Poseidonibacter ostreae sp. nov., Isolated from the Gut of Ostrea from the Seomjin River
Abstract
1. Introduction
2. Material and Methods
2.1. Isolation and Ecology
2.2. 16S rRNA Phylogeny
2.3. Genome Features
2.4. Morphological, Physiological, and Biochemical Characterization
2.5. Chemotaxonomic Characterization
3. Results and Discussion
3.1. Isolation of Strains
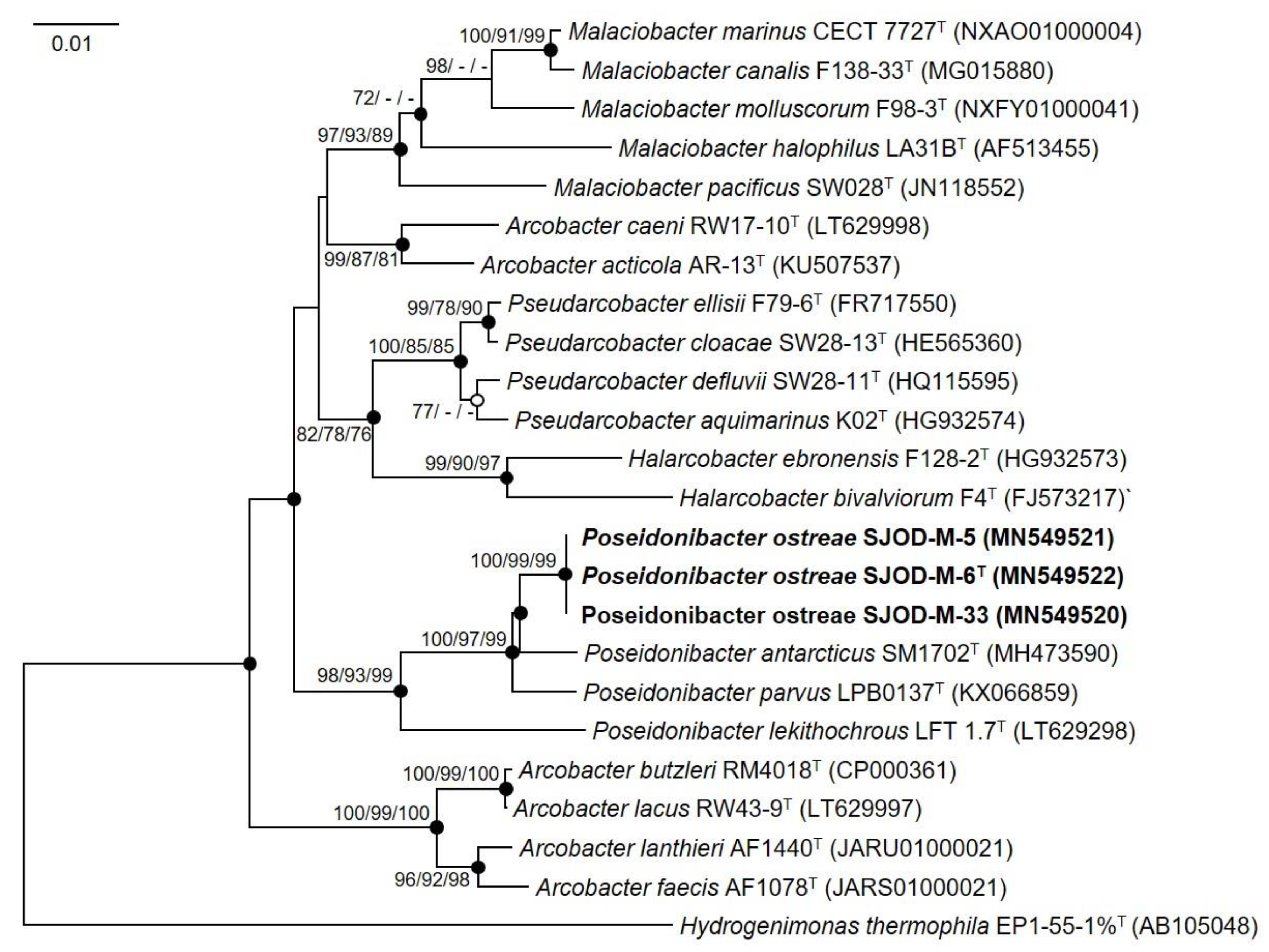
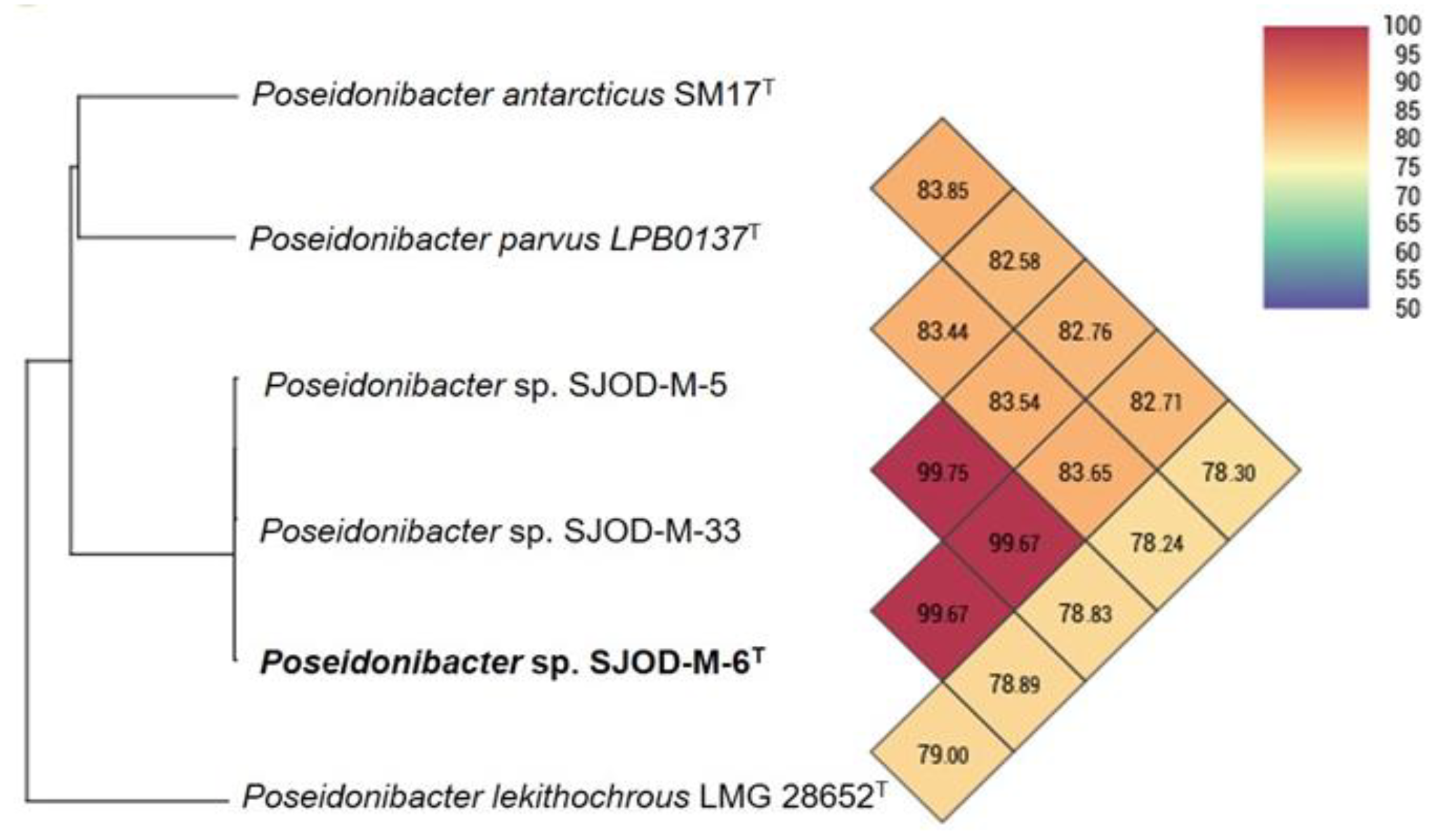
3.2. 16S rRNA Phylogeny
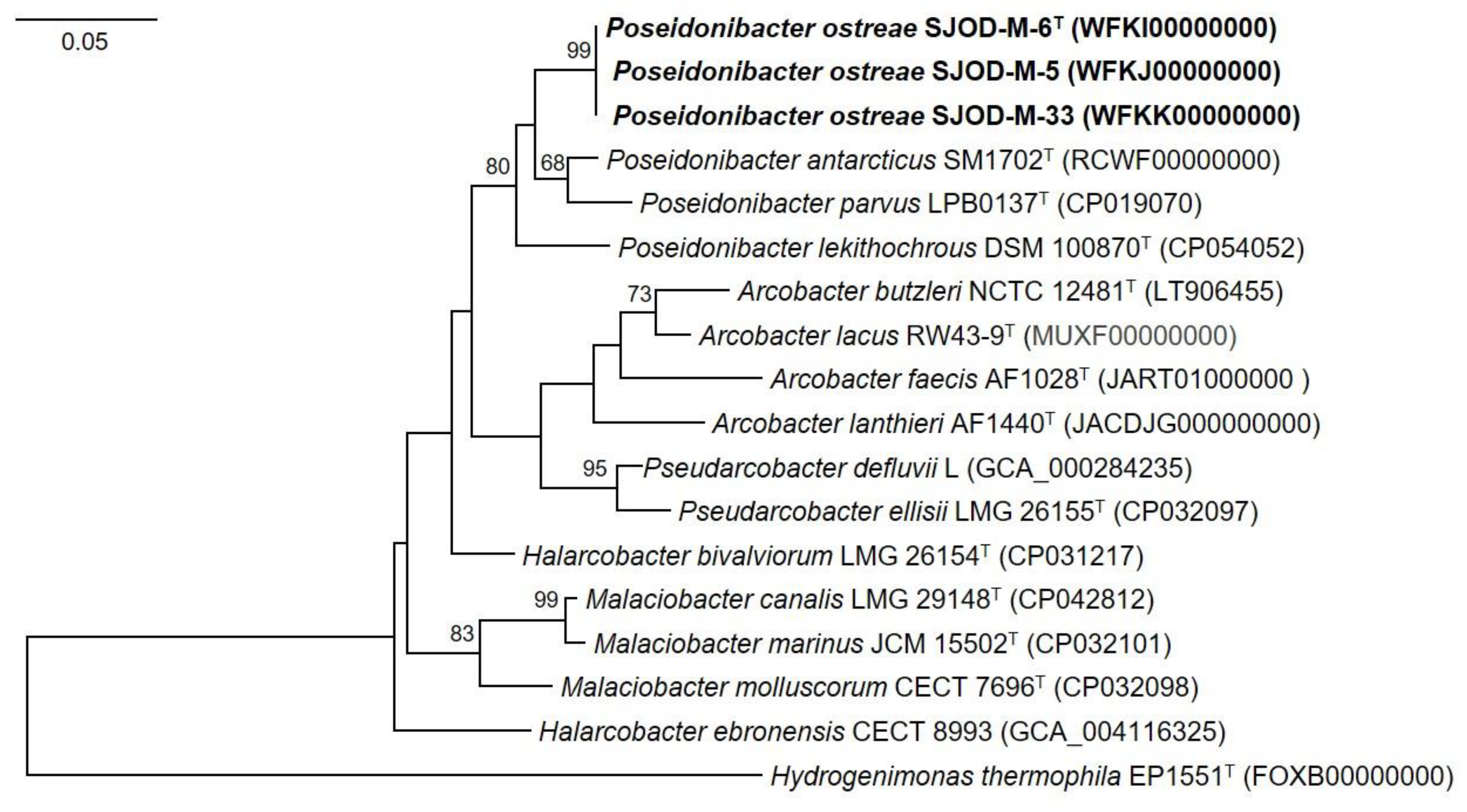
3.3. Genome Features
3.4. Morphological, Physiological, and Biochemical Characteristics
3.5. Chemotaxonomic Characterization
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
References
- Diéguez, A.L.; Balboa, S.; Magnesen, T.; Romalde, J.L. Arcobacter lekithochrous sp. nov., isolated from a molluscan hatchery. Int. J. Syst. Evol. Microbiol. 2017, 67, 1327–1332. [Google Scholar] [CrossRef]
- Diéguez, A.L.; Pérez-Cataluña, A.; Figueras, M.J.; Romalde, J.L. Arcobacter haliotis Tanaka et al. 2017 is a later heterotypic synonym of Arcobacter lekithochrous Diéguez et al. 2017. Int. J. Syst. Evol. Microbiol. 2018, 68, 2851–2854. [Google Scholar] [CrossRef] [PubMed]
- Guo, X.H.; Wang, N.; Yuan, X.X.; Zhang, X.Y.; Chen, X.L.; Zhang, Y.Z.; Song, X.Y. Poseidonibacter antarcticus sp. nov., isolated from Antarctic intertidal sediment. Int. J. Syst. Evol. Microbiol. 2019, 69, 2717–2722. [Google Scholar] [CrossRef] [PubMed]
- Kim, M.J.; Baek, M.-G.; Shin, S.-K.; Yi, H. Poseidonibacter parvus sp. nov., isolated from a squid. Int. J. Syst. Evol. Microbiol. 2021, 71, 004590. [Google Scholar] [CrossRef] [PubMed]
- Fisher, T.R.; Carlson, P.R.; Barber, R.T. Carbon and nitrogen primary productivity in three Noreth Carolina estuaries. Estuar. Coast. Shelf Sci. 1982, 15, 621–644. [Google Scholar] [CrossRef]
- Barlow, J.P. Physical and biological processes determining the distribution of zooplankton in a tidal estuary. Biol. Bull. 1955, 109, 211–225. [Google Scholar] [CrossRef]
- Min, D.K.; Lee, J.S.; Koh, D.B.; Je, J.G. Mollusks in Korea; Hangul Graphics: Busan, Republic of Korea, 2004; pp. 410–415. [Google Scholar]
- Noseworthy, R.G.; Lee, H.-J.; Choi, S.-D.; Choi, K.-S. Unique substrate preference of Ostrea denselamellosa Lischke, 1869 (Mollusca: Ostreidae) at Haechang Bay, on the south coast of Korea. Korean J. Parasitol. 2016, 32, 31–36. [Google Scholar] [CrossRef][Green Version]
- Lane, D.J. 16S/23S rRNA sequencing. In Nucleic Acid Techniques in Bacterial Systematics; Stackebrandt, E., Goodfellow, M., Eds.; John Wiley and Sons: New York, NY, USA, 1991; pp. 115–175. [Google Scholar]
- Yoon, S.H.; Ha, S.M.; Kwon, S.; Lim, J.; Kim, Y.; Seo, H.; Chun, J. Introducing EzBioCloud: A taxonomically united database of 16S rRNA gene sequences and whole-genome assemblies. Int. J. Syst. Evol. Microbiol. 2017, 67, 1613–1617. [Google Scholar] [CrossRef]
- Jeon, Y.S.; Lee, K.; Park, S.C.; Kim, B.S.; Cho, Y.J.; Ha, S.M.; Chun, J. EzEditor: A versatile sequence alignment editor for both rRNA- and protein-coding genes. Int. J. Syst. Evol. Microbiol. 2014, 64, 689–691. [Google Scholar] [CrossRef] [PubMed]
- Saitou, N.; Nei, M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987, 4, 406–425. [Google Scholar]
- Felsenstein, J. Evolutionary trees from DNA sequences: A maximum likelihood approach. J. Mol. Evol. 1981, 17, 368–376. [Google Scholar] [CrossRef] [PubMed]
- Fitch, W.M. Toward defining the course of evolution: Minimum change for a specific tree topology. Syst. Zool. 1971, 20, 406–416. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [PubMed]
- Felsenstein, J. Confidence limits on phylogenies: An approach using the bootstrap. Evolution 1985, 39, 783–791. [Google Scholar] [CrossRef] [PubMed]
- Kimura, M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 1980, 16, 111–120. [Google Scholar] [CrossRef]
- William, S.; Feil, H.; Copeland, A. Bacterial genomic DNA Isolation Using CTAB. Joint Genome Institute, Walnut Creek, CA, USA, 2012. Joint Genome Institute. Bacterial Genomic DNA Isolation Using CTAB. 2012. Available online: https://jgi.doe.gov/wp-content/uploads/2014/02/JGI-Bacterial-DNA-isolation-CTAB-Protocol-2012.pdf (accessed on 7 July 2023).
- Chun, J.; Oren, A.; Ventosa, A.; Christensen, H.; Arahal, D.R.; da Costa, M.S.; Rooney, A.P.; Yi, H.; Xu, X.W.; De Meyer, S.; et al. Proposed minimal standards for the use of genome data for the taxonomy of prokaryotes. Int. J. Syst. Evol. Microbiol. 2018, 68, 461–466. [Google Scholar] [CrossRef]
- Wick, R.R.; Judd, L.M.; Gorrie, C.L.; Holt, K.E. Unicycler: Resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput. Biol. 2017, 13, e1005595. [Google Scholar] [CrossRef]
- Aziz, R.K.; Bartels, D.; Best, A.A.; DeJongh, M.; Disz, T.; Edwards, R.A.; Formsma, K.; Gerdes, S.; Glass, E.M.; Kubal, M.; et al. The RAST server: Rapid annotations using subsystems technology. BMC Genom. 2008, 9, 75. [Google Scholar] [CrossRef]
- Tatusova, T.; DiCuccio, M.; Badretdin, A.; Chetvernin, V.; Nawrocki, E.P.; Zaslavsky, L.; Lomsadze, A.; Pruitt, K.D.; Borodovsky, M.; Ostell, J. NCBI prokaryotic genome annotation pipeline. Nucleic Acids Res. 2016, 44, 6614–6624. [Google Scholar] [CrossRef]
- Lee, I.; Chalita, M.; Ha, S.M.; Na, S.I.; Yoon, S.H.; Chun, J. Contest16S: An algorithm that identifies contaminated prokaryotic genomes using 16S rRNA gene sequences. Int. J. Syst. Evol. Microbiol. 2017, 67, 2053–2057. [Google Scholar] [CrossRef]
- Blin, K.; Shaw, S.; Augustijn, H.E.; Reitz, Z.L.; Biermann, F.; Alanjary, M.; Fetter, A.; Terlouw, B.R.; Metcalf, W.W.; Helfrich, E.J.N.; et al. antiSMASH 7.0: New and improved predictions for detection, regulation, chemical structures and visualization. Nucleic Acids Res. 2023, 51, W46–W50. [Google Scholar] [CrossRef] [PubMed]
- Kanehisa, M.; Goto, S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000, 28, 27–30. [Google Scholar] [CrossRef] [PubMed]
- Lee, I.; Kim, Y.O.; Park, S.-C.; Chun, J. OrthoANI: An improved algorithm and software for calculating average nucleotide identity. Int. J. Syst. Evol. Microbiol. 2016, 66, 1100–1103. [Google Scholar] [CrossRef] [PubMed]
- Auch, A.F.; von Jan, M.; Klenk, H.-P.; Göker, M. Digital DNA–DNA hybridization for microbial species delineation by means of genome-to-genome sequence comparison. Stand Genom. Sci. 2010, 2, 117–134. [Google Scholar] [CrossRef] [PubMed]
- Na, S.I.; Kim, Y.O.; Yoon, S.H.; Ha, S.M.; Baek, I.; Chun, J. UBCG: Up-to-date bacterial core gene set and pipeline for phylogenomic tree reconstruction. J. Microbiol. 2018, 56, 280–285. [Google Scholar] [CrossRef]
- Price, M.N.; Dehal, P.S.; Arkin, A.P. FastTree: Computing large minimum evolution trees with profiles instead of a distance matrix. Mol. Biol. Evol. 2009, 26, 1641–1650. [Google Scholar] [CrossRef]
- Choo, Y.-J.; Lee, K.; Song, J.; Cho, J.-C. Puniceicoccus vermicola gen. nov., sp. nov., a novel marine bacterium, and description of Puniceicoccaceae fam. nov., Puniceicoccales ord. nov., Opitutaceae fam. nov., Opitutales ord. nov. and Opitutae classis nov. in the phylum ‘Verrucomicrobia’. Int. J. Syst. Evol. Microbiol. 2007, 57, 532–537. [Google Scholar] [CrossRef]
- Kim, H.; Choo, Y.-J.; Cho, J.-C. Litoricolaceae fam. nov., to include Litoricola lipolytica gen. nov., sp. nov., a marine bacterium belonging to the order Oceanospirillales. Int. J. Syst. Evol. Microbiol. 2007, 57, 1793–1798. [Google Scholar] [CrossRef]
- Smibert, R.M.; Krieg, N.R. Phenotypic characterization. In Methods for General and Molecular Bacteriology; Gerhardt, P., Murray, R.G.E., Wood, W.A., Krieg, N.R., Eds.; American Society for Microbiology: Washington, DC, USA, 1994; pp. 607–654. [Google Scholar]
- Tindall, B.J.; Sikorski, J.; Smibert, R.A.; Krieg, N.R. Phenotypic Characterization and the Principles of Comparative Systematics. In Methods for General and Molecular Microbiology; Reddy, C.A., Ed.; ASM Press: Washington, DC, USA, 2007; pp. 330–393. [Google Scholar]
- Sasser, M. Identification of bacteria by gas chromatography of cellular fatty acids. Technol. Note 2001, 101. [Google Scholar]
- Minnikin, D.E.; O’Donnell, A.G.; Goodfellow, M.; Alderson, G.; Athalye, M.; Schaal, A.; Parlett, J. An integrated procedure for the extraction of bacterial isoprenoid quinones and polar lipids. J. Microbiol. Methods 1984, 2, 233–241. [Google Scholar] [CrossRef]
- Jain, C.; Rodriguez-R, L.M.; Phillippy, A.M.; Konstantinidis, K.T.; Aluru, S. High throughput ANI analysis of 90K prokaryotic genomes reveals clear species boundaries. Nat. Commun. 2018, 9, 5114. [Google Scholar] [CrossRef] [PubMed]
- Richter, M.; Rosselló-Móra, R. Shifting the genomic gold standard for the prokaryotic species definition. Proc. Natl. Acad. Sci. USA 2009, 106, 19126–19131. [Google Scholar] [CrossRef] [PubMed]
| Characteristic | 1 | 2 | 3 | 4 | 5 | 6 |
|---|---|---|---|---|---|---|
| Isolation source | Oyster | Oyster | Oyster | Squid † | Sediment ‡ | Scallop § |
| Colony color on MA | Ivory | Ivory | Ivory | Pinkish-brown † | Creamy yellow ‡ | Brown § |
| Temperature range (Optimum) (°C) | 10–30 (25) | 10–30 (25) | 10–30 (25) | 10–25 (25) † | 0–30 (10–15) ‡ | 15–25 (25) § |
| NaCl tolerance (%, w/v) | 1.0–5.0 | 1.0–5.0 | 1.0–5.0 | 2.0–4.5 † | 0.5–5.0 ‡ | 0.0–8.0 § |
| API 20NE: | ||||||
| Arginine dihydrolase | + | + | + | − | + | + |
| Indole production | − | − | − | + | − | + |
| glucose fermentation | − | − | − | − | + | |
| Enzyme activity of: | ||||||
| Esterase (C4) | + | + | + | + | + | |
| Acid phosphatase | − | − | − | + | + | + |
| Alkaline phosphatase | − | − | − | + | − | + |
| Leucine arylamidase | − | − | − | − | + | − |
| Utilization of carbon sources (Biolog GN2): | ||||||
| D-sorbitol | + | + | + | + | + | − |
| α-D-lactose, malonic acid, propionic acid, succinic acid, L-aspartic acid, L-glutamic acid, glycerol | + | + | + | + | − | + |
| L-rhamnose | + | + | + | − | + | + |
| D-arabitol | + | + | + | + | − | − |
| D-mannitol | + | + | + | − | + | − |
| D-cellobiose, D-mannose, D-melibiose | + | + | + | − | − | + |
| β-methyl-D-glucoside, quinic acid, glycyl-L-aspartic acid, L-pyroglutamic acid | + | + | + | − | − | − |
| L-arabinose, L-alanine | − | − | − | + | + | − |
| Sucrose, D-trehalose, L-leucine, L-serine | − | − | − | + | − | + |
| α-D-glucose, maltose, D-alanine | − | − | − | − | + | + |
| Lactulose | − | − | − | + | − | − |
| N-acetyl-D-glucosamine | − | − | − | − | + | − |
| D-fructose, α-hydroxybutyric acid, D-saccharic acid, L-histidine, γ-amino butyric acid | − | − | − | − | − | + |
| Hydrolysis of: | ||||||
| Starch | + | + | + | − | − | − |
| DNA | − | − | − | − | + | + |
| DNA G + C content * (mol%) | 27.5 | 27.5 | 27.6 | 27.7 | 27.1 | 28.5 |
| Genome size (Mb) | 2.86 | 2.96 | 3.21 | 2.87 | 2.92 | 3.57 |
| Fatty Acid | 1 | 2 | 3 | 4 | 5 | 6 |
|---|---|---|---|---|---|---|
| C12:0 | 1.2 | 1.1 | Tr | 2.3 | 4.3 | Tr |
| C12:0 3-OH | Tr | Tr | 1.0 | - | 5.6 | - |
| C14:0 | 2.6 | 2.7 | 3.0 | 8.2 | 7.4 | 3.1 |
| C16:0 | 11.7 | 11.9 | 11.9 | 8.5 | 11.9 | 12.0 |
| C16:1 ω5c | 2.8 | 2.6 | 2.8 | Tr | Tr | 1.4 |
| Summed features * | ||||||
| 2 (C14:0 3-OH and/or C16:1 iso-I) | 6.2 | 6.3 | 6.4 | 11.2 | 5.9 | 7.5 |
| 3 (C16:1 ω7c and/or C16:1 ω6c) | 56.7 | 57.2 | 56.8 | 56.8 | 45.1 | 54.8 |
| 8 (C18:1 ω7c and/or C18:1 ω6c) | 17.9 | 17.3 | 17.9 | 12.1 | 18.8 | 15.6 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Baek, K.; Jang, S.; Chung, E.J.; Ryu, S.H.; Choi, A. Poseidonibacter ostreae sp. nov., Isolated from the Gut of Ostrea from the Seomjin River. Diversity 2023, 15, 920. https://doi.org/10.3390/d15080920
Baek K, Jang S, Chung EJ, Ryu SH, Choi A. Poseidonibacter ostreae sp. nov., Isolated from the Gut of Ostrea from the Seomjin River. Diversity. 2023; 15(8):920. https://doi.org/10.3390/d15080920
Chicago/Turabian StyleBaek, Kiwoon, Sumin Jang, Eu Jin Chung, Shi Hyun Ryu, and Ahyoung Choi. 2023. "Poseidonibacter ostreae sp. nov., Isolated from the Gut of Ostrea from the Seomjin River" Diversity 15, no. 8: 920. https://doi.org/10.3390/d15080920
APA StyleBaek, K., Jang, S., Chung, E. J., Ryu, S. H., & Choi, A. (2023). Poseidonibacter ostreae sp. nov., Isolated from the Gut of Ostrea from the Seomjin River. Diversity, 15(8), 920. https://doi.org/10.3390/d15080920







