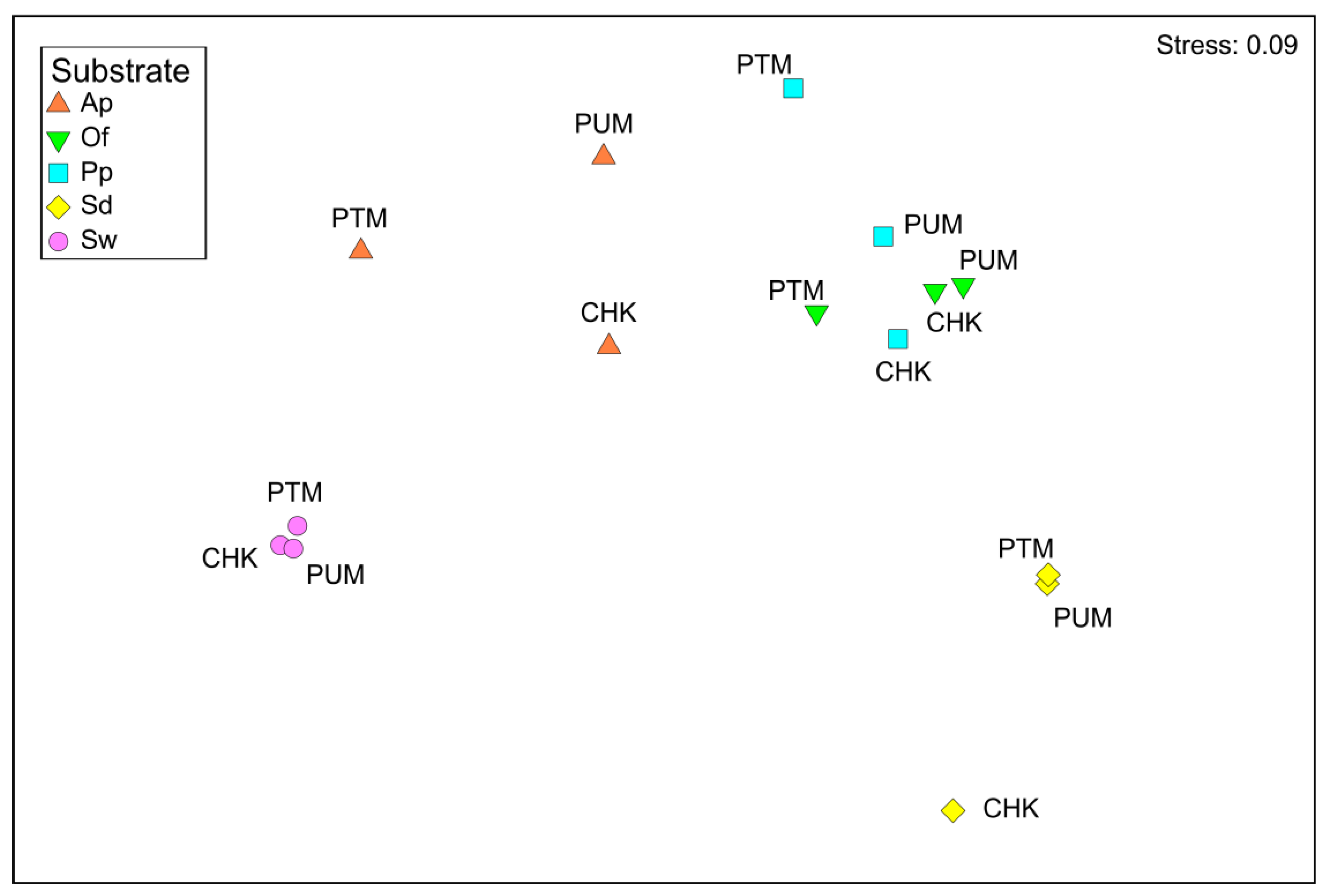
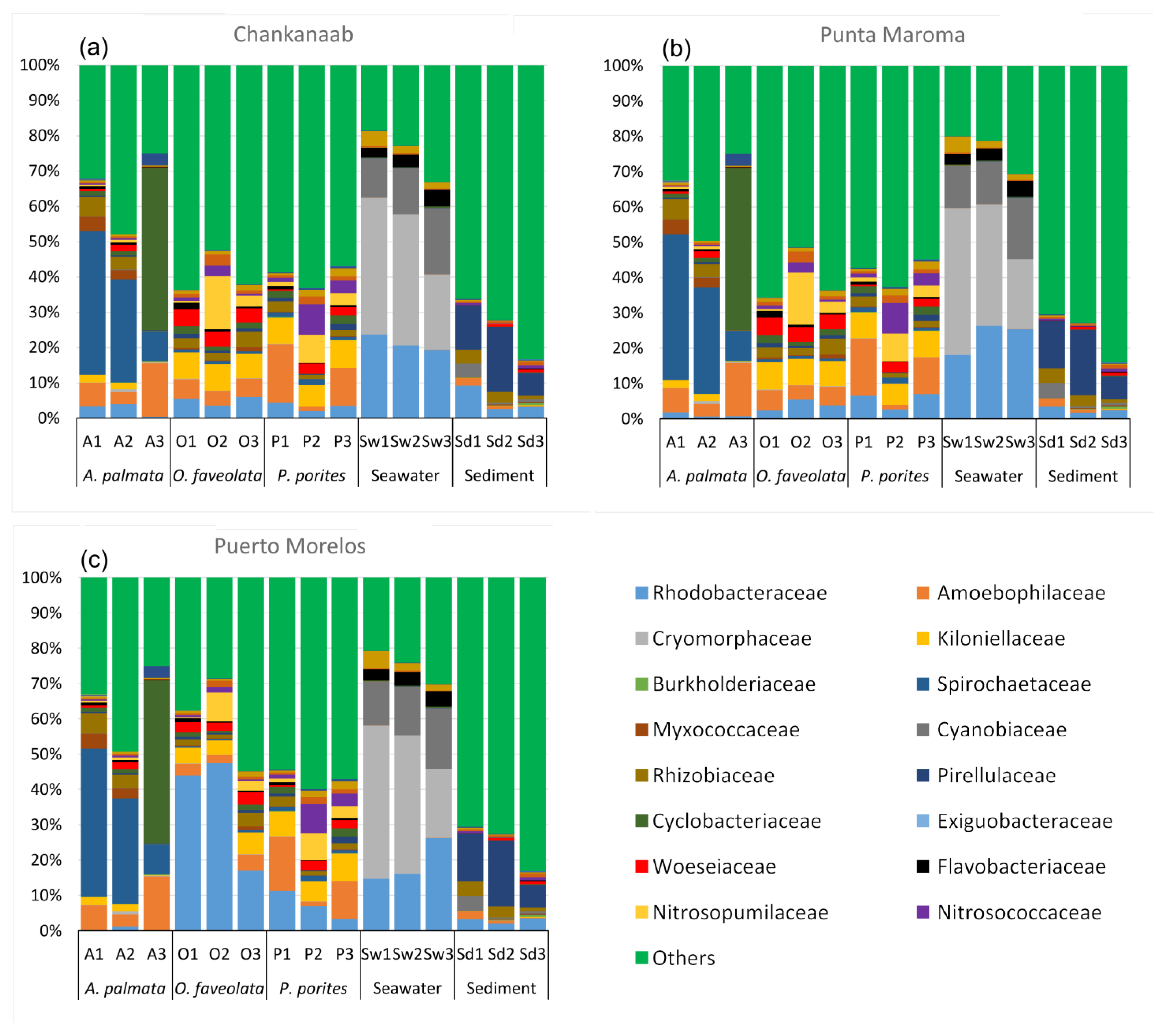
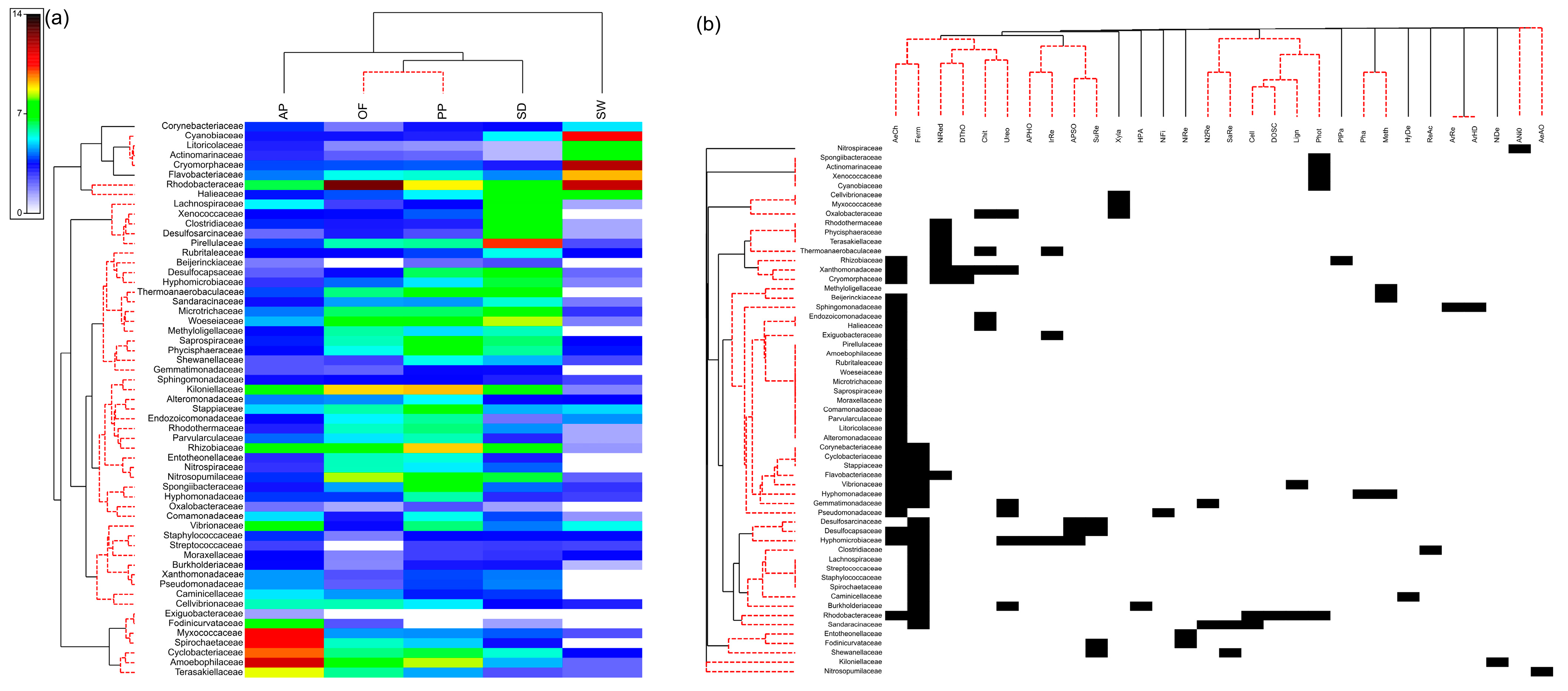
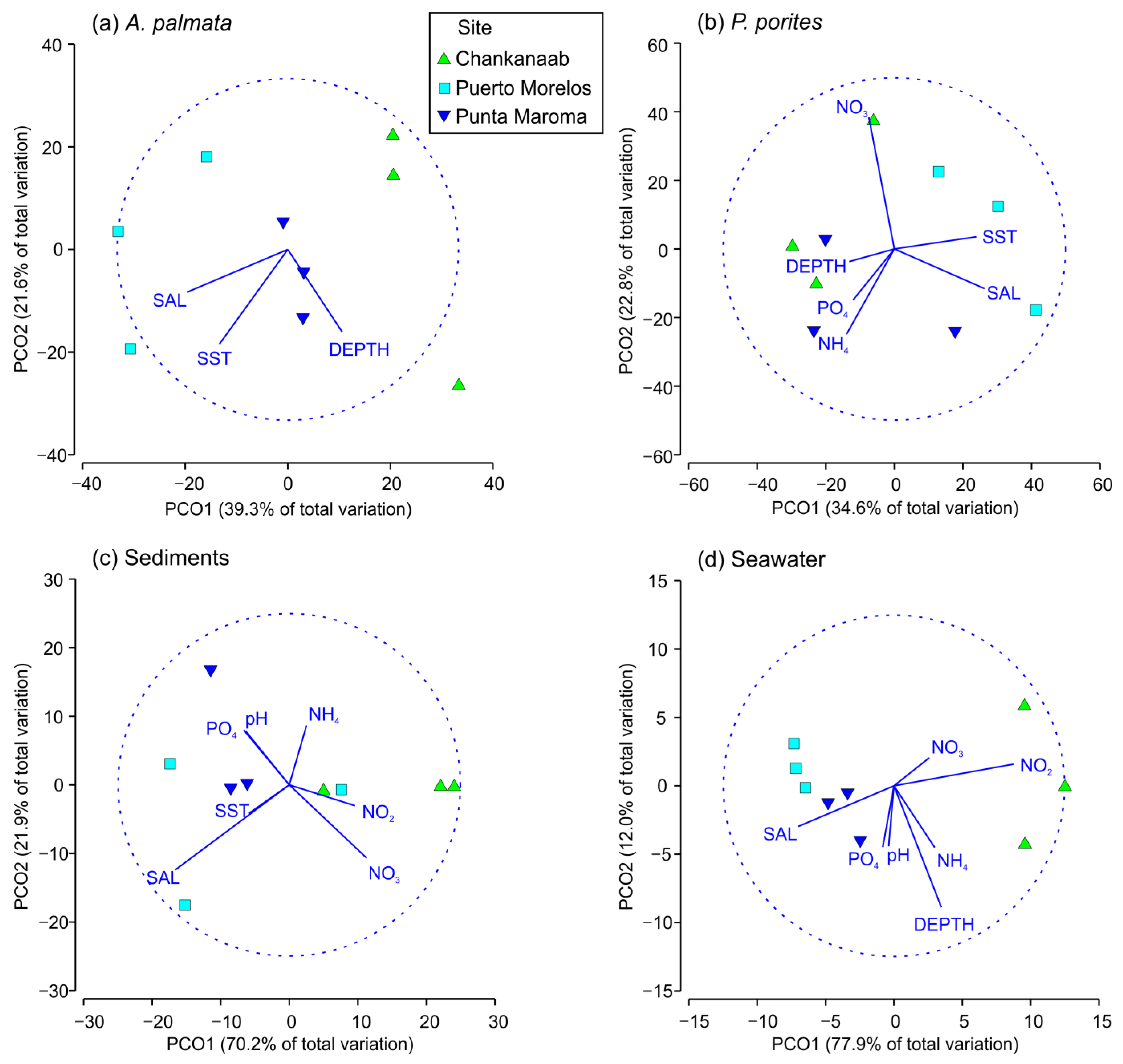
3. Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Jackson, J.B.; Kirby, M.X.; Berger, W.H.; Bjorndal, K.A.; Botsford, L.W.; Bourque, B.J.; Bradbury, R.H.; Cooke, R.; Erlandson, J.; Estes, J.A.; et al. Historical overfishing and the recent collapse of coastal ecosystems. Science 2001, 293, 629–637. [Google Scholar] [CrossRef] [PubMed]
- Woodhead, A.J.; Hicks, C.C.; Norström, A.V.; Williams, G.J.; Graham, N.A.J. Coral reef ecosystem services in the Anthropocene. Funct. Ecol. 2019, 33, 1023–1034. [Google Scholar] [CrossRef]
- Carpenter, K.E.; Abrar, M.; Aeby, G.; Aronson, R.B.; Banks, S.; Bruckner, A.; Chiriboga, A.; Cortés, J.; Delbeek, J.C.; Devantier, L.; et al. One-third of reef-building corals face elevated extinction risk from climate change and local impacts. Science 2008, 321, 560–563. [Google Scholar] [CrossRef]
- Rohwer, F.; Breitbart, M.; Jara, J.; Azam, F.; Knowlton, N. Diversity of bacteria associated with the Caribbean coral Montastraea franksi. Coral. Reefs 2001, 20, 85–91. [Google Scholar]
- Ziegler, M.; Grupstra, C.G.; Barreto, M.M.; Eaton, M.; BaOmar, J.; Zubier, K.; Al-Sofyani, A.; Turki, A.J.; Ormond, R. Coral bacterial community structure responds to environmental change in a host-specific manner. Nat. Commun. 2019, 10, 3092. [Google Scholar] [CrossRef]
- Bourne, D.G.; Webster, N.S. Coral reef bacterial communities. In The Prokaryotes, 4th ed.; Rosenberg, E., DeLong, E.F., Lory, S., Stackebrandt, E., Thompson, F., Eds.; Springer: Berlin/Heidelberg, Germany, 2013; pp. 163–187. [Google Scholar]
- Vanwonterghem, I.; Webster, N.S. Coral reef microorganisms in a changing climate. iScience 2020, 23, 100972. [Google Scholar] [CrossRef]
- Li, J.; Chen, Q.; Long, L.J.; Dong, J.D.; Yang, J.; Zhang, S. Bacterial dynamics within the mucus, tissue and skeleton of the coral Porites lutea during different seasons. Sci. Rep. 2014, 4, 7320. [Google Scholar] [CrossRef]
- Bergman, J.L.; Shaw, T.; Egan, S.; Ainsworth, T.D. Assessing the coral microbiome at the scale of tissue-specific habitats within the coral meta-organism. Front. Mar. Sci. 2022, 9, 985496. [Google Scholar] [CrossRef]
- van Oppen, M.; Medina, M. Coral evolutionary responses to microbial symbioses. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2020, 375, 20190591. [Google Scholar] [CrossRef]
- Peixoto, R.S.; Rosado, P.M.; Leite, D.C.; Rosado, A.S.; Bourne, D.G. Beneficial Microorganisms for Corals (BMC): Proposed Mechanisms for Coral Health and Resilience. Front. Microbiol. 2017, 8, 341. [Google Scholar] [CrossRef] [PubMed]
- Bourne, D.G.; Morrow, K.M.; Webster, N.S. Insights into the coral microbiome: Underpinning the health and resilience of reef ecosystems. Annu. Rev. Microbiol. 2016, 70, 317–340. [Google Scholar] [CrossRef]
- Zhang, Y.; Yang, Q.; Ling, J.; Long, L.; Huang, H.; Yin, J.; Wu, M.; Tang, X.; Lin, X.; Zhang, Y.; et al. Shifting the microbiome of a coral holobiont and improving host physiology by inoculation with a potentially beneficial bacterial consortium. BMC Microbiol. 2021, 21, 130. [Google Scholar]
- Carlos, C.; Torres, T.T.; Ottoboni, L. Bacterial communities and species-specific associations with the mucus of Brazilian coral species. Sci. Rep. 2013, 3, 1624. [Google Scholar] [CrossRef] [PubMed]
- Hernández-Zulueta, J.; Araya, R.; Vargas-Ponce, O.; Díaz-Pérez, L.; Rodríguez-Troncoso, A.P.; Ceh, J.; Ríos-Jara, E.; Rodríguez-Zaragoza, F.A. First deep screening of bacterial assemblages associated with corals of the Tropical Eastern Pacific. FEMS Microbiol. Ecol. 2016, 92, fiw196. [Google Scholar] [CrossRef] [PubMed]
- Morrow, K.M.; Muller, E.; Lesser, M.P. How Does the Coral Microbiome Cause, Respond to, or Modulate the Bleaching Process? In Coral Bleaching. Ecological Studies, 1st ed.; van Oppen, M., Lough, J., Eds.; Springer: Berlin/Heidelberg, Germany, 2018; Volume 233, pp. 153–188. [Google Scholar]
- van Oppen, M.J.H.; Blackall, L.L. Coral microbiome dynamics, functions and design in a changing world. Nat. Rev. Microbiol. 2019, 17, 557–567. [Google Scholar] [CrossRef]
- Voolstra, C.R.; Quigley, K.M.; Davies, S.W.; Parkinson, J.E.; Peixoto, R.S.; Aranda, M.; Baker, A.C.; Barno, A.R.; Barshis, D.J.; Benzoni, F.; et al. Consensus Guidelines for Advancing Coral Holobiont Genome and Specimen Voucher Deposition. Front. Mar. Sci. 2021, 8, 701784. [Google Scholar] [CrossRef]
- Ainsworth, T.D.; Krause, L.; Bridge, T.; Torda, G.; Raina, J.B.; Zakrzewski, M.; Gates, R.D.; Padilla-Gamiño, J.L.; Spalding, H.L.; Smith, C.; et al. The coral core microbiome identifies rare bacterial taxa as ubiquitous endosymbionts. ISME J. 2015, 9, 2261–2274. [Google Scholar] [CrossRef]
- Hernandez-Agreda, A.; Gates, R.D.; Ainsworth, T.D. Defining the core microbiome in corals’ microbial soup. Trends Microbiol. 2017, 25, 125–140. [Google Scholar] [CrossRef] [PubMed]
- Hernandez-Agreda, A.; Leggat, W.; Bongaerts, P.; Herrera, C.; Ainsworth, T.D. Rethinking the coral microbiome: Simplicity exists within a diverse microbial biosphere. mBio 2018, 9, e00812-18. [Google Scholar] [CrossRef] [PubMed]
- Ostria-Hernández, M.; Hernández-Zulueta, J.; Vargas-Ponce, O.; Díaz-Pérez, L.; Araya, R.; Rodríguez-Troncoso, A.P.; Ríos-Jara, E.; Rodríguez-Zaragoza, F.A. Core microbiome of corals Pocillopora damicornis and Pocillopora verrucosa in the northeastern tropical Pacific. Mar. Ecol. 2022, 43, e12729. [Google Scholar] [CrossRef]
- Hernández-Zulueta, J.; Díaz-Pérez, L.; García-Maldonado, J.Q.; Nava-Martínez, G.G.; García-Salgado, M.A.; Rodríguez-Zaragoza, F.A. Bacterial assemblages associated with Acropora palmata affected by white band disease in the Mexican region of the Caribbean and Gulf of Mexico. J. Sea Res. 2022, 185, 102230. [Google Scholar] [CrossRef]
- McKew, B.A.; Dumbrell, A.J.; Daud, S.D.; Hepburn, L.; Thorpe, E.; Mogensen, L.; Whitby, C. Characterization of geographically distinct bacterial communities associated with coral mucus produced by Acropora spp. and Porites spp. Appl. Environ. Microbiol. 2012, 78, 5229–5237. [Google Scholar] [CrossRef] [PubMed]
- Closek, C.; Sunagawa, S.; DeSalvo, M.K.; Piceno, Y.M.; DeSantis, T.Z.; Brodie, E.L.; Weber, M.X.; Voolstra, C.R.; Andersen, G.L.; Medina, M. Coral transcriptome and bacterial community profiles reveal distinct Yellow Band Disease states in Orbicella faveolata. ISME J. 2014, 8, 2411–2422. [Google Scholar] [CrossRef] [PubMed]
- Barranco, L.M.; Carriquiry, J.D.; Rodríguez-Zaragoza, F.A.; Cupul-Magaña, A.L.; Villaescusa, J.A.; Calderón -Aguilera, L.E. Spatiotemporal variations of live coral cover in the northern Mesoamerican Reef System, Yucatan Peninsula, Mexico. Sci. Mar. 2016, 80, 143–150. [Google Scholar]
- Loreto-Viruel, R.M.; García-Beltrán, G.; Bezaury-Creel, J. Caracterización de los Arrecifes Coralinos de Isla Cozumel, Quintana Roo, México. Amigos De Sian Ka’an Ser. Doc. 2017, 7, 11–40. [Google Scholar]
- Rioja-Nieto, R.; Álvarez-Filip, L. Coral reef systems of the Mexican Caribbean: Status, recent trends and conservation. Mar. Pollut. Bull. 2018, 140, 616–625. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez-Zaragoza, F.A.; Arias-González, J.E. Coral biodiversity and bio-construction in the northern sector of the mesoamerican reef system. Front. Mar. Sci. 2015, 2, 13. [Google Scholar] [CrossRef]
- Caballero-Aragón, H.; Perera-Valderrama, S.; Cerdeira-Estrada, S.; Martell-Dubois, R.; Rosique-de la Cruz, L.; Álvarez-Filip, L.; Pérez-Cervantes, E.; Estrada-Saldívar, N.; Ressl, R. Puerto Morelos Coral Reefs, Their Current State and Classification by a Scoring System. Diversity 2020, 12, 272. [Google Scholar] [CrossRef]
- Blanchon, P.; Richards, S.; Bernal, J.P.; Cerdeira-Estrada, S.; Ibarra, M.S.; Corona-Martínez, L.; Martell-Dubois, R. Retrograde Accretion of a Caribbean Fringing Reef Controlled by Hurricanes and Sea-level Rise. Front. Earth Sci. 2017, 5, 78. [Google Scholar] [CrossRef]
- Bourne, D.G.; Munn, C.B. Diversity of bacteria associated with the coral Pocillopora damicornis from the Great Barrier Reef. Environ. Microbiol. 2005, 7, 1162–1174. [Google Scholar] [CrossRef]
- Caporaso, J.; Lauber, C.; Walters, W.; Berg-Lyons, D.; Huntley, J.; Fierer, N.; Owens, S.; Betley, J.; Fraser, L.; Bauer, M.; et al. Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. ISME J. 2012, 6, 1621–1624. [Google Scholar] [CrossRef]
- Bolyen, E.; Rideout, J.R.; Dillon, M.R.; Bokulich, N.A.; Abnet, C.; Al-Ghalith, G.A.; Alexander, H.; Alm, E.J.; Arumugam, M.; Asnicar, F.; et al. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 2019, 37, 852–857. [Google Scholar] [CrossRef]
- Callahan, B.J.; Mcmurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.; Holmes, S.P. DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef]
- Bokulich, N.A.; Kaehler, B.D.; Rideout, J.R.; Dillon, M.; Bolyen, E.; Knight, R.; Huttley, G.A.; Gregory Caporaso, J. Optimizing taxonomic classification of marker-gene amplicon sequences with QIIME 2′s q2-feature-classifier plugin. Microbiome 2018, 6, 90. [Google Scholar] [CrossRef] [PubMed]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glöckner, F.O. The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 2013, 41, D590–D596. [Google Scholar] [CrossRef] [PubMed]
- Anderson, M.J.; Gorley, R.N.; Clarke, K.R. Permanova for Primer Guide to Software and Statistical Methods; PRIMER-E: Plymouth, UK, 2008; p. 214. [Google Scholar]
- Clarke, K.R.; Gorley, R.N. Primer v7: User Manual/Tutorial; Primer-E Ltd.: Plymouth, UK, 2015; p. 192. [Google Scholar]
- Louca, S.; Parfrey, L.W.; Doebeli, M. Decoupling function and taxonomy in the global ocean microbiome. Science 2016, 353, 1272–1277. [Google Scholar] [CrossRef]
- Sweet, M.J.; Croquer, A.; Bythell, J.C. Bacterial assemblages differ between compartments within the coral holobiont. Coral Reefs 2011, 30, 39–52. [Google Scholar] [CrossRef]
- Wild, C.; Huettel, M.; Klueter, A.; Kremb, S.G.; Rasheed, M.Y.; Jørgensen, B.B. Coral mucus functions as an energy carrier and particle trap in the reef ecosystem. Nature 2004, 428, 66–70. [Google Scholar] [CrossRef]
- Beltrán, Y.; Cerqueda-García, D.; Taş, N.; Thomé, P.E.; Iglesias-Prieto, R.; Falcón, L.I. Microbial composition of biofilms associated with lithifying rubble of Acropora palmata branches. FEMS Microbiol. Ecol. 2016, 92, fiv162. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Sánchez-Quinto, A.; Falcón, L.I. Metagenome of Acropora palmata coral rubble: Potential metabolic pathways and diversity in the reef ecosystem. PLoS ONE 2019, 14, e0220117. [Google Scholar] [CrossRef]
- de Voogd, N.J.; Cleary, D.F.; Polónia, A.R.; Gomes, N.C. Bacterial community composition and predicted functional ecology of sponges, sediment and seawater from the thousand islands reef complex, West Java, Indonesia. FEMS Microbiol. Ecol. 2015, 91, fiv019. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.Y.; Ling, J.; Yang, Q.S.; Wang, Y.S.; Sun, C.C.; Sun, H.Y.; Feng, J.B.; Jiang, Y.F.; Zhang, Y.Z.; Wu, M.L.; et al. The diversity of coral associated bacteria and the environmental factors affect their community variation. Ecotoxicology 2015, 24, 1467–1477. [Google Scholar] [CrossRef] [PubMed]
- Cleary, D.F.R.; Polónia, A.R.M.; Becking, L.E.; de Voogd, N.J.; Purwanto; Gomes, H.; Gomes, N.C.M. Compositional analysis of bacterial communities in seawater, sediment, and sponges in the Misool coral reef system, Indones. Mar. Biodivers. 2018, 48, 1889–1901. [Google Scholar] [CrossRef]
- Huggett, M.J.; Apprill, A. Coral microbiome database: Integration of sequences reveals high diversity and relatedness of coral-associated microbes. Environ. Microbiol. Rep. 2019, 11, 372–385. [Google Scholar] [CrossRef] [PubMed]
- Luo, D.; Wang, X.; Feng, X.; Tian, M.; Wang, S.; Tang, S.L.; Ang, P., Jr.; Yan, A.; Luo, H. Population differentiation of Rhodobacteraceae along with coral compartments. ISME J. 2021, 15, 3286–3302. [Google Scholar] [CrossRef]
- Roder, C.; Arif, C.; Daniels, C.; Weil, E.; Voolstra, C.R. Bacterial profiling of White Plague Disease across corals and oceans indicates a conserved and distinct disease microbiome. Mol. Ecol. 2014, 23, 965–974. [Google Scholar] [CrossRef]
- Eaton, K.R.; Landsberg, J.H.; Kiryu, Y.; Peters, E.C.; Muller, E.M. Measuring Stony Coral Tissue Loss Disease induction and lesion progression within two intermediately susceptible species, Montastraea cavernosa and Orbicella faveolata. Front. Mar. Sci. 2021, 8, 717265. [Google Scholar] [CrossRef]
- Alvarez-Filip, L.; González-Barrios, F.J.; Pérez-Cervantes, E.; Molina-Hernández, A.; Estrada-Saldívar, N. Stony coral tissue loss disease decimated Caribbean coral populations and reshaped reef functionality. Commun. Biol. 2022, 5, 440. [Google Scholar] [CrossRef]
- Pootakham, W.; Mhuantong, W.; Yoocha, T.; Putchim, L.; Jomchai, N.; Sonthirod, C.; Naktang, C.; Kongkachana, W.; Tangphatsornruang, S. Heat-induced shift in coral microbiome reveals several members of the Rhodobacteraceae family as indicator species for thermal stress in Porites lutea. Microbiologyopen 2019, 8, e935. [Google Scholar] [CrossRef]
- Alves, L.M.; Marcondes de Souza, J.A.; Varani, A.M.; de Macedo Lemos, E.G. The Family Rhizobiaceae. In The Prokaryotes: Alphaproteobacteria and Betaproteobacteria, 4th ed.; Rosenberg, E., DeLong, E.F., Lory, S., Stackebrandt, E., Thompson, F., Eds.; Springer: Berlin/Heidelberg, Germany, 2014; pp. 419–437. [Google Scholar]
- Rosales, S.M.; Huebner, L.K.; Clark, A.S.; McMinds, R.; Ruzicka, R.R.; Muller, E.M. Bacterial Metabolic Potential and Micro-Eukaryotes Enriched in Stony Coral Tissue Loss Disease Lesions. Front. Mar. Sci. 2022, 8, 776859. [Google Scholar] [CrossRef]
- Frade, P.R.; Glasl, B.; Matthews, S.A.; Mellin, C.; Serrão, E.A.; Wolfe, K.; Mumby, P.J.; Webster, N.S.; Bourne, D.G. Spatial patterns of microbial communities across surface waters of the Great Barrier Reef. Commun. Biol. 2020, 3, 442. [Google Scholar] [CrossRef] [PubMed]
- Glasl, B.; Robbins, S.; Frade, P.R.; Marangon, E.; Laffy, P.W.; Bourne, D.G.; Webster, N.S. Comparative genome-centric analysis reveals seasonal variation in the function of coral reef microbiomes. ISME J. 2020, 14, 1435–1450. [Google Scholar] [CrossRef] [PubMed]
- Apprill, A.; Weber, L.; Santoro, A. Distinguishing between microbial habitats unravels ecological complexity in coral microbiomes. mSystems 2016, 1, e00143-16. [Google Scholar] [CrossRef]
- Albataineh, H.; Stevens, D.C. Marine Myxobacteria: A few good halophiles. Mar. Drugs 2018, 16, 209. [Google Scholar] [CrossRef]
- Baker, B.J.; Lazar, C.S.; Teske, A.P.; Dick, G.J. Genomic resolution of linkages in carbon, nitrogen, and sulfur cycling among widespread estuary sediment bacteria. Microbiome 2015, 3, 14. [Google Scholar] [CrossRef]
- Lilburn, T.G.; Kim, K.S.; Ostrom, N.E.; Byzek, K.R.; Leadbetter, J.R.; Breznak, J.A. Nitrogen fixation by symbiotic and free-living spirochetes. Sci. Rep. 2015, 292, 2495–2498. [Google Scholar] [CrossRef]
- Latysheva, N.; Junker, V.L.; Palmer, W.J.; Codd, G.A.; Barker, D. The evolution of nitrogen fixation in cyanobacteria. Bioinformatics 2012, 28, 603–606. [Google Scholar] [CrossRef]
- Fuerst, J.; Sagulenko, E. Beyond the bacterium: Planctomycetes challenge our concepts of microbial structure and function. Nat. Rev. Microbiol. 2011, 9, 403–413. [Google Scholar] [CrossRef]
- Marshall, A.J.; Longmore, A.; Phillips, L.; Tang, C.; Hayden, H.L.; Heidelberg, K.B.; Mele, P. Nitrogen cycling in coastal sediment microbial communities with seasonally variable benthic nutrient fluxes. Aquat. Microb. Ecol. 2021, 86, 1–19. [Google Scholar] [CrossRef]
- Rivett, D.W.; Bell, T. Abundance determines the functional role of bacterial phylotypes in complex communities. Nat. Microbiol. 2018, 3, 767–772. [Google Scholar] [CrossRef]
- Guibert, I.; Bourdreux, F.; Bonnard, I.; Pochon, X.; Dubousquet, V.; Raharivelomanana, P.; Berteaux-Lecellier, V.; Lecellier, G. Dimethylsulfoniopropionate concentration in coral reef invertebrates varies according to species assemblages. Sci. Rep. 2020, 10, 9922. [Google Scholar] [CrossRef] [PubMed]
- Lima, L.F.O.; Alker, A.T.; Papudeshi, B.; Morris, M.M.; Edwards, R.A.; de Putron, S.J.; Dinsdale, E.A. Coral and seawater metagenomes reveal key microbial functions to coral health and ecosystem functioning shaped at reef scale. Microb. Ecol. 2023, 86, 392–407. [Google Scholar] [CrossRef] [PubMed]
- Guo, A.; Li, J.; Wang, L.; Ju, H.; Li, Q.; Ren, L.; Zhang, S. Temperature affects antagonism among coral-associated bacteria. Front. Mar. Sci. 2022, 9, 840384. [Google Scholar] [CrossRef]
- Van Woesik, R.; McCaffrey, K.R. Repeated thermal stress, shading, and directional selection in the Florida reef tract. Front. Mar. Sci. 2017, 4, 182. [Google Scholar] [CrossRef]
- Lee, O.O.; Yang, J.; Bougouffa, S.; Wang, Y.; Batang, Z.; Tian, R.; Al-Suwailem, A.; Qian, P.Y. Spatial and species variations in bacterial communities associated with corals from the Red Sea as revealed by pyrosequencing. Appl. Environ. Microbiol. 2012, 78, 7173–7184. [Google Scholar] [CrossRef]
- Jessen, C.; Villa Lizcano, J.F.; Bayer, T.; Roder, C.; Aranda, M.; Wild, C.; Voolstra, C.R. In-situ effects of eutrophication and overfishing on physiology and bacterial diversity of the Red Sea coral Acropora hemprichii. PLoS ONE 2013, 8, e62091. [Google Scholar] [CrossRef]
- Rusch, A.; Gaidos, E. Nitrogen-cycling bacteria and archaea in the carbonate sediment of a coral reef. Geobiology 2013, 11, 472–484. [Google Scholar] [CrossRef]
- Thome, P.E.; Rivera-Ortega, J.; Rodríguez-Villalobos, J.C.; Cerqueda-García, D.; Guzmán-Urieta, E.O.; García-Maldonado, J.Q.; Carabantes, N.; Jordán-Dahlgren, E. Local dynamics of a white syndrome outbreak and changes in the microbial community associated with colonies of the scleractinian brain coral Pseudodiploria strigosa. PeerJ 2021, 9, e10695. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hernández-Zulueta, J.; Díaz-Pérez, L.; Echeverría-Vega, A.; Nava-Martínez, G.G.; García-Salgado, M.Á.; Rodríguez-Zaragoza, F.A. An Update of Knowledge of the Bacterial Assemblages Associated with the Mexican Caribbean Corals Acropora palmata, Orbicella faveolata, and Porites porites. Diversity 2023, 15, 964. https://doi.org/10.3390/d15090964
Hernández-Zulueta J, Díaz-Pérez L, Echeverría-Vega A, Nava-Martínez GG, García-Salgado MÁ, Rodríguez-Zaragoza FA. An Update of Knowledge of the Bacterial Assemblages Associated with the Mexican Caribbean Corals Acropora palmata, Orbicella faveolata, and Porites porites. Diversity. 2023; 15(9):964. https://doi.org/10.3390/d15090964
Chicago/Turabian StyleHernández-Zulueta, Joicye, Leopoldo Díaz-Pérez, Alex Echeverría-Vega, Gabriela Georgina Nava-Martínez, Miguel Ángel García-Salgado, and Fabián A. Rodríguez-Zaragoza. 2023. "An Update of Knowledge of the Bacterial Assemblages Associated with the Mexican Caribbean Corals Acropora palmata, Orbicella faveolata, and Porites porites" Diversity 15, no. 9: 964. https://doi.org/10.3390/d15090964
APA StyleHernández-Zulueta, J., Díaz-Pérez, L., Echeverría-Vega, A., Nava-Martínez, G. G., García-Salgado, M. Á., & Rodríguez-Zaragoza, F. A. (2023). An Update of Knowledge of the Bacterial Assemblages Associated with the Mexican Caribbean Corals Acropora palmata, Orbicella faveolata, and Porites porites. Diversity, 15(9), 964. https://doi.org/10.3390/d15090964







