Abstract
(1) Background: Trichomycterinae represent 60% of the species in the family and, while seven genera comprise 1–3 species each, Trichomycterus and Cambeva have over 180 known species between them. Although integrative studies aimed to clarify the relationships within the subfamily, the diversity of species of Trichomycterus remains an open question. Herein, we explored an unprecedented sample to investigate the divergence in the lineages of Trichomycterus. (2) Methods: we recovered the phylogenetic relationships of the subfamily using 566 sequences (999 bp) of the mitochondrial gene cytochrome b, calculated intra- and intergroup distance percentages, and estimated divergence times. (3) Results: we recovered 13 highly supported and geographically structured lineages; intergenus divergence was 11–20%, while interspecies divergence was 3–11%; Trichomycterus, Cambeva, Scleronema, Hatcheria, Eremophilus, and Ituglanis were recovered as monophyletic, with three other highly divergent clades: Guiana Shield, Magdalena basin, and Tapajós basin. (4) Conclusions: We propose that the trans-Andean austral clades be allocated into Hatcheria, and the Guiana clade supports a new genus. We also observed that the headwaters nearest the Magdalena and Orinoco basins showed a high diversity and endemism of Trichomycterinae lineages. We discussed the role of geomorphological events and the climatic features which may explain cladogenesis events in Trichomycterinae.
1. Introduction
The Trichomycteridae are an assemblage of small-sized Neotropical freshwater catfishes with an adult size of less than 150 mm, inhabiting a diverse variety of habitats. The group is divided into eight subfamilies, of which the Trichomycterinae is the most diverse, with 60% of all taxa in the family [1]. So far, nine genera are recognized in Trichomycterinae: Bullockia Arratia, Chang, Menu-Marque and Rojas, 1978; Eremophilus Humboldt, 1805; Hatcheria Eigenmann, 1909; Ituglanis Costa and Bockmann, 1993; Rhizosomichthys Miles, 1942; Scleronema Eigenmann, 1917; Sylvinichthys Arratia, 1998; Trichomycterus Valenciennes, 1832 (De Pinna, 1998, Ochoa et al., 2017, sensu stricto Katz et al., 2018); and Cambeva (Katz et al., 2018). They are widely distributed in the neotropical region, from Costa Rica to Patagonia, and on both sides of the Andes [2,3]; while most genera comprise 1–3 species each, with restricted geographic distributions, Trichomycterus and Cambeva have over 180 known species between them [4].
The presence of odontodes outside the oral cavity organized in patches in the opercular region allows these animals to anchor to the substrate while in strong currents. These dermal teeth may be responsible for the presence of these fish in lotic environments in the very high portions of river headwaters [2,3,5].
This group of catfishes has been widely investigated using ecological, morphological, and, more recently, molecular approaches, with integrative studies clarifying the relationships within the subfamily. Recently, a multilocus molecular analysis was employed to reconstruct the phylogenetic relationships of Trichomycteridae with an unprecedented level of sampling [6]. The authors recovered the nonmonophyletic status of Trichomycterus, showing that some groups were more related to Eremophilus, Bullockia, and Scleronema. However, their analysis did not include the type species of the genus, T. nigricans, nor did it consider the diversity of species of Trichomycterus from Atlantic coastal drainages.
Later, one study used 50 terminal taxa of Trichomycterus and 21 terminal taxa belonging to other Trichomycterinae genera (including Bullockia, Eremophilus, Ituglanis, and Scleronema) to perform the most comprehensive taxonomic revision of Trichomycterus to date based on phylogenetic analyses of molecular and morphological characters [7] and confirmed the structuring of two well-supported lineages and the northern lineage (which includes the type species T. nigricans) corresponding to Trichomycterus sensu stricto. Meanwhile, the authors assigned the southern lineage (species from Paraná, São Francisco, Ribeira de Iguape, and Uruguay river basins) to the new genus Cambeva. The species from the Guianas and the Andes were not investigated in the previous studies [6,7], but rather indicated as an opportunity for future research efforts.
The species of Trichomycterus from the Pakaraima Mountains in the Guiana Shield were investigated [8], and the authors found a high diversity of species (T. conradi; T. guianense; T. cf. guianense; and Trichomycterus sp., a spotted form different from the others) living in sympatry and syntopy in a restricted area.
Nevertheless, uncertainties and confusion remain concerning the morphological recognition of the species within Trichomycterus and the validity of the classification of certain species. The large number of species associated with high endemism or restricted distributions, sympatry among species, and incipient diagnoses [2,9] hamper efforts to resolve species classifications and their interrelationships.
Given that cladogenic events like geomorphological transformations may imprint genetic signatures in the lineages, it may be possible to determine whether the family’s geographical structure is associated with the cladogenesis of lineages. Because Trichomycterus arose very recently (~18 mya) [6] and the species tend to occupy the headwaters of mountain watersheds, with frequent syntopy and geographically restricted species [2,3,5], speciation processes may be heavily affected by such events.
Given the group’s recent cladogenesis [6], there is a high species diversity, with most species geographically restricted. Meanwhile, headland capture events may have greatly influenced their distribution. We therefore aimed to explore an unprecedented collection of specimens to investigate the divergence in the lineages of representatives of Trichomycterus in a wide geographic area. We expect our study to deepen efforts to understand the structural patterns of species distribution within Trichomycterinae in South America.
2. Materials and Methods
2.1. Mapping Patterns of Species Distribution
A survey of the type locality of Trichomycterus nigricans Valenciennes 1832 was performed to understand the general pattern of distribution across altitude regions. For the species survey, the FishBase–Catalogue of Life [1] was consulted. To obtain information on the type localities of species in Trichomycterinae, the original descriptions of the species and complementary studies were utilized to enhance the accuracy of the type locality, particularly in cases where the geographic data of the holotype was not provided or in very old descriptions. If a study did not contain the geographic coordinates, these data were obtained by referencing Google Earth® and following the description of the locality. For each species, the basin, city, state, country, latitude, longitude, holotype code, and species authority were recorded (Table S1). Finally, a map that correlates to the species distribution to the region’s altitude was created.
2.2. Sampling
Our analysis involved georeferenced 566 sequences (see Table S2 for Genbank acession number), being 33 species of Trichomycterinae, and 364 sequences of Trichomycterus sensu stricto (including sequences of the type species of the genus T. nigricans Valenciennes 1832). Of the 324 Brazilian sequenced catfishes (323 from Trichomycterus and one from Cambeva), 182 were collected by our team, and 142 were obtained from fish collections. We used four sequences as an external group, two from Listrura spp. (Glanapteryginae), and two from Trichogenes longipinnis Britski and Ortega 1983 (Trichogeninae). We also used sequences from Genbank® of cis- and trans-Andean species of Trichomycterinae, the locations of which ranged from the Isthmus of Panama in the north to Patagonia at the extreme southern limit of the subfamily’s range (Table S2).
The samples from the Brazilian Atlantic coast comprised 13 sequences of Cambeva spp., four of Scleronema spp., and 14 of Ituglanis spp. The Andean samples consisted of 125 sequences of Trichomycterus areolatus Valenciennes 1846, 10 of Hatcheria macraei Girard 1855, and 12 of Bullockia maldonadoi Eigenmann 1928. The sequences from the northern Amazon basin consisted of 14 sequences of the T. guianense group Eigenmann 1909 from the Guiana Shield, eight from the Magdalena basin, and one Trichomycterus sp. from the Tapajós basin.
Fieldwork was carried out between June 2013 and October 2014 on expeditions that revisited the type localities of the eastern Brazilian species between the Jequitinhonha River basin in the north and the Itabapoana River basin in the south. Specimen sampling procedures followed a standard protocol [10], mainly using trawl nets. Captured specimens were anesthetized by submersion in eugenol solution [11] and the guidelines for fish euthanasia [12]. Tissue samples of the right flank muscle were obtained from specimens fixed in 90% ethanol and preserved in absolute ethanol for molecular analyses. For morphological comparisons, specimens were fixed in formalin for 15 days and then transferred to 70% ethanol. All specimens were registered in the fish collection at the Instituto Nacional da Mata Atlântica (INMA), formerly the Museu de Biologia Professor Mello Leitão, in Santa Teresa, Espírito Santo, Brazil.
2.3. Sequencing and Phylogenetic Analysis
DNA samples were extracted [13] and quantified using a NanoDrop ND-1000 spectrophotometer (Thermo Scientific™). For the in vitro amplification of the mitochondrial gene cytochrome b (cytb), we used the primers CytbSiluF and CytbSiluR [14]. Amplicons were purified by adding 1 μL Exosap to each 10 μL of PCR product. Sequencing reactions were performed using the Big Dye Terminator Cycle Sequencing Kit, following the manufacturer’s protocol. The 332 samples were sequenced in an ABI310 automatic sequencer (Applied Biosystems) at the Núcleo de Biodiversidade Genética Luiz Paulo de Souza Pinto (Nubigen), at the Universidade Federal do Espírito Santo (UFES). The sequences were aligned using Geneious v. 7.1.3 [15]. The saturation test and concatenation of the sequences with identical haplotypes were performed in DAMBE5 [16,17]. The maximum likelihood (ML) tree was generated in MEGA version 7 [18], with 1000 bootstrap replications, “General Time Reversible” model, using gamma distribution and invariant sites, according to results suggested by jModelTest [19].
The Bayesian inference (BI) analysis was conducted in BEAST 2.5 [20], programmed with a relaxed lognormal model and uncorrelated with estimated rate with the ucld.mean parameter set (prior distribution lognormal, initial value of 0.009, mean of 0.009, and log-stdev of 0.0001) and the 0.5 interval uniform distribution, while the other parameters used the default settings. The length of the Markov chain Monte Carlo (MCMC) was 10 million generations, with sampling every 1000 generations. ESS (>200) values were checked using Tracer v. 1.7.1 [21], with the initial 2000 trees discarded as a burn-in period, and the evolution model was found using jModelTest [19]. The main clades recovered by previous studies [6,7,8,22] were selected as monophyletic in the analyses: T. guianense, T. cf. guianense, and T. conradi [6,8]; genus Cambeva [7]; Scleronema + Cambeva [6], and Trichomycterinae [6,22]. Clade robustness was assessed through a bootstrap analysis and clade reliability was measured, considering strong support as >70% and moderate support as 50–70% [23]. Branch support was obtained through posterior probabilities, while clade confidence was defined as either strong (>0.95) or moderate (0.85–0.95). We edited the final trees in FigTree v1.4 [21] and generated the map in ArcMap10.1 [24]. The intra- and intergroup distance percentages were calculated in MEGA version 7 [18] using the Kimura-2-parameters model [25], with rates among sites set to “gamma distributed”, gaps/missing data treatment set as “pairwise deletion”, and other parameters set to default. The boxplot graphic was performed with R 4.2.1 [26]. The molecular phylogenetic analysis was carried out to estimate divergence times using BI in BEAST 2.5 [20] with the same parameter as provided above. The dating of the branches was calibrated following the values previously known [6].
3. Results
Through a survey of the type localities of nominal species of 178 taxa of Trichomycterinae (including 168 of Trichomycterus and 10 of phylogenetically related genera), we observed that these catfishes occur mainly in fast-flowing mountain watersheds that flow into the Pacific and Atlantic Oceans (Figure 1) and that the catfishes were rare (or unrecorded) in the lentic habitats of neotropical wetlands such as those of the Amazon and Pantanal regions (Table S1).
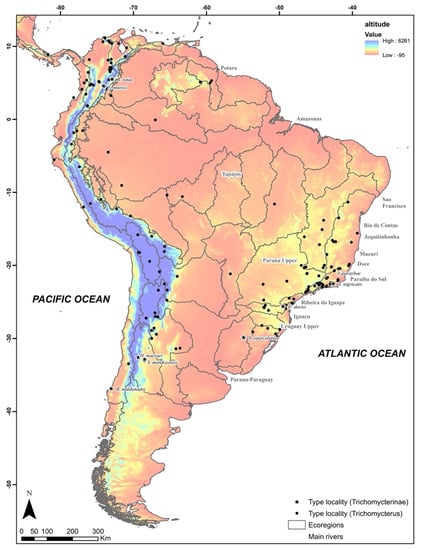
Figure 1.
Type localities of 168 species of Trichomycterinae. Distribution of type localities of the 168 species nominally allocated in Trichomycterus in relation to altitude in the neotropical region. Stars indicate the type locality of each type of species of the genera to Trichomycterinae. Detailed localities are shown in Table S1, available only in the online version.
The present study presents well-supported evidence of genetic partitioning with geographic structuring among representatives of Trichomycterus, with an unprecedented level of sampling of specimens across geographical locations, using the data from the specimens sequenced in the present study (n = 324) and Trichomycterinae sequences from other molecular studies [6,7,8,27]. These sequences comprise thirty-three species, with twenty-four belonging to Trichomycterus sensu stricto, seven to Cambeva, two to Scleronema, and three to Hatcheria. We propose seven additional clades that are phylogenetically distinct from the known genera of Trichomycterinae.
3.1. Relationships among Major Clades
The dataset of 570 georeferenced sequences of Trichomycteridae with the 999-bp fragment of the cytb gene generated 409 haplotypes, with no saturation. We used the “General Time Reversible” method, with gamma distribution and invariant sites (GTR + I + G), as an evolution model.
We found divergence values from 17.8 to 20.5% between the three subfamilies (Trichomycterinae, Glanapteryginae, and Trichogeninae), with the lowest divergence between Trichomycterinae and Glanapteryginae, and the highest between Trichogeninae and Glanapteryginae.
The ML and BI trees were identical for Trichomycterinae, recovering ten strongly supported clades (clades A–J), structured geographically into two east-west major clades (EW major clade; Figure 2 and Figures S1–S3), with a divergence of 12.5% (intraeast = 7%; intrawest = 5%). The distances between clades A and J ranged from 9.7 to 15.7% (Table 1).
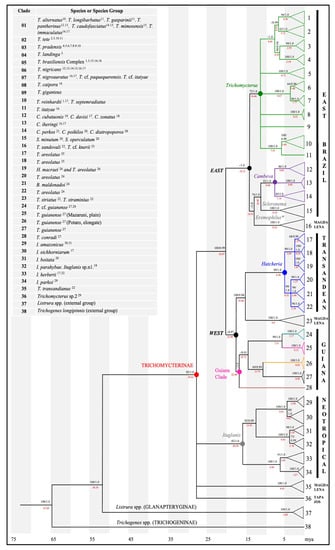
Figure 2.
Phylogenetic tree of Trichomycterinae based on 566 cytochrome b sequences (405 haplotypes) with 999-bp. Topology of the tree refers to the analysis generated by Bayesian inference. Values above each branch correspond to the branch consistency index (ML bootstrap/BPP). The minus signal (-) corresponds to support lower than 60 in ML and 0.90 in BI. The red numbers (below the branch) represent the time of clade divergence (in millions of years ago). Inset box: superscript numbers refer to species by clade and their distribution by ecoregion: 1São Francisco; 2Rio de Contas; 3Jequitinhonha; 4Buranhém; 5Jucuruçu; 6Mucuri; 7Itaúnas; 8São Mateus; 9Barra Seca; 10Doce; 11Piraquê-Açu; 12Santa Maria da Vitória; 13Jucu; 14Itapemirim; 15Itabapoana; 16Paraíba do Sul; 17Paraná-Paraguay; 18Ribeira do Iguape; 19Itapocu; 20Jacui; 21Uruguay; 22Magdalena (MAG); 23Orinoco; 24Central region of trans-Andean Chile basins; 25South region of trans-Andean Chile basins; 26Southern end of trans-Andean Chile basins; 27Potaro. Guiana; 28Mazaruni, Guiana; 29Tapajós (Amazonas; TAP); 30Madeira (Amazonas); 31Jari (Amazonas); and 32Araguaia.

Table 1.
Percentage of genetic distance pairwise between genera of Trichomycteridae. Bolded values on the diagonal are intraclade distances. Values below the diagonal (dark grey) represent the genetic distance between the respective clades. Values above the diagonal (blue) are the standard error of the distance indices.
For the east major clade, Trichomycterus sensu stricto (clade A), Cambeva (clade B), Scleronema (clade C), and T. sandovali + T. knerii (clade D) were recovered, with genetic distances of 10.3 to 12.9%, and intrageneric distances of 5.1 to 7.2%. For the west major clade, Hatcheria macraei + Bullockia maldonadoi + T. alternatus (clade E: genus Hatcheria), T. striatus + T. straminius (clade F), and T. guianense + T. conradi (clade G) were found, with genetic distances of 9.7 to 10.1%, and intrageneric distances of 3.2 to 6.4% (Table 1). Ituglanis spp. (clade H) was recovered as a monophyletic genus, in addition to two other clades less related to the other Trichomycterinae: T. transandianus (clade I) and Trichomycterus sp. (clade J: from Tapajós basin, sensu [6]). Based on the genetic divergence, geographic structure, and distribution (Figure 3), we analyzed these 10 clades (A–J) as potential genera or potential new genera arrangements.
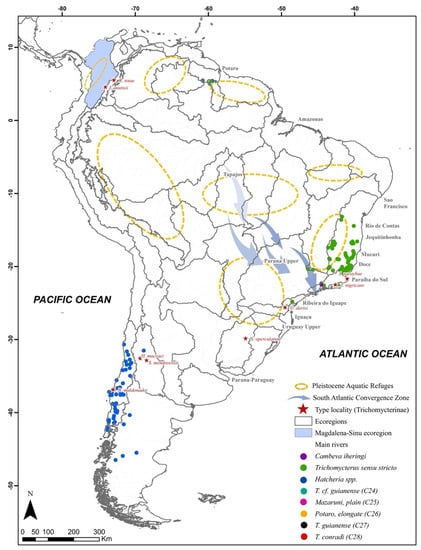
Figure 3.
Distribution of the georeferenced lineages of the Trichomycterinae subfamily in the neotropical region. Geographic area divided in ecoregions, sensu [28]. Stars indicate the type locality of each type species within each subfamily. Orange dotted circles representing aquatic Pleistocene refugia [29]. Note: many areas do not have the coordinate of the sample. Instead, authors cited the basin of occurrence (for details see Table S1). This is the case of highlights on the map of Magdalena-Sinu ecoregion.
3.2. East Major Clade
Included nominal species: Trichomycterus alternatus, T. longibarbatus, T. pantherinus, T. caudofasciatus, T. immaculatus, T. mimosensis, T. tete, T. pradensis, T. landinga, T. brasiliensis, T. brunoi, T. candidus, T. macrotrichopterus, T. pirabitira, T. rubiginosus, T. nigricans, T. nigroauratus, T. cf. paquequerensis, T. cf. itatyae, T. caipora, T. giganteus, T. reinhardti, T. septemradiatus, T. itatyae, Cambeva cubataonis, C. davisi, C. zonatus, C. iheringi, C. perkos, C. poikilos, C. diatropoporos, Scleronema minutum, S. operculatum, T. sandovali, and T. cf. knerii.
The clade comprising Trichomycterus, Scleronema, Cambeva, and Eremophilus was recovered as monophyletic, with Trichomycterus as sister to Cambeva + Scleronema (TCS), and Eremophilus as sister to the TCS clade.
The clade Trichomycterus sandovali and T. knerii (subclade 16) from the Magdalena Ecoregion was recovered as sister to the Cambeva + Scleronema clade, the unique lineage from the north included in the lineages from eastern Brazil, comprising Trichomycterus sensu stricto, Cambeva, Scleronema, and subclade 16.
Beyond the distinct genus status, the Trichomycterinae from eastern Brazil were grouped into four endemic and phylogenetically related genera and subdivided into 15 subclades (1–15; Figure 2), as follows: Trichomycterus sensu stricto (subclades 1–11), Cambeva (subclade 12–14), and Scleronema (subclade 15). Cambeva and Scleronema were recovered as sister genera (ac. 16.05 mya), with a genetic distance of 12.7%. The clade Trichomycterus sensu stricto showed a genetic distance of 13.1% to the clade Cambeva + Scleronema. Among the four genera, Trichomycterus has the northernmost distribution within the subfamily and coalesced from the others at approximately 18.02 mya.
The distribution of Scleronema lies to the south of Cambeva. Trichomycterus and Cambeva are sympatric in the headwaters of the Paraíba do Sul, São Francisco, and Paraná basins, where Trichomycterus subclades 10–11 are endemics.
The main eastern Brazilian clades (Trichomycterus sensu stricto) + (Cambeva + Scleronema), sister of clade D (subclade 16, comprising T. sandovali from Magdalena and T. cf. knerii from Orinoco), show an estimated coalescence time of 27.57 mya. Subclade 16 has an intrageneric distance of 8.8%, differing from the eastern Brazilian clade by 13.4%.
The relationship of the other genera was not recovered because they were recovered as polyphyletic. The clade Ituglanis was recovered with high support and widely distributed in the neotropical region.
3.2.1. Trichomycterus Sensu Stricto—Clade A
Included species: T. alternatus, T. longibarbatus, T. pantherinus, T. caudofasciatus, T. immaculatus, T. mimosensis, T. tete, T. pradensis, T. landinga, T. brasiliensis, T. brunoi, T. candidus, T. macrotrichopterus, T. pirabitira, T. rubiginosus, T. nigricans, T. nigroauratus, T. cf. paquequerensis, T. cf. itatyae, T. caipora, T. giganteus, T. reinhardti, T. septemradiatus, and T. itatyae.
We recovered Trichomycterus sensu stricto within two groupings from eastern Brazil with high genetic divergence of 9.8%. Across all 11 subclades in the clade Trichomycterus sensu stricto (subclades 1–11, Figure 2), pairwise genetic distances ranged between 4.3% and 11.3% (Table S3). One grouping (subclades 1–9) includes the type species T. nigricans; the second (subclades 10–11) is located at the southern limit of Trichomycterus sensu stricto.
The representatives of the clade occur throughout the coastal drainages of the Brazilian Atlantic coast and the Upper Paraná (ACUP) watershed of the southern Brazilian Shield. Its range is bordered by the Rio de Contas basin to the north and Ribeira do Iguape to the south, extending through ecoregions fully or partially within the Atlantic Forest. The western limits of the distribution correspond to the divide with the Rio São Francisco basin. Species of Trichomycterinae are generally limited to the headwaters and tributaries of river basins.
Subclade 1 has an intraclade distance of 2.9% and comprises seven taxa from the southern distribution of Trichomycterus. The subclade is only present in the Rio Doce basin; it comprises T. alternatus (Doce), T. longibarbatus (Piraquê-Açu), T. pantherinus (Santa Maria da Vitória and Jucu), T. caudofasciatus (Itapemirim and Itabapoana), T. mimosensis (Itabapoana), Trichomycterus sp. (Paraíba do Sul), and T. zonatus (Ribeira do Iguape). Subclade 2 is represented by T. tete from the Rio de Contas basin and T. jequitinhonhae from the Jequitinhonha basin in the Espinhaço mountains. Subclade 2 diverged from subclade 1 with a genetic distance of 6.9%. Subclade 3 consists of only one species, T. pradensis, which is widely distributed from the northern Doce basin to the southern Jucuruçu watershed. Subclade 4 diverged from subclade 3 with a genetic distance of 5.6%. Subclade 4 comprises T. landinga, endemic to the Jequitinhonha River basin. Subclades 1 to 4 form a clade and cover the entire distribution of Trichomycterus sensu stricto. Subclade 5 comprises the Trichomycterus brasiliensis species complex with an intraclade distance of 3%. The subclade’s distribution extends to the São Francisco, Jequitinhonha, Itabapoana, and Paraíba do Sul basins, reaching the Atlantic Forest biome in its western limits. Subclade 6 diverged from subclade 7 with a genetic distance of 4.3% and occurs in sympatry in the Paraíba do Sul and Paraná–Paraguay basins. Subclade 6 consists exclusively of the type species of the genus, T. nigricans, which occurs in the Santa Maria da Vitória, Jucu, Itapemirim, Itabapoana, and Paraíba do Sul basins, overlapping with the distribution of subclade 1 at its southernmost extent. Subclade 7 consists of T. nigroauratus, T. cf. paquequerensis, and T. cf. itatyae, with a distribution in the Paraíba do Sul and Upper Paraná basins. Subclade 8 consists of T. cf. caipora from the Paraíba do Sul basin. Subclade 9 is represented by T. giganteus, occurring in Guanabara Bay, Rio de Janeiro.
Subclades 10 and 11 coalesced from subclades 1–9 of Trichomycterus sensu stricto at an estimated 14 mya, with the highest genetic distance from the other subclades in the genus.
Subclades 10 and 11 comprise endemic species from the upper Paraná (T. septemradiatus), upper São Francisco (T. reinhardti), and upper Paraíba do Sul (T. itatyae) watersheds, each of which is geographically isolated. The main overlapping area of sympatry among the subclades is the Paraíba do Sul basin (subclades 1, 6, 7, 8, and 11), a sympatric region between Trichomycterus sensu stricto and Cambeva, as it represents the southern limit of Trichomycterus and the northern limit of Cambeva.
3.2.2. Cambeva—Clade B
Included species: Cambeva cubataonis, C. davisi, C. zonatus, C. iheringi, C. perkos, C. poikilos, and C. diatropoporos.
Cambeva is represented by three subclades (subclades 12–14) with genetic distances between 7.5% and 9.7% and an intrageneric distance of 6.5% (Table 1). Subclade 12 comprises C. cubataonis, C. davisi, and C. zonatus, with a distribution that stretches across the Itapocu, Ribeira do Iguape, and Paranapanema (Paraná) basins, with a genetic distance of 3.9% within the subclade (Table S3). Subclade 13 is represented by C. iheringi with a range in the Paraíba do Sul and Paraná basins, in sympatry with Trichomycterus (subclades 1, 5, 6, 7, 8, 10, and 11). Subclade 14 is formed of C. perkos, C. poikilos, and C. diatropoporos, with a distribution including the Jacui and Upper Uruguay basins, representing the southern limit of the genus, a region in which it occurs in sympatry with Scleronema.
3.2.3. Scleronema—Clade C
Included species: Scleronema minutum and S. operculatum.
Scleronema (clade C, subclade 15) is represented by S. minutum and S. operculatum with an intraclade genetic distance of 6.1% (Table 1). Its distribution is limited to the Lower Uruguay basin, the southern limit of the eastern Brazilian clade. Subclade 15 was recovered as sister to the clade Cambeva, with a genetic distance of 12.7%. Scleronema occurs in sympatry with Cambeva in the Uruguay basin, at the southern edge of the latter’s range.
3.2.4. Eremophilus—Clade D
Included species: Trichomycterus sandovali and T. cf. knerii.
Clade D (subclade 16) represented by T. sandovali and T. cf. knerii is known as the clade Eremophilus due to the previous recovered phylogenetic relationship [6], which includes E. mutisii as a member of the clade with the species mentioned above. The clade Eremophilus is distributed in the Orinoco and Magdalena basins and is sister to Cambeva + Scleronema, both endemic genera to eastern Brazil.
3.3. West Major Clade
Included species: Trichomycterus areolatus, Hatcheria macraei, Bullockia maldonadoi, T. striatus, T. straminius, T. cf. guianense, T. guianense, and T. conradi.
The west major clade is represented by the genus Hatcheria (clade E, subclades 17–22), comprising H. macraei, B. maldonadoi, and T. areolatus from the central and southern regions of trans-Andean Chile; T. striatus and T. straminius (clade F, subclade 23), both from the Magdalena basin, and the Guiana clade formed by T. guianense, T. cf. guianense, and T. conradi (clade G, subclades 24–28) from the Potaro basin of the Essequibo ecoregion, corresponding to the Guiana Shield portion.
Hatcheria—Clade E
Included species: Trichomycterus areolatus, Hatcheria macraei, and Bullockia maldonadoi.
The trans-Andean Austral clade was recovered as a monophyletic clade with a reference intraclade distance of 3.8%, coalescing in two subclades (17–19 and 20–22) at an estimated 18.59 mya, with an intraclade genetic distance of 3.2% and an intersubclade genetic divergence of 6%. Representatives of Trichomycterus areolatus, Hatcheria macraei, and Bullockia maldonadoi are grouped in subclade 17–19 from the southernmost area of Chile (Trichomycterus areolatus and Hatcheria macraei), with an intrasubclade divergence of 2.5%. Subclade 20–22 is represented by catfishes from the central region of trans-Andean Chile and comprises T. areolatus and Bullockia maldonadoi, with an intrasubclade divergence of 3.2%. Specimens identified as T. areolatus from clades 17 and 18 diverged by 6.1% from the T. areolatus individuals from clades 20 and 22.
With respect to intrageneric divergence, it is reasonable to assume that T. areolatus and B. maldonadoi (subclade 21) should be allocated to Hatcheria. The divergence levels in subclades 17–22 are compatibles with distinct species of the same genus. Furthermore, the divergence and cladogenesis findings do not corroborate the validity of three genera. In our analysis, Bullockia is recognized as a junior synonym to Hatcheria. Because we found that Trichomycterus sensu stricto is restricted to species inhabiting eastern Brazil (subclades 1–11), Trichomycterus areolatus Valenciennes 1846 is therefore recognized as Hatcheria areolata n. comb. In our analysis, Hatcheria is recognized as a single clade and the only genus of trichomycterids inhabiting austral freshwater environments. From the delimitations of the lineages obtained through data on genetic divergence and phylogenetic cladogenesis, we identify six species of Hatcheria. From these same criteria, we recover H. aerolata as polyphyletic (subclades 17, 18, 20, and 22), but it was not possible to determine the clade to which the species should belong. Thus, H. areolata can belong to any of these four subclades, which can also include previously described southern species or even new species. However, as a previous study aimed to understand the biogeography of T. areolatus [30], and the most representative subclade is 17, it is reasonable to assume that the species H. areolata belongs to this clade.
3.4. Striatus—Clade F
Included species: Trichomycterus striatus and T. straminius.
Clade F (subclade 23) comprises T. striatus and T. straminius, with an intrageneric distance of 5%, endemic to the Magdalena basin. Clade striatus is sister to the clade Hatcheria, comprising cis-trans Andean species (H. macraei, B. maldonadoi, and T. areolatus), with a genetic distance of 9.8% in relation to this clade.
3.5. Guiana—Clade G
Included species: Trichomycterus cf. guianense, T. guianense, and T. conradi.
The clade Guiana is formed of T. guianense, T. cf. guianense, and T. conradi (clade G, subclades 24 to 28), with an intraclade genetic distance of 6.4%. It is distributed in the Potaro basin of the Essequibo ecoregion, corresponding to the Guiana Shield portion. Interclade genetic distances with other genera vary from 9.7% (clade E: Hatcheria from trans-Andean portion) to 15.7% (clade J from Tapajós). These distances are compatible with genus-level differences, suggesting that this clade is a distinct genus.
3.6. Ituglanis—Clade H
Included species: Ituglanis parkoi, I. herberti, I. boitata, I. parahybae, Ituglanis sp. n1, I. amazonicus, and I. eichhorniarum.
Based on our analysis, Ituglanis is represented by six subclades (29–34) distributed throughout the neotropical region: I. amazonicus from Madeira and Jari in the Amazon basin (subclade 29), sister to I. eichhorniarum from Taquari and Aquidauana in the Paraguay and Paraná basins (subclade 30); I. boitata from Jacui in the Laguna dos Patos basin (subclade 31), sister to I. parahybae in the Macabu and Ribeira do Iguape basins (subclade 32); and I. herberti from Araguaia in the Tocantins basin and Piraí in the Paraná basin (subclade 33), sister to I. parkoi from Tapajós in the Amazonas basin (subclade 34; Table S2). The clade Ituglanis has an intrageneric distance of 9.5%. Interspecific distances vary between 4.3% and 13.1%, while intraspecific distances range from 1.6% to 4.6%. These high divergence rates among congeneric species indicate a need for revision since Ituglanis may represent more than one genus. Although the genus was recovered as monophyletic, it is distributed throughout the entire neotropical region, explaining the high divergence between more geographically distant species (Table S3).
3.7. Transandianus—Clade I
Included species: Trichomycterus transandianus.
Based on the genetic distance and geographic distribution of the monophyletic clades, two additional clades that have not been included in any known genus already delimited in the phylogeny were recovered: one from the Amazon and the other from the Magdalena basin (Figure S2). The two distinct lineages of Trichomycterinae herein recovered are termed clades I and J. Clade I (subclade 35), comprising T. transandianus from trans-Andean Magdalena, has a genetic distance to other genera of Trichomycterinae that ranges from 12% (clade E) to 16.5% (clade J) and an intraclade distance of 3.1%.
3.8. Tapajós—Clade J
Included species: Trichomycterus sp.2.
The second distinctive clade (J, subclade 36) comprises Trichomycterus sp.2 as defined previously [6] and consists of a single unnamed specimen, a catfish from the Tapajós basin (Amazonas). It has the greatest genetic distance from all the other clades, ranging from 13.1% (clade E) to 16.5% (clade I).
3.9. Referential Indices of Molecular Divergence for Trichomycterinae
Intrageneric divergences for Trichomycterinae, Glanapteryginae, and Trichogeninae ranged from 3.1% to 8.1% (except Ituglanis, where it was 10%). The genetic distance between the subfamilies varied from 17.8% to 20.5%, while it was between 9.7% and 15.7% across genera of Trichomycterinae (Table 1 and Table 2; Figure 4). The divergence between species or species groups within genera ranged from 2.5% to 11.3%, except for those of Ituglanis, with higher values ranging from 4.3 to 13.1% (Table 2, Figure 4, Table S3).

Table 2.
Percentage (%) of maximum, minimum, and mean genetic divergence between species, genera, and subfamilies of Trichomycterinae.
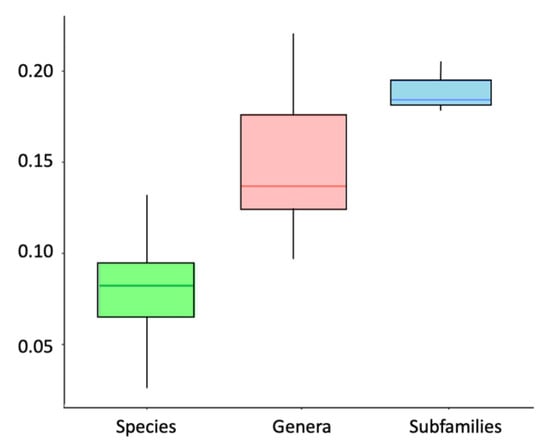
Figure 4.
Boxplot of the genetic distance percentage interval in species (green), genera (red), and subfamilies (blue).
The lowest pairwise distance across genera of Trichomycterinae was 9.7%, between Hatcheria spp., from the trans-Andean region (clade E) and T. guianense + T. conradi from the Guiana Shield (clade G); while the highest distance (15.7%) was obtained between Ituglanis spp. (clade H) and Trichomycterus sp. from Tapajós (clade J).
Intergeneric indices within Trichomycterinae ranged from 9.7% to 16.5%, with the lowest value between Hatcheria (clade E) and species from Guiana (clade G) and the highest values between T. transandianus from Magdalena (clade I) and Trichomycterus sp. from Tapajós (clade J) (Table 1). We also noted that the most divergent lineages within Trichomycterinae were T. transandianus (Magdalena, trans-Andean), Trichomycterus sp. (Tapajós), and Ituglanis, with similar values (between 11.7% and 16.5%).
The distances between the main clades (A–J) suggest that they may represent different genera. However, more research is required to support such decisions, even though we have recovered clades with high support, high genetic distances (compatible with distances from previously defined and named genera), and geographic structuring. Our decisions were based on phylogenetic lineage support as well as the intraclade (A–J) divergences between 3.2% and 9.5% and interclade divergence from 9.7 to 15.7% of recognized genera.
The interclade genetic distances that grouped species into the same genus of Trichomycterinae varied between 2.5% and 11.5%, except in the case of Ituglanis species, which had values between 4.3% and 13.1% (Table S3). The pairwise genetic divergence of each genus with more than one species varied as follows: 10.3–15.6% for Trichomycterus sensu stricto; 10.5–15.3% for Cambeva; and 11.7–15.1% for Ituglanis. Since the genetic distances found in clade E (9.7–13.1%), comprising Hatcheria macraei, Bullockia maldonadoi, and Trichomycterus areolatus, and clade G (9.7–14.7%), comprising the species from Guiana, Trichomycterus guianense and T. conradi, are compatible with the genus level, these clades may represent different genera.
The divergences among the subclades 17–22 of Hatcheria (as herein proposed) ranged from 2.5 to 5.7% (Table S3). The intrageneric values between clades in Trichomycterus sensu stricto ranged from 4.3% to 11.3%, with the highest genetic divergence values found between subclades 10 and 11 and all other subclades (mean pairwise divergence: 9.0% and 9.4%, respectively).
In each genus, interspecific divergence ranges were as follows: Trichomycterus sensu stricto from 4.3 to 11.3% (mean: 8.2%); Cambeva from 6.3 to 10.9% (mean: 8.6%); Ituglanis from 4.3 to 13.1% (mean: 10.4%); Hatcheria from 2.5 to 11.5% (mean: 6.3%), and clade Guiana from 4.8 to 11% (mean: 9.2%). Notably, the genetic distances in clades E and G are compatible with the genus level, suggesting that they may represent new genera. Specifically, clade E, comprising Hatcheria macraei, Bullockia maldonadoi, and Trichomycterus areolatus, had interspecific genetic divergence between 2.5% and 11.5% (mean: 6.3%), while clade G, comprising species from Guiana, as Trichomycterus guianense and T. conradi, had genetic distances ranging from 4.8 to 11% (mean: 9.2%).
We therefore propose that distances as high as 17.8–20.5% should be used to delimit different subfamilies, while values of 9.7–15.7% would separate genera within the same subfamily (Table 1 and Table 2), and interspecific variation within a genus would have values of 2.5–11% (Table S3).
It is important to mention that genetic divergence per se is not sufficient to delimit taxa. However, since we found that the genetic divergence was also associated with geographically structured clades, our findings indicate a need to revisit species classification and occurrence. Furthermore, they suggest a considerable impact of landscape factors on the genetic signatures of these lineages. Our reference values of intra- and interspecific variation do not encompass the ranges found within Ituglanis since its range represents an outlier in relation to the other genera of Trichomycterinae.
Our study established reference values of genetic diversity for one mitochondrial gene at the subfamily, genus, and species levels representing the first evaluation of its kind for Trichomycterus. Subfamilies in the Trichomycteridae diverged by 21–24%, genera in Trichomycterinae by 11–20%, and congeneric species by 3–11%. Intraspecific distances ranged between 0.4% and 8.8%.
4. Discussion
All eastern Brazilian genera have been recovered as monophyletic, as follows: Trichomycterus sensu stricto as the sister clade of Cambeva + Scleronema and T. sandovali + T. cf. knerii (subclade 16 or Eremophilus mutisii) as the sister clade to the former group, T. cf. knerii, which occurs in the Magdalena and Orinoco basins (cis-Andean portion). This finding provides valuable evidence for the phylogenetic relationships between species from eastern Brazil and those from Magdalena and Orinoco, a highly diverse region for Trichomycterinae. Also, the monophyly of Trichomycterinae, Ituglanis, and Hatcheria was recovered, and due to the number and location of samples, the geographic distribution of the subfamily’s genetic diversity throughout South America was characterized, with particular emphasis on the distribution limits of lineages from eastern Brazil.
Our potential confirmation of the delimitation of Trichomycterus sensu stricto to species from eastern Brazil underscores the urgency and necessity to revise the classification of species that are still nominally considered as being representatives of Trichomycterus, recorded in other geographical ranges (Table 3).

Table 3.
Revisiting the classification of the species included in the genus. See Table S2 for detailed information on the number of GenBank specimens used in each reference. * Eremophilus mutisii multilocus sequences were recovered from the clade with T. sandovali and T. cf. knerii in previous papers [6,7].
4.1. The Major Clades in the Trichomycterinae
Given that Trichomycterus sensu stricto, Cambeva, and Scleronema have been recognized as phylogenetically close but distinct genera, supported by ample morphological and genetic evidence [7], we anticipated that the divergences between other genera within the subfamily would be comparable to the values of 12.4% to 15.5% calculated for the clade. We also calculated the between-species distances within the Trichomycterus sensu stricto clades, ranging from 5.0% to 11.2%. In contrast, the genetic differences we observed between the trans-Andean Austral taxa were between 3.1% and 6.8%, which is incompatible to the notion of distinct genera.
4.1.1. Clade Trichomycterus Sensu Stricto
Trichomycterus sensu stricto is recognized as an assemblage with species from the northeastern Atlantic Forest ecoregion and adjacent basins. The intraclade relationships between species of Trichomycterus are out of the scope of the present study. Trichomycterus sensu stricto is a highly supported monophyletic group, geographically structured in eastern Brazil with a range bounded in the north by the Rio de Contas basin in Bahia state, the São Francisco basin in the west, and the Ribeira do Iguape river basin in the south. This range represents a distribution from 10° S to 25° S and from 40° W to 50° W. Trichomycterus nigricans is the type of species, and the type of locality is in a drainage of Rio de Janeiro state, Brazil [31]. The genus Trichomycterus is attributed solely to species occurring in eastern Brazil.
Until recently, species from southeastern and southern Brazil were considered part of Trichomycterus. Nevertheless, Cambeva has now been identified as a distinct genus with a geographic distribution that overlaps with Trichomycterus sensu stricto in the upper São Francisco, Paraná, and Paraíba do Sul basins [6]. The three clades from the Brazilian Shield [6] were also recovered: one corresponding to Trichomycterus with a northern distribution covering the Paraná, Paraíba do Sul, São Francisco, Itabapoana, Jucuruçu, and Doce basins; another to Cambeva in the southeastern portion of the distribution of Trichomycterus; and the third as Scleronema in lower Uruguay. In our analysis, we found that Trichomycterus sensu stricto and Cambeva occur in sympatry in the Paraíba do Sul, Upper Paraná, and upper São Francisco basins, highlighting the significance of this region in the biogeographic history and cladogenesis of the lineages within the eastern Brazilian Shield. The discovery of a potential new endemic sister genus of Trichomycterus sensu stricto underscores the remarkable diversity of Trichomycterinae in this area. Additionally, the coexistence of the established sister genera Cambeva and Scleronema in the lower Paraná–Uruguay further emphasizes the region’s significance in hosting diverse Trichomycterinae species.
The genus Trichomycterus is still cited in the literature as having 166 species [29]. These numbers, however, do not consider the reallocation of species to Cambeva. Further efforts are required to diagnose and classify approaches since future studies aiming to understand the phylogeny of Trichomycterus or Cambeva must deal with samples whose identification has not been updated in the databases. This is particularly important given that studies are often based on samples with few specimens and low representativeness in terms of species and geographic areas.
In relation to Trichomycterus alternatus species group (Clade 1), we found that T. alternatus is part of a species complex we refer to as subclade 1 of Trichomycterus sensu stricto, comprising T. longibarbatus, T. gasparinii, T. pantherinus, T. caudofasciatus, and T. mimosensis. However, the molecular differences between the species within the complex are unclear. For example, T. gasparinii is indistinguishable from T. longibarbatus, and T. mimosensis remains indistinguishable from T. caudofasciatus. The low intraclade genetic distance within the species complex warrants future analysis to assess its validity.
The low genetic diversity associated with many valid species highlights the problem of the genus. Therefore, using a 2% genetic distance threshold may overestimate the number of species in Trichomycterinae, which cannot always be distinguished morphologically. By applying a conservative lumper view to delimit species genetically [32], we propose a threshold level of interspecific molecular divergence of at least 3%, as this value is sufficient to reveal different monophyletic species without creating morphological problems in distinguishing them. If a 2% threshold is used to validate molecular divergence in T. alternatus, T. longibarbatus, T. gasparinii, T. pantherinus, T. caudofasciatus, T. immaculatus, and T. mimosensis, we could likewise propose at least 13 new morphologically indistinguishable species.
In a previous study [22] the clade immaculatus was recovered as sister to the clade brasiliensis, which correspond to subclades 1 and 5 in the present study, respectively. We expanded the understanding of the relationship of the clade immaculatus sensu [22], here referred to as subclade 1 (T. alternatus, T. longibarbatus, T. gasparinii, T. pantherinus, T. caudofasciatus, T. immaculatus, and T. mimosensis), with other clades, positioning it as part of the same clade as subclades 2–4, sister clade to subclades 2 (T. tete) and 3 + 4 (T. pradensis + T. landinga, respectively). However, we did not recover the phylogenetic relationship of the clade brasiliensis sensu [22] or subclade 5.
The clade 2 of Trichomycterus sensu stricto (subclades 10–11) is endemic to the headwaters of the São Francisco, Paraíba do Sul, and Paraná–Paraguay basins, where it is sympatric with the first clade (subclades 1–9). However, based on a combined analysis of molecular and morphological characteristics, it is suggested that the species represents a distinct clade of Trichomycterus sensu stricto [22]. Our findings reconfigure the extent of Trichomycterus sensu stricto proposed previously [7] to exclude T. septemradiatus, T. pauciradiatus, T. reinhardti, and T. itatyae. Recently, it was proposed that these species represent an endemic clade [31]. According to these authors, this clade is represented by 10 species (T. reinhardti, T. funebris, T. humboldti, T. ingaiensis, T. pauciradiatus, T. piratymbara, T. sainthilairei, T. septemradiatus, T. anaisae, and T. luetkeni), endemic to the Rio São Francisco basin, Rio Doce headwaters, Espinhaço mountain range, and Paraná (Rio Grande) basin between the Mantiqueira and Canastra mountain ranges. Also, it is attributed the high endemism of these species to the uncommon regional relief [31], with small subdrainages crossed by transversal hills due to geological gaps, supporting a complex paleogeographic scenario favoring isolation of taxa in small areas. This evidence may support Clade 2 (subclades 10–11) as a new genus. However, in keeping to a conservative “lumper” perspective to delimit diversity [32], we have chosen not to propose another genus until evidence beyond what has been explored in the present study comes to light.
4.1.2. Clades Cambeva and Scleronema
The genus Cambeva was recovered as the monophyletic and sister clade to Scleronema, both sisters to Trichomycterus sensu stricto. Cambeva, Scleronema, and Trichomycterus are endemic genera to the eastern neotropical region, occurring in sympatry in São Francisco, Paraíba do Sul, São Francisco, Paraná, and Uruguay basins. This phylogenetic relationship is congruent with previous studies [6,7,22].
4.1.3. Clade Eremophilus
Subclade 16, represented by T. sandovali (Magdalena) and T. cf. knerii (Orinoco), was recovered as a sister clade to the eastern Brazilian species [(Cambeva + Scleronema) + (Trichomycterus sensu stricto)]. This subclade exhibits a disjunct distribution since it was not phylogenetically close to the nearby lineages. Subclade 16 occurs in sympatry in the Magdalena basin with subclades 23 (T. striatus and T. straminius) and 35 (T. transandianus). However, despite this sympatry, it shows significant genetic distance values and lacks any close phylogenetic relationship to the other taxa in its vicinity. Previous studies had recovered T. sandovali and T. cf. knerii with Eremophilus mutisii from the Magdalena basin [6,22]. These results provide an insight into the high diversity and endemism in these regions, mainly in the right arm of the headwaters of the Magdalena River [33].
4.1.4. Clade Hatcheria
The genus Hatcheria Eigenmann 1927 was originally proposed to include Hatcheria maldonadoi [3]. Trichomycterus areolatus is a more senior name than B. maldonadoi and H. macraei. We hereby propose the senior name Hatcheria areolata n. comb. to encompass the senior name of T. areolatus, while designating Hatcheria macraei and Bullockia maldonadoi as junior synonyms. Consequently, we recognize the Andean clade as the monotypic Hatcheria areolata, morphologically distinctive by the presence of a flattened head with the largest width at the opercular patch of odontodes; wide maxilla; dorsally placed eyes; dorsal fin rays 13–15 branched, and a caudal peduncle strongly compressed laterally [34,35]. Hatcheria is easily distinguished from both Trichomycterus and Cambeva by the dorsal position of eyes (vs. lateral or dorsolateral in Trichomycterus and Cambeva) and the number of branched dorsal fin rays (13–15) vs. 6–7 in Trichomycterus and 7–8 in Cambeva.
As a result of our analysis, we have identified a monophyletic trans-Andean group of Trichomycterinae, comprising the austral species Trichomycterus areolatus, Bullockia maldonadoi, and Hatcheria macraei. This group should be recognized as a distinct genus, separate from Trichomycterus, Cambeva, and Scleronema. The phylogenetic topology, associated with the geographic distribution of the lineages and genetic distances between clades, supports the reclassification of T. areolatus and B. maldonadoi under Hatcheria. In this context, our study represents an initial step towards clarifying the taxonomic status of the mentioned species. Based on phylogenetic and molecular evidence, we propose that both T. areolatus and Bullockia maldonadoi should be reclassified and named as Hatcheria areolata n. comb. and H. maldonadoi n. comb., respectively.
Hatcheria macraei was first described in 1855 in the vicinity of Uspullata, Mendoza, Argentina, on the cis-Andean side, but has also been recorded in the southern Chilean province of Aysén [30]. Hatcheria areolata n. comb. is one of the most well-distributed species of Trichomycterinae in austral drainages, and it was described in 1846 in the San Jago River in Santiago and is known to occur in the rivers of the western slopes of central Chile [3]. Hatcheria maldonadoi is endemic to trans-Andean drainages in the south-central provinces of Chile [34,35], and it was first described in 1928 in the Nonguen River.
The phylogenetic trees obtained in this study revealed two lineages showing 6% divergence. One lineage includes H. areolate + H. maldonadoi, while the other comprises the southern Chilean populations of H. areolate grouped with H. macraei, resulting in H. areolatus being polyphyletic. This paraphyly of H. macraei and H. areolate was also reported in a previous phylogenetic study using cytb sequences [30]. Our analysis indicates that the close genetic relationship between these species does not support their classification into separate genera.
An examination of the morphological characters shared by these three Andean species reveals several character states that are not shared with Trichomycterus sensu stricto. The cephalic laterosensory system is an uninterrupted supraorbital canal reaching the sensory pore s1 in the trans-Andean species [36,37] vs. s1 interrupted from s3 in the Trichomycterus sensu stricto species of the eastern Brazilian clade. The difference also includes a pectoral fin with absent or rudimentary filament vs. a pectoral fin filament always present in Trichomycterus sensu stricto and a flattened caudal peduncle with low depth vs. rounded with high depth in Trichomycterus sensu stricto [38].
In a prior study [39], an attempt was made to resolve the trans-Andean species complex by analyzing Trichomycterinae specimens in the British Museum. The study proposed that Hatcheria should be considered synonymous with the trans-Andean species of Trichomycterus, owing to the challenges in distinguishing between these two groups. Another analysis focused on morphological characters, revealing that the highest retention of plesiomorphic traits within Trichomycteridae occurs among species from trans-Andean Patagonia. This finding implies that the family’s ancestry is linked to this particular region [40].
4.1.5. Clade Striatus
The clade Striatus comprises the west major clade and is the sister group to the clade Hatcheria. It is endemic to the Magdalena basin, which harbors the highest diversity of lineages of Trichomycteridae in the region.
4.1.6. Clade Guiana
The Guiana clade (subclades 24–28) is represented by T. cf. guianense (clade 24), the Mazaruni region (clade 25), the Potaro region (clade 26), T. guianense (clade 27), and T. conradi (clade 28), which are endemics and occur in sympatry and nearby the Essequibo ecoregion. We obtained the same topology, confirming the monophyly of the Guiana species, as well as the internal relationships within species and groups, which have been previously reported [6,8,22]. These findings further underscore the region’s high diversity and endemism, aligning with the Pleistocene refuges identified for Characiformes [33] and Trichomycterus [8].
Other clades are also distributed in the areas surrounding the Guiana ecoregion, mainly in the Magdalena ecoregion, where most of the diversity of the main clades is found. In a previous study [6], a monophyletic group comprising T. guianense and other neotropical species from both sides of the Andes in the northern reaches of the Magdalena and Amazon basins was recovered. These included T. banneaui Eigenmann, 1912 (Magdalena basin, Colombia); T. cachiraensis Ardila-Rodríguez, 2008 (Magdalena basin, Colombia); T. ruitoquensis Ardila-Rodriguez, 2007 (Magdalena basin, Colombia); T. spilosoma Regan, 1913 (San Juan basin, Colombia); T. striatus Meek and Hilbebrand, 1913 (Cana River, Panama); and T. transandianus Steindachner, 1915 (Magdalena basin, Colombia). Also, a high species diversity was found in the Pakaraima Mountains of Guyana [8], including new species occurring in sympatry in a restricted region.
4.1.7. Clade Ituglanis
We recovered six subclades of Ituglanis spp. structured geographically in three clades: (1) I. amazonicus (Madeira: Amazonas basin) and I. eichhorniarum (Paraná–Paraguay basin) more to the west of the sampling distribution; (2) I. boitata (Laguna dos Patos basin) and I. parahybae (Ribeira do Iguape basin) more to the south of the distribution, and (3) I. herberti (Tocantins and Paraná basins) and I. parkoi (Tapajós: Amazonas basin) more to the east of the sampling area.
4.1.8. Clade Transandianus
Trichomycterus transandianus (clade I, subclade 35), endemic to the trans-Andean portion of the Magdalena basin, was very distinct from the other Trichomycterinae in the region or any other included in the analysis. Even so, this result provided additional support to the high diversity and endemism of the Magdalena ecoregion, highlighting the area’s importance for the diversification of Trichomycterinae, supporting its identification [33] as an important area of endemism and aquatic diversity.
4.1.9. Clade Tapajós
Trichomycterus sp2. ([6]; clade J, subclade 36) is endemic to the Tapajós basin, and we did not recover any relationship of this species with the others of the subfamily. However, in previous study [6] this species was related to the clade herein called Hatcheria (subclades 17–22), meaning that it may be related to the west major clade.
4.2. Efficiency of Cytochrome b
There might be a question regarding whether data from a single locus is sufficient to unveil the evolutionary history of Trichomycterus. Despite using only one mitochondrial gene with 999 bp, we successfully confirmed the monophyly of Trichomycterus sensu stricto, Cambeva, Scleronema, and Ituglanis, along with their interrelationships. Furthermore, we expanded the scope of our study by utilizing a comprehensive set of georeferenced data, covering a larger sampling area with higher representativeness compared to previous research [6,7,8,22].
Our data were consistent with previous studies that used five genes ([6], 3.284 bp for 16S, COI, cytb, Myh6 and Rag2) and six genes ([7], 4.380 bp for COI, cytb, Glyt, Myh6, Rag2 and Sh3px), providing additional support for the high discriminatory potential of cytb gene sequences. Several characteristics of cytb have enabled the evaluation of recent phylogenetic relationships with robustness. These characteristics include a high number of copies per cell, maternal inheritance, low ancestral polymorphism, and the absence of recombination. Moreover, its evolutionary rates facilitate species-level distinctions. However, caution is advised, as a study of Cichlidae relationships [41] discovered at genetic distances exceeding 15%, saturation levels might pose challenges in the analyses, particularly when assessing the phylogenetic relationships between less closely related genera.
4.3. The Magdalena–Sinu Diversity
Our study provides support for the identification of seven additional clades, with seven of them geographically associated with the Amazon region, particularly the Magdalena–Sinu ecoregion. We observed the relationship between two major lineages as follows: (1) Eastern/Magdalena (cis-Andean; subclades 1–16): {[Trichomycterus + (Cambeva + Scleronema)] + subclade 16 (T. sandovali and T. knerii; Eremophilus mutisii)} and (2) Western/Magdalena (cis and trans-Andean; subclades 17–28): [Hatcheria + subclade 23 (T. striatus and T. straminius)] + [Guiana clade (T. guianense and T. conradi)]. This topology suggests that cladogenesis occurred within the Magdalena ecoregion, as both eastern and western lineages are present in this region. Furthermore, the area exhibits high diversity, serving as Pleistocene refuges to Characiformes [33], which may also explain the high diversity of Trichomycterinae lineages.
4.4. The Cladogenesis Processes under a Phylogeographic Perspective
Our analyses brought significant contributions to our understanding of how lineages are organized within a phylogenetic context. Additionally, we established reference values of genetic divergence, aiding in the classification of taxa and serving as a valuable reference for barcoding purposes. Furthermore, we assessed the phylogeographic structure and examined geological evidence to infer processes that have influenced the vicariant pattern. This comprehensive approach enables a better understanding of the current distribution of lineages and sheds light on the ecological and phylogeographic history that underlies the cladogenic processes.
We classified South American species of Trichomycterinae into two geographic regions: (1) Eastern Brazil (clades Trichomycterus sensu stricto; Cambeva; Scleronema; and T. sandovali + T. knerii + E. mutisii); and (2) trans-Andean Austral (clade Hatcheria, which includes H. macraei, H. maldonadoi, and H. areolata; T. striatus + T. straminus; and clade Guiana, which includes T. guianense and T. conradi). While prior studies propose similar topologies [6,7], they lack an in-depth discussion of the trans-Andean Austral and Guiana Shield clades. Moreover, these studies do not delve into the phylogeographic perspective regarding the boundaries of the geographic distributions of neotropical lineages.
The Pleistocene period coincides with the great diversification of the main Trichomycterinae lineages. It is plausible that the processes that facilitated the allopatric speciation of Characiformes, with their refugees in the highlands during the glaciations and subsequent colonization of the lowlands, along with interactions between marine incursions, uplift of the paleoarches, and historical connections allowing cross-drainage dispersal, may also account for the speciation processes of Trichomycterinae in the Paraná–Paraguay, Guianas, and Magdalena–Orinoco refuges [33].
The first proposed partition of eastern Brazilian Trichomycterus into Cambeva and Trichomycterus sensu stricto [7] referred to the Andean and northwestern neotropical species as “other Trichomycterinae”. Our analyses go further on this topic, discussing the organization of Andean lineages and their biogeographic history.
Based on [42], species richness hotspots include the Chocó region of Colombia, the Pantanal wetlands of the Upper Paraguay basin (central Brazil and Paraguay), and the Atlantic Forest of southeastern Brazil. These regions were also recognized as areas of endemism [33]. Furthermore, our study identifies these areas as significant for comprehending the cladogenesis between Trichomycterus sensu stricto and Cambeva + Scleronema.
Considering the intrinsic characteristics of these catfish, commonly associated with freshwater headlands [2,3], altitude is a limiting factor for the Trichomycterinae, which may explain their high structuring even in geographically close areas. The main highland areas in the neotropical region, namely the Andes, the Guiana Shield, and the Atlantic coastal drainages, are separated by depressions between the shields [43]. Multiple river systems drain the high plateau of the Guiana Shield in all directions [8]. The Andes, Guiana, and Eastern Brazil regions are situated in the three highest neotropical regions, which were geographically isolated by the Paranaense and Amazon Seas during the Pleistocene glacial maximums [44]. This isolation has significantly influenced the distribution of freshwater fishes, particularly rheophilic fish, which thrive in high-energy waters and are commonly found in the highland regions [45]. In contrast, these fish taxa are typically scarce in lowland plains or floodplains [46], explaining the near absence of species of Trichomycterinae in such environments (except for Ituglanis). The different lineages of Trichomycterinae are structured in various neotropical high plains, a pattern that can be attributed to recent events—particularly the formation of the Austral Andean high plains—or to older features like the Guiana and Brazilian Shields [47].
We can posit that these three neotropical highlands function as “lineage islands” (as we termed them) due to the unique life habits of these catfish. The valleys that separate these highlands serve as barriers to dispersal, imposing constraints of gene flow between these “islands”. Consequently, understanding vicariant patterns between lineages necessitates a comprehensive examination of geomorphological processes.
Due to significant neotectonic activity in the eastern portion of the Brazilian Shield, the landscape dynamics in the region, particularly in the area of sympatry between Trichomycterus sensu stricto and Cambeva at the headwaters of the Paraíba do Sul, São Francisco, and Paraná–Paraguay basins, have undergone considerable change. A study focusing on the geomorphological analysis of South America [48] revealed that this region is divided at the 125° Azimuth lineament, which separates the Amazon region from Paraná–Paraguay. The presence of interbasin arches in the landscape, associated with intrusive magmatism, contributed to the formation of divides between the Amazon, São Francisco (draining northward), and Paraná–Paraguay basins (draining southward). These geomorphological events have led to high environmental heterogeneity in the region, which is likely linked to the high genetic diversity observed in the taxa of Trichomycterinae found in the limits of the 125° Azimuth lineament. Moreover, this area served as a refuge for many aquatic species during the glaciations of the late Miocene for Characiformes, acting as a center of endemism and dispersion [33].
In a previous study [49], the authors presented compelling evidence of a prominent denudational escarpment that separates the São Francisco, Paraná, Doce, and Paraíba do Sul basins, leading to a gradual shift of the lower plateaus towards the interior of the continent. The occurrence of escarpment events renders the headwaters of these rivers more susceptible to denudation, potentially resulting in their isolation or connection over time. Therefore, the diversity within this region may hold greater significance, as it likely facilitates the exchange of diversity events among different portions that were previously isolated.
Also, a study that examined the role of paleodrainages on the distribution pattern of the genetic diversity of T. alternatus was conducted [50], and their findings indicated that the intricate distribution patterns of this species were influenced by factors, including paleoriver dispersion, headwater capture, and geographical barriers like the Cabo Frio Magmatic Lineament. These events would explain the complexity and dynamism observed in the Serra do Mar region and the recurrent examples of biogeographic breaks identified in this area, which align with the results we observed in Trichomycterinae.
The South Atlantic Convergence Zone, characterized by extensive cloud cover across the Amazon–Atlantic region, exhibits its most intense influence in the headwaters of the São Francisco, Paraná, Paraíba do Sul, and Doce basins. This can lead to distinct precipitation compared to the other regions and cause climatic anomalies, particularly during austral summer [51]. The interplay of these factors collectively influences the dynamics of the Trichomycterinae diversity in the area.
Neotropical freshwater fish display primary diversity patterns driven by erosion-induced river capture and climate-induced eustatic (global) sea-level oscillations. Landscape evolution processes can alter the spatial dynamics and course of rivers [42], which could account for the disjunct distribution of sister lineages, such as T. sandovali + T. cf. knerii from the Magdalena and Orinoco basins, relative to (Trichomycterus) + (Cambeva + Scleronema) from eastern Brazil.
An interesting finding is the strong support found between the two major Trichomycterinae lineages, with one predominantly distributed in the eastern neotropical region and the second in the western region. However, both lineages also occur in the Magdalena ecoregion, suggesting its significant role in the eastern–western cladogenesis of the subfamily. This observation gains further support when considering the high diversity of lineages in the Magdalena region and neighboring areas (e.g., Guianas), with representation of most of the subfamily’s lineages and high species endemism. Furthermore, a distinct pattern emerges, with the western lineages showing connections with the trans-Andean portion, including the Magdalena, while the eastern lineages predominate in the cis-Andean portion, even within that ecoregion. The cladogenesis of these two major lineages was estimated at 28 mya, aligning with the active period of the Andes uplift (40 to 4 mya). The isolation of Orinoco and the Guianas during the Miocene marine incursions [33] likely contributed to the significant difference in fish species from these regions, despite their geographic proximity.
Finally, our findings highlight the remarkable complexity within the Trichomycterinae subfamily. Comprising a highly diverse and widely distributed group, studying their evolutionary history presents unique challenges. The relationships between species of Trichomycterus are quite complex and will require the inclusion of the greatest possible representativeness of the genus, particularly considering the characteristics associated with several taxa that were still not yet explored in this study. Nonetheless, our results constitute a significant step towards unraveling monophyletic groupings within the genus. Incorporating additional species previously not included in genetic studies may further refine the clades identified in our investigation. Additionally, an important avenue for further investigation involves identifying species from the central neotropics in relation to the eastern, western, and northeastern groups, necessitating the inclusion of more species from the genus and the subfamily in phylogenetic analyses.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/d15080929/s1. Figure S1. Phylogenetic tree original of Trichomycterinae based on 566 cytochrome b sequences (405 haplotypes) with 999 bp. Bayesian Inference with values above each branch correspond to the branch consistency index (BPP); Figure S2. Phylogenetic tree original of Trichomycterinae based on 566 cytochrome b sequences (405 haplotypes) with 999 bp. Bayesian Inference with values above each branch correspond to age for clades. as well as the margin of error demonstrated in node bars; Figure S3. Phylogenetic tree original of Trichomycterinae based on 566 cytochrome b sequences (405 haplotypes) with 999 bp. Maximum Likelihood with values above each branch correspond to the branch consistency index (ML bootstrap); Table S1. Type locality of the species nominally allocated to Trichomycterus and of the type species of the nine genera of Trichomycterinae. Type species in asterisk; Table S2. Specimens used in the present study, with their respective locations (basin, latitude, and longitude), the collection where specimen is deposited (only for sample exclusive from this study), GenBank accession number of all sequences used on the phylogenetic analysis, and the source that generated the sequences (present study or previous studies). Clade numbers are in according to the phylogenetic tree in Figure 2; Table S3A. Table with percentages of K2P pairwise genetic distances between the subclades of Trichomycterus sensu stricto. Values in diagonal (below, dark grey) correspond to the genetic distance between clades by category. Values above (above, blue) refer to the respective standard errors. Bold values on the diagonal refer to the intraclade distance (1 to 31, as referenced in Tables S1 and S3C); References [52,53,54,55,56,57,58,59,60,61,62,63,64,65,66,67,68,69,70,71,72,73,74,75,76,77,78,79,80,81,82,83,84,85,86,87,88,89,90,91,92,93,94,95,96,97,98,99,100,101,102,103,104,105,106,107,108,109,110,111,112,113] are cited in the supplementary materials. Table S3B. Genetic divergence range, by category, and occurrence within subclades, genera, and subfamily; Table S3C. List of alphabetical clade code and respective genus/species and numerical subclade code with respective species.
Author Contributions
Conceptualization: T.d.A.V., L.M.S.-S. and V.F.; methodology: T.d.A.V. and V.F.; software: T.d.A.V.; validation: T.d.A.V. and V.F.; formal analysis: T.d.A.V.; investigation: T.d.A.V.; resources: V.F.; data curation: T.d.A.V., M.M., L.M.S.-S. and V.F.; writing-original draft preparation: T.d.A.V., M.M. and V.F.; writing-review and editing: T.d.A.V., M.M., L.M.S.-S. and V.F.; supervision: L.M.S.-S. and V.F.; project administration: V.F.; funding acquisition: V.F. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES) for the doctoral bursary granted to T.d.A.V. (1170780/2012-6); Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq) for the research bursary granted to L.M.S.S. (PCI-E1 grant 465023/2014-2); and CNPq and Fundação de Amparo à Pesquisa e Inovação do Espírito Santo (FAPES) under Pronem Program to V.F. (CNPq/FAPES 80600417/17).
Institutional Review Board Statement
Ethical review and approval were waived for this study due to most of the samples being donated from the museum collections. The Instituto Chico Mendes de Conservação da Biodiversidade (ICMBio) granted collecting license number 41225-1. In this study the collecting and processing specimens as well as access to the genetic heritage was duly registered with SisGen-Sistema Nacional de Gestão do Patrimônio Genético e dos Conhecimentos Tradicionais Associados, as required by Brazilian law.
Data Availability Statement
The data underpinning the analysis reported in this paper are deposited at GBIF, the Global Biodiversity Information Facility, and are available at https://ipt.pensoft.net/resource?r=trichomycteridae-ufes-2023 (accessed on 9 May 2023).
Acknowledgments
We thank the curators and staff of the collections for providing or helping to acquire specimens and/or tissues used in this study: Luiz Fernando Duboc and Leonardo Ferreira da Silva Ingenito (Coleção Zoológica Norte Capixaba, Universidade Federal do Espírito Santo, CZNC/CEUNES-UFES); Instituto Nacional da Mata Atlântica (INMA); Marcelo Ribeiro de Britto (Museu Nacional do Rio de Janeiro. MNRJ); Daniel Cardoso de Carvalho (Pontifícia Universidade Católica de Minas Gerais, PUC-MG); Carlos Alberto Santos Lucena (Pontifícia Universidade Católica do Rio Grande do Sul, PUC-RS); André Teixeira da Silva (Universidade Estadual do Sudoeste da Bahia, UESB); Jorge Dergam (Universidade Federal de Viçosa, UFV), and Sergio Maia Queiroz Lima (Universidade Federal do Rio Grande do Norte, UFRN). We also thank all the collaborators who helped with the field activities, Juliana de Freitas Justino for molecular laboratory support, and Victor Vale for helping with statistical analysis.
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study, in the collection, analyses or interpretation of data, in the writing of the manuscript, or in the decision to publish the results.
References
- FishBase. Available online: https://www.fishbase.org (accessed on 9 May 2023).
- De Pinna, M.C.C. Phylogenetic relationships of Neotropical Siluriformes (Teleostei: Ostariophysi); historical overview and synthesis of hypothesis. In Phylogeny and Classification of Neotropical Fishes; Malabarba, L.R., Reis, R.E., Vari, R.P., Lucena, Z.M., Lucena, C.A.S., Eds.; Edipucrs: Porto Alegre, Brazil, 1998; pp. 279–330. [Google Scholar]
- De Pinna, M.C.C.; Wosiacki, W.B. Family Trichomycteridae (pencil or parasitic catfishes). In Check List of the Freshwater Fishes of South and Central America; Reis, R.E., Kullander, S.O., Ferraris, C.J., Jr., Eds.; Edipucrs: Porto Alegre, Brazil, 2003; pp. 270–290. [Google Scholar]
- Eschmeyer’s Catalog of Fishes: Genera, Species, References. Available online: http://researcharchive.calacademy.org/research/ichthyology/catalog/fishcatmain.asp (accessed on 10 May 2019).
- Sazima, I. Natural history of Trichogenes longipinnis, a threatened trichomycterid catfish endemic to Atlantic Forest streams in Southeast Brazil. Ichthyol. Explor. Freshw. 2004, 15, 49–60. [Google Scholar]
- Ochoa, L.E.; Roxo, F.F.; Do-Nascimiento, C.; Sabaj, M.H.; Datovo, A.; Alfaro, M.; Oliveira, C. Multilocus analysis of the catfish family Trichomycteridae (Teleostei: Ostariophysi: Siluriformes) supporting a monophyletic Trichomycterinae. Mol. Phylogenetics Evol. 2017, 115, 71–81. [Google Scholar] [CrossRef] [PubMed]
- Katz, A.M.; Barbosa, M.A.; Mattos, J.L.O.; Costa, W.J.E.M. Multigene analysis of the catfish genus Trichomycterus and description of a new South American trichomycterine genus (Siluriformes, Trichomycteridae). Zoosystematics Evol. 2018, 94, 557–566. [Google Scholar] [CrossRef]
- Hayes, M.H.; Paz, H.J.; Stout, C.C.; Werneke, D.C.; Armbruster, J.W. A Hotspot Atop: Rivers of the Guiana Highlands Hold High Diversity of Endemic Pencil Catfish (Teleostei: Ostariophysi: Siluriformes). Biol. J. Linn. Soc. 2020, 129, 862–874. [Google Scholar] [CrossRef]
- De Pinna, M.C.C. A new Sarcoglanidine catfish. phylogeny of its subfamily, and an appraisal of the phyletic status of the Trichomycterinae (Teleostei, Trichomycteridae). Am. Mus. Novit. 1989, 2950, 1–39. [Google Scholar]
- Uieda, V.S.; Castro, R.M.C. Coleta e fixação de peixes de riachos. In Ecologia de Peixes de Riachos; Caramaschi, E.P., Mazzoni, R., Peres-Neto, P.R., Eds.; Universidade Federal do Rio de Janeiro (Oecologia Brasiliensis): Rio de Janeiro, Brazil, 1999; Volume 6, pp. 1–22. [Google Scholar]
- Lucena, C.A.; Calegari, B.; Pereira, E.; Dallegrave, E. O uso de óleo de cravo na eutanásia de peixes. Bol. Soc. Bras. Ictiol. 2013, 105, 20–24. [Google Scholar]
- Close, B.; Banister, K.; Baumans, V.; Bernoth, E.-M.; Bromage, N.; Bunyan, J.; Erhardt, W.; Flecknell, P.; Gregory, N.; Hackbarth, H.; et al. Recommendations for euthanasia of experimental animals Part 2 Report of a Working Party. Lab. Anim. 1997, 31, 1–32. [Google Scholar] [CrossRef]
- Bruford, M.W.; Hanotte, O.; Brookfield, J.F.Y.; Burke, T. Molecular Genetic Analysis of Populations: A Practical Approach; IRL Press: Oxford, UK, 1992; 315p. [Google Scholar]
- Villa-Verde, L.; Lazzarotto, H.; Costa, W.J.E.M.; Lima, S.M.Q. A new Glanapterygine catfish of the genus Listrura (Teleostei: Siluriformes: Trichomycteridae) from southeastern Brazil. Neotrop. Ichthyol. 2012, 10, 527–538. [Google Scholar] [CrossRef]
- Kearse, M.; Moir, R.; Wilson, A.; Stones-Havas, S.; Cheung, M.; Sturrock, S.; Buxton, S.; Cooper, A.; Markowitz, S.; Duran, C.; et al. Geneious Basic: An integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 2012, 28, 1647–1649. [Google Scholar] [CrossRef]
- Xia, X.; Xie, Z.; Salemi, M.; Chen, L.; Wang, Y. An index of substitution saturation and its application. Mol. Biol. Evol. 2003, 26, 1–7. [Google Scholar] [CrossRef]
- Xia, X. DAMBE5: A comprehensive software package for data analysis in molecular biology and evolution. Mol. Biol. Evol. 2013, 30, 1720–1728. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef]
- Darriba, D.; Taboada, G.L.; Doallo, R.; Posada, D. jModelTest 2: More models. new heuristics and parallel computing. Nat. Methods 2012, 9, 772. [Google Scholar]
- Bouckaert, R.; Vaughan, T.G.; Barido-Sottani, J.; Duchêne, S.; Fourment, M.; Gavryushkina, A.; Heled, J.; Jones, G.; Kühnert, D.; De Maio, N.; et al. BEAST 2.5: An advanced software platform for Bayesian evolutionary analysis. PLoS Comput. Biol. 2019, 15, e1006650. [Google Scholar] [CrossRef] [PubMed]
- Tracer Version v1. 5.0. Available online: http://tree.bio.ed.ac.uk/software/tracer (accessed on 5 May 2020).
- Ochoa, L.E.; Datovo, A.; Do-Nascimiento, C.; Roxo, F.F.; Sabaj, M.H.; Chang, J.; Melo, B.F.; Silva, G.S.C.; Foresti, F.; Alfaro, M.; et al. Phylogenomic analysis of trichomycterid catfishes (Teleostei: Siluriformes) inferred from ultraconserved elements. Sci. Rep. 2020, 10, 2697. [Google Scholar] [CrossRef]
- Hillis, D.; Bull, J. An empirical test of bootstrapping as a method for assessing confidence in phylogenetic analysis. Syst. Biol. 1993, 42, 182–192. [Google Scholar] [CrossRef]
- ESRI. Environmental Systems Research Institute. ArcGIS Desktop: Release 10. Redlands. CA: Environmental Systems Research Institute 2011. Available online: http://www.esri.com/software/arcgis/index.html (accessed on 10 April 2021).
- Kimura, M. A simple method for estimating evolutionary rate of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 1980, 16, 111–120. [Google Scholar] [CrossRef] [PubMed]
- R Core Team. R: A Language and Environment for Statistical Computing; Version 4.2.1; R Foundation for Statistical Computing: Vienna, Austria, 2022; Available online: https://www.R-project.org (accessed on 20 July 2022).
- Hardman, M. The phylogenetic relationships among non-diplomystid catfishes as inferred from mitochondrial cytochrome b sequences: The search for the ictalurid sister taxon (Otophysi: Siluriformes). Mol. Phylogenetics Evol. 2006, 41, 636–662. [Google Scholar] [CrossRef]
- Abell, R.; Thieme, L.M.; Revenga, C.; Bryer, M.; Kottelat, M.; Bogutskaya, N.; Coad, B.; Mandrak, N.; Balderas, S.C.; Bussing, W.; et al. Freshwater Ecoregions of the World: A New Map of Biogeographic Units for Freshwater Biodiversity Conservation. BioScience 2008, 58, 403–414. [Google Scholar] [CrossRef]
- Species 2000 & ITIS Catalogue of Life. Available online: www.catalogueoflife.org/col (accessed on 24 February 2020).
- Unmack, P.J.; Bennin, A.P.; Habit, E.M.; Victoriano, P.F.; Johnson, J.B. Impact of ocean barriers, topography, and glaciation on the phylogeography of the catfish Trichomycterus areolatus (Teleostei: Trichomycteridae) in Chile. Biol. J. Linn. Soc. Lond. 2009, 97, 876–892. [Google Scholar]
- Costa, W.J.E.M.; Katz, A. Integrative taxonomy supports high species diversity of south-eastern Brazilian mountain catfishes of the T. reinhardti group (Siluriformes: Trichomycteridae). Syst. Biodivers. 2021, 19, 601–621. [Google Scholar]
- Endersby, J. Lumpers and Splitters: Darwin, Hooker, and the Search for Order. Science 2009, 326, 1496–1499. [Google Scholar] [CrossRef] [PubMed]
- Hubert, N.; Renno, J.F. Historical biogeography of South American freshwater fishes. J. Biogeogr. 2006, 33, 1414–1436. [Google Scholar] [CrossRef]
- Arratia-F, G.; Chang-G, A.; Menu-Marque, S.; Rojas-M, G. About Bullockia gen. nov., Trichomycterus mendozensis n. sp, and Revision of the Family Trichomycteridae (Pisces, Siluriformes). Stud. Neotrop. Environ. 1978, 13, 157–194. [Google Scholar] [CrossRef]
- Ferraris, C.J. Checklist of catfishes, recent and fossil (Osteichthyes: Siluriformes), and catalogue of siluriform primary types. Zootaxa 2007, 1418, 1–628. [Google Scholar] [CrossRef]
- Arratia-F, G.; Huaquin, L. Morphology of the lateral line system and of the skin of diplomystid and certain primitive loricarioid catfishes and systematic and ecological considerations. Bonn. Zool. Monogr. 1995, 36, 1–110. [Google Scholar]
- Mesa-S, L.M.; Lasso, C.A.; Ochoa, L.E.; Do-Nascimiento, C. Trichomycterus rosablanca (Siluriformes, Trichomycteridae) a new species of hipogean catfish from the Colombian Andes. Biota Colomb. 2018, 19, 95–116. [Google Scholar]
- Arratia-F, G.; Menu-Marque, S. Revision of the freshwater catfishes of the genus Hatcheria (Siluriformes, Trichomycteridae) with commentaries on ecology and biogeography. Zool. Anz. 1981, 207, 88–111. [Google Scholar]
- Tchernavin, V.V. A revision of some Trichomycterinae based on material preserved in the British Museum (Natural History). Proc. Zool. Soc. Lond. 1944, 114, 234–275. [Google Scholar] [CrossRef]
- Eigenmann, C.H. The Pigidiidae, a family of South American catfishes. Mem. Carnegie Mus. 1918, 7, 259–398. [Google Scholar] [CrossRef]
- Farias, I.P.; Orti, G.; Sampaio, I.; Schneider, H.; Meyer, A. The cytochrome b gene as a phylogenetic marker: The limits of resolution for analyzing relationships among cichlid fishes. J. Mol. Evol. 2001, 53, 89–103. [Google Scholar] [CrossRef] [PubMed]
- Albert, J.S.; Tagliacollo, V.A.; Dagosta, F. Diversification of Neotropical Freshwater Fishes. Annu. Rev. Ecol. Evol. Syst. 2020, 8, 27–53. [Google Scholar] [CrossRef]
- Albert, J.S.; Carvalho, T.P. Neogene Assembly of Modern Faunas. In Historical Biogeography of Neotropical Freshwater Fishes; Albert, J.S., Reis, R.E., Eds.; University of California Press: Berkeley, CA, USA, 2011; pp. 119–136. [Google Scholar]
- Ramos, V.A.; Aleman, A. Tectonic of the Andes. In Tectonic Evolution of South America; Cordani, U.G., Milani, E.J., Thomaz-Filho, A., Campos, D.A., Eds.; Academia Brasileira de Ciências e Departamento Nacional da Produção Mineral (DNPM): Rio de Janeiro, Brazil, 2000; pp. 635–685. [Google Scholar]
- Lujan, N.K.; Conway, K.W. Life in the fast lane: A review of rheophily in freshwater fishes. In Extremophile Fishes; Riesch, R., Tobler, M., Plath, M., Eds.; Springer: Cham, Switzerland; New York, NY, USA, 2015; pp. 107–136. [Google Scholar]
- Jégu, M.; Santos, G.M. Révision du status de Myleus setiger Muller and Troschel, 1844 et de Myleus knerii (Steindachner, 1881) (Teleostei: Characidae: Serrasalminae) avec une description complémentaire des deux espèces. Cybium 2002, 26, 33–57. [Google Scholar]
- Rodríguez-Tribaldos, V.; White, N.J.; Roberts, G.G.; Hoggard, M.J. Spatial and temporal uplift history of South America from calibrated drainage analysis. Geochem. Geophys. Geosystems 2017, 18, 2321–2353. [Google Scholar] [CrossRef]
- Ribeiro, A.C.; Riccomini, C.; Leite, J.A.D. Origin of the largest South American transcontinental water divide. Sci. Rep. 2018, 8, 17144. [Google Scholar] [CrossRef] [PubMed]
- Cherem, L.F.S.; Varajão, C.A.C.; Braucher, R.; Bourlés, D.; Salgado, A.A.R.; Varajão, A.C. Long-term evolution of denudational escarpments in southeastern Brazil. Geomorphology 2012, 173–174, 118–127. [Google Scholar] [CrossRef]
- Lima, S.M.Q.; Berbel-Filho, W.M.; Vasconcellos, A.V.; Lazoski, C.; Volpi, T.A.; Lazzarotto, H.; Russo, C.A.M.; Tatarenkov, A.; Avise, J.C.; Sole-Cava, A.M. Rio de Janeiro and other paleodrainages evidenced by the genetic structure of an Atlantic Forest catfish. J. Biogeogr. 2021, 48, 1475–1488. [Google Scholar] [CrossRef]
- Carvalho, L.M.V.; Jones, C.; Liebmann, B. The South Atlantic convergence zone: Intensity, form, persistence, and relationships with intraseasonal to interannual activity and extreme rainfall. J. Clim. 2004, 17, 88–108. [Google Scholar]
- Fernandez, L.; Osinaga, K. A new Trichomycterus (Siluriformes: Trichomycteridae) from Aguarague National Park of the Bolivian preandean region, with comments on relationships within of the genus. Environ. Biol. Fish. 2006, 75, 385–393. [Google Scholar]
- Costa, W.J.E.M. Description de huit nouvelles espèces du genre Trichomycterus (Siluriformes: Trichomycteridae), du Brésil oriental. Revue Française d’Aquariologie et Herpétologie 1992, 18, 101–110. [Google Scholar]
- Triques, M.L.; Vono, V. Three new species of Trichomycterus (Teleostei: Siluriformes: Trichomycteridae) from the Rio Jequitinhonha basin, Minas Gerais, Brazil. Ichthyol. Explor. Freshw. 2004, 15, 161–172. [Google Scholar]
- Buckup, P.A.; Menezes, N.A.; Ghazzi, M.A. Catálogo das Espécies de Peixes de Água Doce do Brasil; Série Livros 23; Museu Nacional: Rio de Janeiro, Brazil, 2007; 195p. [Google Scholar]
- Reis, V.J.C.; De Pinna, M.C.C. The type specimens of Trichomycterus alternatus (Eigenmann, 1917) and Trichomycterus zonatus (Eigenmann, 1918), with elements for future revisionary work (Teleostei: Siluriformes: Trichomycteridae). Zootaxa 2019, 4585, 100–120. [Google Scholar]
- Fernandez, L.; Vari, R.P. New Species of Trichomycterus from the Andes of Argentina with a redescription of Trichomycterus alterus (Siluriformes: Trichomycteridae). Copeia 2002, 3, 739–747. [Google Scholar] [CrossRef]
- Lezama, A.Q.; Triques, M.L.; Santos, P.S. Trichomycterus argos (Teleostei: Siluriformes: Trichomycteridae), a new species from the Doce River Basin, Eastern Brazil. Zootaxa 2012, 3352, 60–68. [Google Scholar] [CrossRef]
- Ardila Rodríguez, C.A.A. Cinco Nuevas Especies de Peces Trichomycterus Para la Región Caribe-Colombia; Departamento del Atlántico. Barranquilla 2016, 2, 1–26. [Google Scholar]
- Reis, V.J.C.; Santos, S.A.; De Pinna, M.C.C.; Pessali, T.C. An osteological CT-scan and description of a new species of Trichomycterus (Trichomycteridae: Siluriformes) from the Rio Doce drainage with remarkable similarities with Bullockia. J. Fish. Biol. 2019, 95, 918–931. [Google Scholar]
- Ferrer, J.; Malabarba, L.R. Taxonomic review of the genus Trichomycterus Valenciennes (Siluriformes: Trichomycteridae) from the laguna dos Patos system, Southern Brazil. Neotrop. Ichthyol. 2013, 11, 217–246. [Google Scholar]
- Castellanos-Morales, C.A.; Galvis, F. Las especies del género Trichomycterus (Siluriformes: Trichomycteridae) en Colombia. Bol. Cient. Mus. Hist. Nat. 2012, 16, 194–206. [Google Scholar]
- Fernandez, L.A. Redescription of the teleost Trichomycterus barbouri (Eigenmann, 1911), occurrence in Argentina and comparison with related species (Ostariophysi: Siluriformes: Trichomycteridae). Stud. Neotrop. Fauna Environ. 2000, 35, 27–33. [Google Scholar] [CrossRef]
- Arratia, G.; Menu-Marque, S. New catfishes of the genus Trichomycterus from the high Andes of South America (Pisces, Siluriformes) with remarks on distribution and ecology. Zool. Jahrb. Abt. Syst. 1984, 111, 493–520. [Google Scholar]
- Barbosa, M.A.; Costa, W.J.E.M. Seven new species of the catfish genus Trichomycterus (Teleostei: Siluriformes: Trichomycteridae) from southeastern Brazil and redescription of T. brasiliensis. Ichthyol. Explor. Freshw. 2010, 21, 97–122. [Google Scholar]
- Ardila-Rodríguez, C.A.A. Trichomycterus cachiraensis (Siluriformes: Trichomycteridae), nueva especie del rio cachira, cuencia del rio Magdalena, Colombia, Dahlia. Rev. Asoc. Colomb. Ictiol. 2008, 10, 33–41. [Google Scholar]
- Lima, S.M.Q.; Lazzarotto, H.; Costa, W.J.E.M. A new species of Trichomycterus (Siluriformes: Trichomycteridae) from lagoa Feia drainage, southeastern Brazil. Neotrop. Ichthyol. 2008, 6, 315–322. [Google Scholar] [CrossRef]
- Barbosa, M.A.; Costa, W.J.E.M. Validade, relações filogenéticas e redescrição de Eremophilus candidus Ribeiro, 1949 (Teleostei, Siluriformes, Trichomycteridae). Arq. Mus. Nac. 2003, 61, 179–188. [Google Scholar]
- De Pinna, M.C.C. Trichomycterus castroi, a new species of trichomycterid catfish (Teleostei: Silurifomes). Ichthyol. Explor. Freshw. 1992, 3, 89–95. [Google Scholar]
- Fernandez, L.; Vari, R.P. New Species of Trichomycterus (Teleostei: Siluriformes: Trichomycteridae) Lacking a Pelvic Fin and Girdle from the Andes of Argentina. Copeia 2000, 4, 990–996. [Google Scholar] [CrossRef]
- Alencar, A.R.; Costa, W.J.E.M. Description of two new species of the catfish genus Trichomycterus from southeastern Brazil (Siluriformes: Trichomycteridae). Zootaxa 2004, 744, 1–8. [Google Scholar]
- Lasso, C.A.; Provenzano, F. Dos nuevas especies de bagres del género Trichomycterus (Siluriformes: Trichomycteridae) de la Gran Sabana, Escudo de las Guayanas, Venezuela. Rev. Biol. Trop. 2003, 50, 1139–1149. [Google Scholar]
- Wosiacki, W.B.; De Pinna, M.C.C. Trichomycterus igobi, a new catfish species from the rio Iguaçu drainage: The largest head in Trichomycteridae (Siluriformes: Trichomycteridae). Neotrop. Ichthyol. 2008, 6, 17–23. [Google Scholar] [CrossRef]
- Rizzato, P.P.; Costa, E.P.D., Jr.; Trajano, E.; Bichuette, M.E. Trichomycterus dali: A new highly troglomorphic catfish (Silurifomes: Trichomycteridae) from Serra da Bodoquena, Mato Grosso do Sul State, Central Brazil. Neotrop. Ichthyol. 2011, 9, 477–491. [Google Scholar] [CrossRef]
- Bockmann, F.A.; Casatti, L.; De Pinna, M.C.C. A new species of trichomycterid catfish from the Rio Paranapanema basin, southeastern Brazil (Teleostei: Siluriformes), with comments on the phylogeny of the family. Ichthyol. Explor. Freshw. 2004, 15, 225–242. [Google Scholar]
- Barbosa, M.A. Description of two new species of the catfish genus Trichomycterus (Teleostei: Siluriformes: Trichomycteridae) from the coastal river basins, southeastern Brazil. Vertebr. Zool. 2013, 63, 269–275. [Google Scholar]
- Lima, S.M.Q.; Costa, W.J.E.M. Trichomycterus giganteus (Siluriformes: Loricarioidea: Trichomycteridae): A new catfish from the Rio Guandu basin, southeastern Brazil. Zootaxa 2005, 761, 1–6. [Google Scholar] [CrossRef]
- Fernández, L.; Schaefer, S.A. New Trichomycterus (Siluriformes: Trichomycteridae) from an offshore Island of Colombia. Copeia 2005, 1, 68–76. [Google Scholar]
- Do-Nascimiento, C.; Prada-Pedreros, S.; Guerrero-Kommritz, J. Trichomycterus venulosus (Steindachner, 1915), a junior synonym of Eremophilus mutisii Humboldt, 1805 (Siluriformes: Trichomycteridae) and not an extinct species. Neotrop. Ichthyol. 2014, 12, 707–715. [Google Scholar]
- Wosiacki, W.B. A new species of Trichomycterus (Siluriformes: Trichomycteridae) from south Brazil and redescription of T. iheringi (Eigenmann). Zootaxa 2005, 1040, 49–64. [Google Scholar]
- Fernández, L.; Vari, R.P. New species of Trichomycterus from the Andean Cordillera of Argentina (Siluriformes: Trichomycteridae). Copeia 2009, 1, 195–202. [Google Scholar]
- Caramaschi, E.P.; Caramaschi, U. Taxonomic Status of the Trichomycterid Catfish Trichomycterus itatiayae. Copeia 1991, 1, 222–224. [Google Scholar] [CrossRef]
- Barbosa, M.A.; Costa, W.J.E.M. Description of a new species of catfish from the upper rio Paraíba do Sul basin, south-eastern Brazil (Teleostei: Siluriformes: Trichomycteridae) and re-description of Trichomycterus itatiayae. Int. J. Ichthyol. 2008, 14, 176–186. [Google Scholar]
- Wosiacki, W.B.; Oyakawa, O.T. Two new species of the catfish genus Trichomycterus (Siluriformes: Trichomycteridae) from the rio Ribeira de Iguape Basin, Southeastern Brazil. Neotrop. Ichthyol. 2005, 3, 465–472. [Google Scholar]
- Barbosa, M.A.; Costa, W.J.E.M. Trichomycterus macrophthalmus (Teleostei: Siluriformes: Trichomycteridae), a new species of catfish from the Paraíba do Sul River basin, southeastern Brazil. Vertebr. Zool. 2012, 62, 79–82. [Google Scholar] [CrossRef]
- Barbosa, M.A.; Costa, W.J.E.M. Description of a new species of the catfish genus Trichomycterus (Teleostei: Siluriformes: Trichomycteridae) from the rio Paraíba do Sul basin, southeastern Brazil. Vertebr. Zool. 2010, 60, 193–197. [Google Scholar] [CrossRef]
- Bockmann, F.A.; Sazima, I. Trichomycterus maracaya, a new catfish from the upper rio Paraná, southeastern Brazil (Siluriformes: Trichomycteridae), with notes on the T. brasiliensis species complex. Neotrop. Ichthyol. 2004, 2, 61–74. [Google Scholar]
- Wosiacki, B.W.; Garavello, J.C. Five new species of Trichomycterus from the rio Iguaçu (rio Paraná Basin), southern Brazil (Siluriformes: Trichomycteridae). Ichthyol. Explor. Freshw. 2004, 15, 1–16. [Google Scholar]
- Fernandez, L.; Chuquihuamani, R.Q. Species of Trichomycterus (Siluriformes: Trichomycteridae) from the Andean Cordillera of Perú, with comments on relationships within the genus. Zootaxa 2007, 1545, 49–57. [Google Scholar]
- Reis, V.J.C.; dos Santos, S.A.; Britto, M.R.; Volpi, T.A.; De Pinna, M.C.C. Iterative taxonomy reveals a new species of Trichomycterus Valenciennes 1832 (Siluriformes, Trichomycteridae) widespread in rio Doce basin: A pseudocryptic of T. immaculatus. J. Fish Biol. 2020, 97, 1607–1623. [Google Scholar]
- Fernandez, L.; Vari, R.P. New Species of Trichomycterus (Teleostei: Siluriformes) from the Andean Cordillera of Argentina and the Second Record of the Genus in Thermal Waters. Copeia 2012, 4, 631–636. [Google Scholar] [CrossRef]
- Ardila-Rodriguez, C.A. Trichomycterus nietoi sp. nov. (Siluriformes: Trichomycteridae) Una nueva especie de pez del Río Guachaca Sierra Nevada de Santa Marta, Departamento del Magdalena, Colombia. Barranquilla 2014, 4, 1–24. [Google Scholar]
- Costa, W.J.E.M.; Katz, A.M.; Mattos, J.L.O.; Amorim, P.F.; Mesquita, B.O.; Vilardo, P.J.; Barbosa, M.A. Historical review and redescription of three poorly known species of the catfish genus Trichomycterus from south-eastern Brazil (Siluriformes: Trichomycteridae). J. Nat. Hist. 2020, 53, 2905–2928. [Google Scholar]
- Ardila Rodriguez, C.A. Trichomycterus maldonadoi (Siluriformes, Trichomycteridae), especie nueva de la cuenca alta del rio Sinú, Colombia. Dahlia 2011, 11, 13–21. [Google Scholar]
- Alencar, A.R.; Costa, W.J.E.M. Trichomycterus pauciradiatus, a new catfish species from the upper rio Paraná basin. southeastern Brazil (Siluriformes: Trichomycteridae). Zootaxa 2006, 1269, 43–49. [Google Scholar]
- Sarmento-Soares, L.M.; Zanata, A.M.; Martins-Pinheiro, R.F. Trichomycterus payaya, new catfish (Siluriformes: Trichomycteridae) from headwaters of rio Itapicuru. Bahia. Brazil. Neotrop. Ichthyol. 2011, 9, 261–271. [Google Scholar]
- Datovo, A.; Carvalho, M.; Ferrer, J. A new species of the catfish genus Trichomycterus from the La Plata River basin, southern Brazil, with comments on its putative phylogenetic position (Siluriformes: Trichomycteridae). Zootaxa 2012, 3327, 33–44. [Google Scholar]
- Barbosa, M.A.; Azevedo-Santos, V.M. A new species of the catfish genus Trichomycterus (Teleostei: Siluriformes: Trichomycteridae) from the rio Paraná basin, southeastern Brazil. Vertebr. Zool. 2012, 62, 357–362. [Google Scholar]
- Barbosa, M.A.; Costa, W.J.E.M. Trichomycterus potschi (Siluriformes: Trichomycteridae): A new species of trichomycterid catfish from coastal streams of southeastern Brazil. Ichthyol. Explor. Freshw. 2003, 14, 281–287. [Google Scholar]
- Sarmento-Soares, L.M.; Martins-Pinheiro, R.F.; Aranda, A.T.; Chamon, C.C. Trichomycterus pradensis. a new catfish from southern Bahia coastal rivers. northeastern Brazil (Siluriformes: Trichomycteridae). Ichthyol. Explor. Freshw. 2005, 16, 289–302. [Google Scholar]
- Fernandez, L.; Vari, R.P. New species of Trichomycterus from mid elevation localities of northwestern Argentina (Siluriformes: Trichomycteridae). Copeia 2004, 4, 876–882. [Google Scholar]
- Barbosa, M.A.; Costa, W.J.E.M. Trichomycterus puriventris (Teleostei: Siluriformes: Trichomycteridae), a new species of catfish from the rio Paraíba do Sul basin, southeastern Brazil. Vertebr. Zool. 2012, 62, 155–160. [Google Scholar] [CrossRef]
- Bichuette, M.E.; Rizzato, P.P. A new species of cave catfish from Brazil, Trichomycterus rubbioli sp.n., from Serra do Ramalho karstic area, São Francisco River basin, Bahia State (Silurifomes: Trichomycteridae). Zootaxa 2012, 3480, 48–66. [Google Scholar]
- Ardila-Rodriguez, C.A.A. Trichomycterus ruitoquensis (Siluriformes: Trichomycteridae) una nueva especie de pez de la cuenca alta del río Lebrija, Departamento de Santander-Colombia. Barranquilla 2007, 3, 1–20. [Google Scholar]
- Castellanos-Morales, C.A. Trichomycterus sketi: A new species of subterranean catfish (Siluriformes: Trichomycteridae) from the Andean Cordillera of Colombia. Biota Colombiana 2011, 11, 33–41. [Google Scholar]
- Barbosa, M.A.; Costa, W.J.E.M. Description of a new species of the catfish genus Trichomycterus (Teleostei: Siluriformes: Trichomycteridae) from the rio de Contas basin, northeastern Brazil. Vertebr. Zool. 2011, 61, 307–312. [Google Scholar]
- Fernandez, L.; Miranda, G. A catfish of the genus Trichomycterus from a thermal stream in southern South America (Teleostei. Siluriformes. Trichomycteridae), with comments on relationships within the genus. J. Fish. Biol. 2007, 71, 1303–1316. [Google Scholar] [CrossRef]
- Wosiacki, W.B. New species of the catfish Trichomycterus (Siluriformes: Trichomycteridae) from the headwaters of the rio São Francisco basin, Brazil. Zootaxa 2004, 592, 1–12. [Google Scholar] [CrossRef]
- Ferrer, J.; Malabarba, L.R. A new Trichomycterus lacking pelvic fins and pelvic girdle with a very restricted range in Southern Brazil (Siluriformes: Trichomycteridae). Zootaxa 2011, 2912, 59–67. [Google Scholar]
- Castellanos-Morales, C.A. Trichomycterus uisae: A new species of hypogean catfish (Siluriformes: Trichomycteridae) from the northeastern Andean Cordillera of Colombia. Neotrop. Ichthyol. 2008, 6, 307–314. [Google Scholar] [CrossRef][Green Version]
- Fernandez, L.; Schaefer, S.A. Trichomycterus yuska, a new species from high elevations of Argentina (Siluriformes: Trichomycteridae). Ichthyol. Explor. Freshw. 2003, 14, 353–360. [Google Scholar]
- Ferrer, J.; Malabarba, L.R. Systematic revision of the Neotropical catfish genus Scleronema (Siluriformes:Trichomycteridae), with descriptions of six new species from Pampa grasslands. Neotrop. Ichthyol. 2020, 18, e190081. [Google Scholar] [CrossRef]
- Lima, S.M.Q.; Vasconcellos, A.V.; Berbel-Filho, W.M.; Lazoski, C.; Russo, C.A.; Sazima, I.; Solé-Cava, A.M. Effects of Pleistocene climatic and geomorphological changes on the population structure of the restricted-range catfish Trichogenes longipinnis (Siluriformes: Trichomycteridae). Syst. Biodivers. 2016, 14, 155–170. [Google Scholar]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).