Abstract
In wild boar, Sus scrofa, from Europe, domestic pig-typical ancestry is traced at varying levels. We hypothesised wild boar with pig-typical gene pool characteristics, i.e., “introgression”, congregate more in peri-urban habitats, because of less shyness and better adaptation to anthropogenic stress. We used 16 microsatellites to study introgression levels of 375 wild boar from peri-urban Vienna, Austria, and rural regions in comparison to commercial slaughter pigs, Mangaliza, and Turopolje pigs. We also expected more introgression in locations of warmer climates and lower precipitation. Despite discrimination of wild boar and pigs with 99.73% and 97.87% probability, respectively, all wild boars exhibited pig-typical gene pool characteristics, mostly at a very low level. Recent hybridisation was suspected in only 0.53% of wild boar, corresponding to the current largely indoor pig breeding/rearing in the region, with no chance of natural gene exchange between pigs and wild boar. Rather, pig ancestry in wild boar stems from incomplete gene pool differentiation during domestication and/or historical introgressions, when free-ranging pig farming was common. Individual introgression levels were lower in wild boar from peri-urban habitats, possibly reflecting the largely historical absence of pig farms there. Moreover, a marginal precipitation effect, but no temperature effect on introgression was observed. The latter, however, needs to be explored further by a more comprehensive data set.
1. Introduction
In recent years, wild boar, Sus scrofa L., 1758, have increased in numbers and have expanded their ranges, e.g., [1], and they are increasingly being considered as pests or lead to conflicts with humans, not only in agricultural or forestry contexts, but also in peri-urban and metropolitan regions where they are seemingly aggregating [2,3] and possibly adapting to urban environments [4].
An earlier review [1] found overall relatively few pig-typical characteristics in gene pools of wild boar from different regions throughout Europe and concluded that, considering studies that have applied different molecular approaches, hybridisation had a minor effect on gene pool variability of wild boar. Nevertheless, hybridisation of wild boar and pigs has been reported from different parts of Europe, e.g., [5] and studies cited therein, at varying levels. The latter study also reported on substantial bi-directional hybridisation between Bulgarian wild boar and the free-ranging East Balkan Pig by MC1R coat colour genes, but very little introgression of pigs (i.e., East Balkan Pig and Commercial Pig) into wild boar as estimated by a microsatellite panel and as evidenced by a genetic structure and admixture analysis. A recent study [6] identified eight out of nine wild boar individuals from two locations in eastern Austria as hybrids, using SNP (i.e., single nucleotide polymorphism)-based screening across large parts of Europe.
Given the notable global increase in ambient temperature over the last decennia resulting in milder and shorter winters in many parts of Europe, e.g., [7], it is conceivable that wild boar with larger portions of pig-typical gene pool characteristics, i.e., “introgressed” hereafter, may have a higher chance of survival—particularly piglets and yearling in regions of central and eastern Europe with relatively high ambient temperature and low precipitation, because of less adaptation to unfavourable weather. Moreover, introgressed wild boar may also be preadapted to tolerate higher anthropogenic stress levels in their habitat and may thus aggregate in peri-urban or urban habitats.
In this study, we use a microsatellite marker system to identify levels of pig-typical gene pool characteristics in wild boar from eastern Austria, with a focus on peri-urban Vienna, with increased human activities in forested areas (especially the “Vienna Forest”), the preferred habitat of wild boar. There, frequent disturbances by people seeking relaxation, particularly on weekends and in the warm season of the year, when piglets and young are developing, may lead to increased stress in wild boars. Introgressed individuals may thus have a better chance of survival and aggregate in such peri-urban habitats, possibly due to their higher stress tolerance and reduced shyness. Introgressed sows may also carry advantageous gene variants for various reproductive traits in local wild boar populations [8], possibly in combination with lower aggressiveness, a behavioural characteristic of domestic pigs compared to wild boar. In fact, according to an experimental study [9], reduced maternal aggressiveness under favourable feeding conditions led to reduced juvenile mortality in wild boar. Pig-typical gene pool characteristics associated with better reproductive success could therefore accumulate in peri-urban habitats or with improved food availability and favourable climatic conditions.
However, given that pig breeding and rearing systems in eastern Austria nowadays represent largely commercial agro-industrial production units practically with no chance of gene exchange between pigs and wild boar, we examine if and to what extent pig-typical gene pool characteristics in wild boar can be attributed to recent hybridisation or to historical introgressions and incomplete gene pool differentiation between wild boar and pigs, e.g., [10]. Historical introgressions could have occurred when pig rearing was common on small family farms in eastern Austria, with pigs under semi-free ranging conditions, probably, at least occasionally, until the second half of the 20th century, after many generations of pig pannage in medieval times and later.
We also considered possible climatic effects (ambient temperature and precipitation) on the spatial distribution of introgression levels, as particularly the modern breeds are clearly less robust against cold or hot and wet weather, e.g., [11], than their wild counterparts (or older races such as Mangaliza and Turopolje pigs that were usually reared under free- or semi-free conditions in historical times). In particular, piglets and young during their growth phase need favourable weather for survival, especially in the face of their high energy demands for growth and immune defence, e.g., [12].
Specifically, we tested the following hypotheses:
- Signals of historical introgression of pigs or of incomplete gene pool differentiation between wild boar and pigs are prevailing rather than ones of current hybridisation among “introgressed wild boar”, and current hybridisation does not strongly affect or increase the genetic diversity, i.e., allelic richness, of wild boar;
- Introgression occurs more frequently and individually to a greater extent in habitats of peri-urban Vienna than in rural habitats, as a high proportion of pig-typical gene pool characteristics may be associated with reduced shyness of an individual and introgressed wild boar may generally be more resistant to anthropogenic stress;
- Introgression occurs more often and at higher individual levels at locations of relatively high ambient temperature and low precipitation, as favourable climate increases the chance specifically of piglets and young wild boar harbouring many pig-typical gene variants.
2. Material and Methods
2.1. Samples, Molecular Markers, and Laboratory Analyses
This study is based on a total of 375 individual genetic samples of georeferenced free-ranging wild boars from 50 locations (hunting grounds) or local sample areas in eastern Austria, including peri-urban and rural habitats of the larger environs of Vienna, and a few from transitional locations (Figure 1). No samples from the “Lainzer Tiergarten”, a 2450 ha wooded enclosure of wild boar and other wildlife in peri-urban Vienna, were used. Only 366 wild boar samples were used for our statistical models of the individual proportions of pig-typical gene pool characteristics, because some individuals had more than five loci with uncertain/missing genotypes; this might have distorted the estimation of the introgression proportions too much.
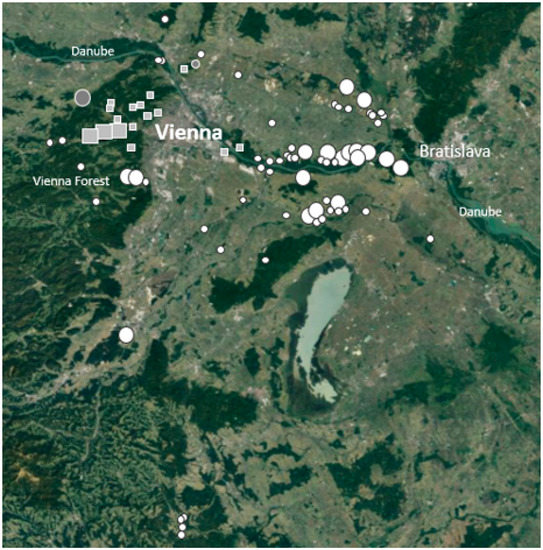
Figure 1.
Sample distribution of wild boar. Grey squares—peri-urban Vienna, close to areas of relatively dense human population and activities; white circles—rural locations; grey circles—transitional location; small symbols—1–5 individuals and big symbols—>5 individuals.
In addition, 57 domestic pig samples purchased as packages of commercial pork from supermarkets in Vienna, Austria (n = 7), Znojmo and Břeclav, Czech Republic (n = 14), Dresden, Germany (n = 8), Mosonmagyaróvár, Hungary (n = 9), Ljubljana, Slovenia (n = 12), and Sofia, Bulgaria (n = 7) were included, as well as muscle samples of Hungarian (n = 4) and Serbian (n = 21) Mangaliza pigs and four samples of Turopolje pigs from a breeder in the province of Lower Austria, purchased from one of his distributors (“Regional Bio-Greisslerei“, Gars am Kamp). The commercial pork samples originated from diverse—usually unspecified—industrial indoor pig farms, with no chance of interactions of pigs with free-ranging wild boar. The Mangaliza and Turopolje pig breeds have been reared in free- or semi-free-ranging systems in historical times (Mangaliza mainly in Hungary and Serbia, and Turopolje in Croatia), with particularly the Mangaliza pigs being still under semi-free or free-rearing in several regions of Serbia. To our knowledge, those two breeds are also reared under fenced outdoor conditions at diverse locations in Hungary and Austria, such as the provinces of Lower and Upper Austria, Burgenland, and Styria. The currently studied Hungarian Mangaliza samples were purchased from a butcher (Fleischhauerei Karlo) at Pamhagen, Burgenland, eastern Austria, who imported them with official permits from Hungary by commercial trade. Whether or not those Mangaliza pigs were maintained in a semi-free-ranging farming system could not be determined. All pork samples were purchased in 2017 and 2018.
All wild boar samples were collected from individuals shot during regular hunts or found dead as road casualties between 2011 and 2018. The organ tissue (e.g., liver, muscle, skin, and cartilage) samples were frozen at −20 °C or at −80 °C as soon as possible after collection, which might, however, have taken more than one day in some cases, particularly for those from wild boar brought to our institute for necropsy.
To assess the extent of nuclear gene pool characteristics typical for domestic pigs in wild boar, we employed microsatellite markers following several studies, e.g., [6,13,14,15,16]. Specifically, we scored individual genotypes at the following sixteen dinucleotide microsatellites (Table 1) on a 3130 x l Genetic Analyzer (Applied Biosystems, Fischer Scientific (Austria) GmbH, Vienna, Austria) in six multiplex sets consisting of a maximum of three microsatellite loci with nonoverlapping ranges, respectively, and different dyes: S0002, S0097, S0101, S0155, S0215, SW24, SW72, SW122, SW240, SW461, SW857, SW936, SW1492, SW2021, SW2496, and SW2532. Details of DNA extraction, purification, and STR scoring using the GeneMapper program (v4.0, Fischer Scientific Austria GmbH, Vienna, Austria), as well as null allele and linkage disequilibrium (LD) checks were reported by an earlier study [10,17].

Table 1.
Allelic variation at the sixteen microsatellite loci of wild boars (WB) and pigs (PI), associated indices of genetic variability, and locus-specific inbreeding coefficients (fis). Ho—observed heterozygosity; He—expected heterozygosity, corrected for small sample sizes; AL—number of locus-specific alleles; Arange—range of allele sizes at each locus; Amofr—most frequent allele and its frequency (FrAmofr); Apriv—number of private alleles (i.e., occurring only in wild boars or in pigs; and *—indicates significant departure of fis from zero, based on 1000 permutations (Genetix vers. 4. 05.2) and after strict Bonferroni corrections for multiple testing based on α = 0.05.
2.2. Population Genetic Statistics
We used IDENTIX vers. 1.1.5 as of 3 April 2003 [18] to calculate pairwise Queller and Goodnight rxy indices [19] of relationship, separately for each local sample or regional sample area, to test for unrelatedness of the individuals and to single out the identity of pairs of individual composite genotypes potentially due to sample confusion or identical twins. Low numbers of identical individual composite genotypes would indicate a relatively high power of genetic resolution of individuals, whereas many identical composite genotypes would reduce the level of statistical independence of the data set and lead to distorted population genetic statistics. Specifically for the pork samples, it was important not to use the tissue of identical individuals. We further used GENETIX vers. 4.05.2 as of 5 May 2004 [20] to calculate allele frequencies, linkage disequilibrium, mean numbers of alleles per locus (A), observed (Ho) and expected heterozygosity (He), and F-statistics of wild boar and domestic pigs as well as to run a ten-factorial correspondence analysis (FCA) for wild boar and pigs. The latter multivariate statistics were particularly intended to check for the power of genetic distinction between wild boar and pigs and to identify possible individuals of admixed/hybrid status. The FSTAT vers. 2.1 program [21] was used to calculate population-specific values of allelic richness (Rs) based on a rarefaction approach to account for different sample sizes. Additionally, we used GeneClass2 vers. 2.0.h as of 2 August 2005 [22] to evaluate our microsatellite marker system in terms of its capability to distinguish between wild boars and pigs by estimating the likelihood of each individual genotype being assigned to wild boar or pigs (see Ref. [23]). Specifically, we applied the Bayesian approach of [24] and computed individual likelihood values by using the resampling algorithm of [25] with an assignment threshold of scores of 0.01 and a type-1 error (alpha) of 0.01 for 1000 simulated individuals. We used the same software to identify first-generation migrants between domestic pigs and wild boar in combination with inspection of suspect allele and genotype frequencies. Wild boar individuals with many pig-typical alleles or genotypes might be identified as if they were first-generation migrants from the pig group to the wild boar group, indicating, indeed, hybrids or backcrosses. Specifically, we run likelihoods employing the −log(L_home/L_max) approach based on 10,000 individual simulations at a probability level < 0.01. Finally, we estimated the levels of relative genetic variation between pigs and wild boar, between individuals within each of the two groupings, and within individuals by an analysis of molecular variance (AMOVA) implemented in ARLEQUIN vers. 3.11 (February 2007; see Ref. [26]).
2.3. Genetic Admixture Analyses and Rationale of Quantification of Introgression
We used STRUCTURE vers. 2.3.4 [27,28,29] (http://pritch.bsd.uchicago.edu) to assess the proportion (Q) of pig-typical gene pool characteristics of each individual wild boar and pig. Generally, this software allows for exploring the genetic admixture of an individual’s overall genotype and spatial or group-specific structure in an overall microsatellite genotype data set. Using STRUCTURE, numbers of genetic clusters (K), basically Hardy–Weinberg populations, underlying the overall genetic data can be inferred from the likelihood distributions of model iterations assuming a certain number of K. It also indicates the estimated percentage (Q) of each K occurring in each individual overall genotype. Q values of pig-typical K in wild boar individuals can be used to estimate their level of introgression. Specifically, we run admixture models based on correlated allele frequencies with and without population priors (i.e., with/without information on the location of each wild boar sample and assigning all pig samples to one separate population), with the following specifications: 250,000 Monte Carlo Markov Chain (MCMC) repetitions after 100,000 burn-in lengths, with an initial alpha = 1.0 for K = 1–15 K and 10 iterations per K. We also calculated mean, maximum, minimum, and standard deviation of ln [Pr(XjK)] for each K and the ad hoc statistics of the second order rate of change in the likelihood function with respect to K [30], by using the STRUCTURE HARVESTER online platform [31].
To specifically calculate the level of introgression by pig-typical K for each wild boar individual, we used the averaged (over iterations) sum of Q values from three genetic clusters, typical for domestic pigs as resulting from our structure and admixture analysis described above, based on the most likely number of K inherent to the total data set (i.e., K = 13, see Results).
2.4. Climate Data and Statistical Modeling of Introgression Level in Wild Boar
Following our hypotheses of introgressed wild boar being more common and individual introgression being more pronounced at locations of milder climate, as specifically piglets and young might be less resistant against wet and cold weather conditions [12,32], we focused on ambient temperature and precipitation in our statistical models of individual Q values of pig-typical K in wild boar.
We used the WORLDCLIM data set for 2.5 min intervals (https://www.worldclim.org/ (accessed on 23 November 2022)) to automatically extract all sample location-specific temperature and precipitation variables using DIVA-GIS vers. 7.5 (https://www.diva-gis.org/download (accessed on 23 November 2022)). We performed two principal component analyses (PCA), one for the temperature variables and one for the precipitation variables, to obtain separate principal components (factors) for temperature and precipitation, which we tested in our statistical models for their respective effects on the individual Q values of pig-typical K in wild boars.
Specifically, we used standardised Box-Cox-transformed variables bio 1, 5, 6, 8, 9, 10, and 11 for the correlation-matrix-based PCA to extract temperature factors and standardised Box-Cox-transformed variables bio 12, 13, 14, 16, 17, 18, and 19 for the PCA to extract precipitation factors (for respective climate variables see Table 2). Four temperature variables (bio 2, 3, 4, and 7) and one precipitation variable (bio 15) were excluded from PCAs, as those variables represented merely derived ones from the original panel. The variable transformations were carried out to achieve at least an approximation of the multidimensional normality of residuals and to reduce variances, which is recommended for PCA. The individual scores of the resultant principal components (i.e., factors—see Results) were used in our statistical models of averaged Q values of pig-typical K in wild boar individuals. As PCA aims to reduce the numbers of (correlated or uncorrelated) variables, we used the common option to consider only resultant principal components with eigenvalues above 1.0, and that yielded factors explaining high percentages of the underlying data variability (see Results).

Table 2.
Arithmetic means of the sample location-specific bioclimatic variables and respective loadings of the transformed variables into the extracted principal components (tempfac—general temperature factor; precipfac1—first precipitation factor; and precipfac2—second precipitation factor (see running text for interpretation). For bioclimatic variables see (1).
We used the R platform [33,34] for running generalised additive models (GAM) of individual sums of Q values of the three pig-typical K revealed by STRUCTURE with the individual geographic coordinates and the scores of the temperature and precipitation factors obtained from the PCAs. We used the information theory-based approaches of multi-model inference, i.e., model ranking by the Akaike Information Criterion (AICc, corrected for small sample sizes) and model averaging [35] to determine the best model and the “Relative Variance Importance” (RVI) of the explanatory variables (temperature, precipitation, longitude, and latitude) for the variation of the considered individual Q values. The RVI values are equivalent to the probabilities of the explanatory variables to be present in the best model, and we considered values equal to or above 0.7 as statistically important, i.e., indicating a statistical effect on the dependent variable [33,35]. We run two separate suites of models, one for Q values obtained from STRUCTURE iterations without and one with population priors (i.e., sample locations). Prior to the model runs, we tested for multicollinearity among the independent factors, which was not the case (Tolerance < 10, VIF > 0.1) for all factors, and allowed the inclusion of all factors in the models.
The syntax of our global models was as follows:
where Qtr was the standardised Box-Cox-transformed individual percentage of pig-typical K (i.e., the sum of all three K typical for commercial slaughter pigs, Turopolje, as well as Mangaliza from Serbia and Hungary averaged over all iterations by STRUCTURE; see Results), long and lat were the sampling locality-specific geographical coordinates, tempfac was the individual score of the general temperature factor, and precipfac1 and precipfac2 were the individual scores of the two obtained precipitation factors. For long and lat values, we used a spline for their interaction, i.e., a non-linear function estimate for combined long and lat, to allow for potential non-linear variation across the whole study area.
Qtr~s(long*lat) + tempfac + precipfac1 + precipfac2
3. Results
3.1. Allelic Diversity and Genetic Differentiation between wild Boars and Pigs
All rxy indices of pairwise individual relationships for wild boar were lower than 1.0, which indicated the absence of identical composite genotypes among individuals when calculated separately for regional sample arrays. However, five commercial slaughter pig samples were disregarded for further analyses because of the identity of pieces from single pork packages that strongly suggested samples of identical individuals. Overall, we found 147 alleles at the sixteen loci analysed, with 142 alleles being present in wild boar and 109 alleles in pigs (Table 1). A total of 104 alleles were shared by wild boar and pigs; 38 alleles were recovered exclusively in wild boar, whereas 5 were found exclusively in pigs. However, both the wild boar-specific and the pig-specific alleles occurred at low frequencies, respectively (for wild boars: mean = 3.74%, s.d. = 4.94%, 0.13–21%; for pigs: mean = 3.62%, s.d. = 4.48%, 1.16–11.59%). Overall, there was a moderately positive correlation (rs = 0.444, p < 0.001, and n = 104) between frequencies of alleles shared by wild boar and pigs. Observed (Ho) and unbiased expected (He) heterozygosity amounted to 0.606 and 0.722, respectively, in wild boar and to 0.501 and 0.681 in pigs, respectively, and mean numbers of alleles per locus amounted to 8.875 in wild boar and to 6.813 in pigs. Overall allelic richness (Rs) that accounted for different sample sizes (based on the minimum sample size of n = 76 individuals acc. to the FSTAT algorithm) was significantly (p = 0.011, Wilcoxon matched-pairs signed rank test) higher in the wild boar (mean over all 16 loci = 7.81, min = 4.0, and max = 13.42) than in the pigs (mean = 5.78, min = 3.0, and max = 11.88). Only 24.62% of the allelic variability was conveyed by the ten factors of our FCA, with the first factor conveying only 4.53% and the tenth factor reflecting 1.6% of the allelic variability. Only the individual scores of FCA factors 1 and 4 differed significantly (p < 0.001, respectively, after strict Bonferroni correction of multiple Kruskal–Wallis tests) between wild boar and pigs. Nevertheless, almost all individuals could be correctly identified as wild boar or pigs by those two factors (Figure 2).

Figure 2.
Factorial correspondence analysis (FCA). Plot of individual scores of factors 1 and 4 of the ten-factorial FCA, conveying 7.31% of allelic variability.
Our Geneclass simulations resulted in false classifications of only one single wild boar and two pigs, indicating a 99.73% probability of wild boar and 97.87% probability of pigs being correctly identified. They also indicated two wild boars as “first-generation migrants” from pigs, i.e., their allele/genotype composition fitted better to that of pigs; conversely, two pigs (one Czech pork sample and one Hungarian Mangaliza sample) were identified as “first-generation migrants” from wild boar, i.e., their allele/genotype composition fitted better to those of wild boar. The two wild boars concerned one individual of unrecorded sex and age class from a location close to the Slovak border (Eckartsau) and one older male from Hinterbrühl, south of Vienna. Their respective Ho was 0.625% and 0.563%, in line with the average observed and expected heterozygosity for the total wild boar sample. The former individual harboured several alleles with higher frequencies in pigs than in wild boar and vice versa. The same held for the latter individual, which also harboured a few alleles that were generally absent in pigs or at very low frequency but, on the contrary, also a few alleles that were very rare in wild boar.
No significant linkage disequilibrium was found when correcting for multiple testing, separately in wild boar and pigs. However, both in wild boar and pigs, a significant (p < 0.001) deviation from Hardy–Weinberg expectation of overall genotype frequencies was found. The overall relative genetic differentiation between wild boar and pigs amounted to 0.107% (Fst, p < 0.005, 1000 permutations), and the absolute genetic differentiation amounted to 0.086 (Cavalli-Sforza and Edwards distance, p < 0.005, 1000 permutations). Our AMOVA model indicated that 10.65% of the overall relative allelic variability was due to partitioning into pigs (incl. Mangaliza and Turopolje) and wild boar, whereas 14.0% were partitioned among individuals within the two groupings, and 75.34% were due to within-individual variation.
3.2. Genetic Admixture and Introgression of Wild Boar
Our STRUCTURE model runs based on the individual composite genotypes of both wild boar and pigs suggested K = 13 inherent in the total data set, both for the models without and with population priors (see Figure 3 for an example of individual admixture resulting from an iteration without population priors). Our conclusion of K = 13 was based on the distribution characteristics of the likelihood values (Ln(PD)) for each K and additionally on the comparison of minimum likelihood values per K; it was corroborated by checking for increased biologically relevant information for each additional cluster, as suggested by [28,29]. However, we did not follow the results of the ad hoc statistics according to [30], as it suggested only K = 2 for the overall data set, both for iterations with and without population priors, which was clearly underestimating the underlying pattern of partitioning.

Figure 3.
STRUCTURE analysis of wild boars and pigs. Exemplary plot of genetic admixture of individuals (columns) based on an iteration without population priors, i.e., without information on individual sample locality, and assuming thirteen genetic clusters (K). Coloured segments represent different K at various frequencies (Q) per individual. All columns refer to wild boars, except for those that are specifically indicated as pigs. MP (H)—Mangaliza pigs from Hungary and TP—Turopolje pigs. Even though each wild boar individual showed small portions of pig-typical genetic clusters, they are often not visible in the present example, because of their very small Q values (see current text).
Rather, three genetic clusters were characteristic for pigs, i.e., one for Serbian Mangaliza, one for Hungarian Mangaliza, and only one common to all commercial slaughter pig samples and the few Turopolje pigs, but ten clusters were present in wild boar, both for models without and with population priors. Remarkably, some wild boar individuals were largely uniform in their genetic make-up, i.e., they showed almost one exclusive or at least one clearly predominating K, whereas others were quite admixed, with several Ks at variable Q (Figure 3). A comprehensive analysis of spatial differentiation and gene flow patterns of wild boar in the study region will be published elsewhere.
Basically, all wild boar individuals harboured pig-typical K (i.e., their combined mean Q values of typical K of slaughter pigs, Mangaliza pigs from Serbia and Hungary, and Turopolje pigs were greater than zero), but mostly at low or very low frequency; specifically, Q for iterations without/with population priors were as follows: mean = 3.70%/4.10%, median = 1.97%/2.65%, s.d. = 5.09%/4.31%, min = 0.6%/0.5%, and max = 47.1%/34.6%. In fact, 15.7% of all wild boar harboured mean Q < 1%, and 50.9% of all wild boar had mean Q < 2% of the summed pig-typical K, as estimated by model runs with and without population priors, respectively. The by far dominating pig-typical signal in wild boar was that of commercial slaughter pigs (incl. Turopolje pigs). Similarly, all pigs harboured wild boar-typical K, but also predominantly at low overall Q (Table 3).

Table 3.
Sums of percentages (Q values) of wild boar-typical genetic clusters (K) in domestic pigs; sample sizes of groups are in parentheses. Q values averaged over the STRUCTURE iterations based on runs without/with population priors are given in first/second rows, respectively.
3.3. PCA of local Climate Data
The PCA of the standardised Box-Cox-transformed Bioclim data of ambient temperature for the wild boar sampling localities resulted in only one principal component that explained 95.35% of the variability of the data, with high positive loadings of all variables (Table 2). Hence, that factor could be interpreted as a general temperature factor. The PCA of the precipitation variables yielded two principal components, the first explaining 83.6% of data variability and the second explaining 14.81%, totalling 98.41%. The first principal component (precipfac1) had high loadings of all initial values and could therefore be interpreted as a general precipitation factor. The second principal component (recipfac2), however, had only moderate positive loadings of bio 14, 17, and 19 and negative loadings of bio 12, 13, 16, and 18. Thus, individual precipfac2 scores could be interpreted as conveying to some extent relatively low overall precipitation, specifically during the warm period of the year (when there is relatively much precipitation in the study region), but relatively high precipitation during the cold period of the year (when there is relatively little precipitation in the study region). Relatively high individual precipfac2 scores indicated locations with low annual and specifically low summer precipitation (June–August), but relatively high winter precipitation (i.e., December–February, for Vienna). In contrast, small or negative precipfac2 scores indicated sample locations with higher overall and specifically higher summer precipitation, but relatively low winter precipitation.
3.4. Models of Introgression Levels (Qtr values) in Wild Boars
The explained deviances of the GAMs of Qtr values of pig-typical K based on STRUCTURE runs without and with population priors amounted to 17.5% and 53.1%, respectively. The independent variables s(long*lat), i.e., the spline of the interaction factor longitude by latitude and precipfac2 (precipitation factor 2), were contained in the best models of both approaches, respectively. Equally, the model averaging indicated statistically important RVI values of s(long*lat) and precipfac2 for the models of Qtr values resulting from STRUCTURE interactions both without and with population priors, respectively (see Table 4). The averaged precipfac2 coefficients amounted to 0.00437 for the models based on STRUCTURE iterations without population priors and to 0.011748 for the models with population priors. No simple geographical pattern of the Qtr values of pig-typical K in wild boar was evident, but the modelled Qtr values were clearly higher at rural and the few transitional sample locations than at peri-urban sample locations of Vienna (Figure 4).

Table 4.
Values of relative variable importance (RVI, sum of weights) of the independent factors in the generalised additive models (GAMs) based on Qtr values obtained from STRUCTURE iterations without (RVI-NP) and with (RVI-PP) population priors (see Material and Methods). s(long*lat)—spline of interaction factor longitude by latitude, precipfac2—precipitation factor 2, precipfac1—precipitation factor1, and tempfac—temperature factor (see above). Statistically important RVI values [36] are in bold, indicating statistical effects on the individual level of pig-typical gene pool characteristics in wild boar individuals.

Figure 4.
Predicted Qtr values (dots—means; ranges—95% c.i.) of pig-typical genetic clusters in wild boar individuals for peri-urban, transitional, and rural sample/habitat locations in relation to Vienna, as resulting from our GAM based on the STRUCTURE results with population priors and the statistically relevant fixed factors (i.e., accounting for geographical and precipfac2 effects). Low predicted Qtr values correspond to low pig-typical gene pool characteristics.
4. Discussion
4.1. Introgression in Wild Boar
Our current study revealed pig-typical nuclear gene pool characteristics virtually in all free-ranging wild boar individuals from eastern Austria, a region where the species currently occurs at high densities (5.7–8.2 individuals per km2), as estimated from modelling hunting bags [36]. The wild boar range in our study region is particularly connected to ranges of similarly high density in the northeast, east, and southeast, i.e., the Czech Republic, Slovakia, Hungary, and Slovenia, and coincides with a high stock density of pigs in terms of swine heads and holdings [36]. In conformity with our results, an earlier study [6] identified eight out of nine wild boar individuals from two locations in eastern Austria as hybrids with domestic pigs. That finding was based on a panel of SNPs, applying a threshold of >10% of individual genomes showing domestic pig ancestry. The 10% threshold was chosen because the average wild boar ancestry over all populations studied was >90% in the above-cited study. Although we cannot perform a direct comparison, when applying the same approach and a mean of pig-typical ancestry of Q = 3.7% and Q = 4.1% to our wild boar samples as resulting from our STRUCTURE iterations with and without population priors, we arrived at 24.5% and 31.5% of hybrids, respectively. However, a direct comparison of our estimates with those of the SNP-based study cited above (or any other SNP-based study) is not useful, because of the clearly higher gene pool resolution of SNP-based studies. Furthermore, as the distributions of the summed Q values of wild boar-typical genetic clusters in wild boar individuals (and correspondingly also the summed Q values of pig-typical K) were generally strongly skewed, medians rather than means of the distributions of Q values of pig-typical K would be more adequate for determining the thresholds. Correspondingly, the estimated frequencies of hybrids for our currently studied wild boar amounted to 49.9% and 49.3% when based on medians and the STRUCTURE results without and with population priors, respectively.
Nevertheless, given the generally very low level of pig-typical ancestry signals in our analysed wild boar (by far, most individual Q values were clearly below the 10% level), we consider most of them as not signalling “hybrids”, indicative of very recent hybridisation. Rather, we interpret most of them as indicating common gene pool characteristics that were shared by wild boar and regional pig breeds (“land races”) in historical times, when pigs used to be reared in open or semi-open rather extensive farming systems. In historical times those often free-ranging local breeds had chances over many generations of introgressing widely in wild boar and vice versa, and have likely formed a certain genetic basis of the modern commercial slaughter pigs produced nowadays in large indoor breeding systems. Even though we cannot rule out occasional gene flow from pigs currently reared in outdoor (fenced) farming systems in eastern and southeastern Austria, such as Mangaliza pigs or, e.g., crossbreeds of Duroc and Schwäbisch-Hällische pigs (according to various commercial advertisings on the internet), by far most of the pork being currently produced for diverse food chains, supermarkets, or regional butchers in Austria and the international market stems from industrially organised indoor swine farming units without the chance of contact between pigs and wild boar.
4.2. Historical Gene Flow and Current Pig Rearing
Probably, at least until the early 20th century and possibly even until the 1950s, pig holdings in the study region and adjacent regions were mostly in small family farms and, to some extent, in partly free-ranging systems. In fact, at least during the Austro-Hungarian period throughout the 19th and early 20th centuries, extensive pig farming of free-ranging local pig breeds such as the Szalonta pig east of the River Danube and the Bakony pig in more western parts of the Hungarian Kingdom (which included parts of eastern Austria until 1921, i.e., the area of today’s province of Burgenland) was the prevailing system of rearing pigs [37]. Hungarian Mangaliza resulted probably largely from crossbreeding those two regional Hungarian breeds (from 1833 onward). The Sumadija pig, a Serbian breed, and/or the Syrmia pig from Croatia, usually reared also under free-ranging conditions, most likely contributed to Mangaliza pig gene pools, but to our knowledge, it is unclear to what extent and to which of the currently acknowledged three lines. Mangaliza pigs were commonly also reared under free-ranging conditions, and to our knowledge, “Mangulica” are still under free-ranging conditions in parts of Serbia. Even still today, commercial domestic pigs are at least occasionally reared under semi-free conditions even without swineherds in parts of Romania (own observations by FS; see also [38]). Seemingly, huge numbers of pigs were roaming freely in these areas of eastern central Europe and the Balkans. Statistics on farm animals listed 3,571,728 pigs in 1857 and 6,447,143 pigs in 1895 in the Hungarian Kingdom, and, for instance, in 1894, 73% of Hungarian Mangaliza pigs were exported to central and western Europe, with the intention to improve local pig breeds, e.g., [39,40]. Almost every second pig that was slaughtered in the central slaughterhouse of Vienna at St. Marx in 1889 originated from Hungary [41], in addition to cattle, buffalo, and other farm animals. Hence, we can infer plenty of chances of gene flow between pigs and wild boar over many generations during those times, and most likely later as well, when the local pig breeds of central Europe that were reared in semi-free ranging systems (with or without swineherds) formed the basis of the development of the main commercial breeds that are today industrially bred for pork production (in eastern-central Europe and parts of the Balkans). At least for Austria (and according to our findings, seemingly also for other countries in central and eastern-central Europe as well as the Balkans) that standard commercial breed is a crossbreed between the female German Landrace (Deutsche Veredelte Landrasse) and German Edelschwein (Deutsches Edelschwein) and male Pietrain [42]. The former two breeds were developed by the end of the 19th century in Germany from existing ancestor landraces, while the Pietrain pig originated from Belgium and increased in its commercial importance for industrial pig breeding in central Europe only during the second half of the 20th century. Remarkably, this quite standardised breeding type of commercial slaughter pigs from Austria revealed the same and only one genetic cluster according to our marker system, seemingly very much predominating the pork samples from supermarkets in Austria, the Czech Republic, eastern Germany, Hungary, Slovenia, and Bulgaria, even though we purchased them in different supermarket chains.
Mangaliza-typical genome signals are comparatively rare in commercial slaughter pigs, according to our microsatellite marker system. Indeed, Mangaliza was nearly wiped out and substituted by other breeds during the communistic period in Hungary, attempting to standardise pig breeding for cooperative farming in line with the USSR agro-policy (pers. comm. C. Gedeon, Budapest). On the other hand, despite pigs being isolated in industrialised breeding systems in eastern Austria, our STRUCTURE results indicated wild boar-typical signals in commercial slaughter pigs often at a low level, but nevertheless, on average, still at a higher level than that for pig-typical ancestry signals in wild boars. Given the fair assumption of largely neutral evolution of the currently used markers, this latter finding reinforces our interpretation of pig-typical ancestry signals in wild boar as indicating historical gene pool characteristics common to ancestors of the current commercial pig breeds and wild boar in the study region rather than hybrids of recent origin.
4.3. Possible Recent Hybrids and Genetic Diversity
Still, given the successful distinction between pigs and wild boar, as demonstrated by our FAC, Bayesian assignment, and admixture analysis, we may interpret the results of our Bayesian Geneclass analysis of first-generation migrants as indicating two recent hybrids between wild boar and pigs. That interpretation is corroborated by (1) the fact that the full set of markers could be undoubtedly genotyped for those two individuals—hence, no reduced assignment probability; (2) both individuals revealing a heterozygosity value compatible with the overall observed and expected wild boar-wide heterozygosity—hence, again no reduced assignment probability; (3) the presence of several alleles, which were generally found at low frequency in wild boar but significantly more frequently in pigs and vice versa. Consequently, we infer that the 2 = 0.53% of our currently studied wild boar may indeed represent relatively recent hybrids. However, pig-typical gene pool characteristics could also have accumulated in the ancestry lineages of those two wild boars by random genetic drift. As those two individuals affected the overall frequencies of some alleles only very marginally in the total gene pool composition, we conclude that current hybridisation between wild boar and pigs does not contribute much to the level of genetic diversity of wild boar in the study region. Notably, allelic richness was significantly higher in wild boar compared to domestic pigs, which showed collectively only 74% of the allelic richness of that of wild boar, even though the current wild boar samples originated from a relatively small study area, probably with a drastic population low during parts of the 20th century, particularly during the war times. The high genetic diversity of wild boar in our study area corresponds to earlier findings in Europe, e.g., [1], supposedly attributable to sufficient and continuous gene flow from neighbouring ranges in recent times rather than to ongoing hybridisation with pigs.
4.4. Genetic Differentiation and Admixture
As in other studies, e.g., [13], relative (FST, AMOVA) and absolute (Cavalli-Sforza and Edwards distances) genetic partitioning of wild boar across our study area as well as between wild boar and domestic pigs is at a low to moderate level, essentially at one that may be expected for regional populations of many terrestrial mammals or slightly higher. The low genetic differentiation between wild boar and pigs corresponds to their long-term close evolutionary history with frequent gene flow over many generations before controlled breeding of modern pig races started under increasing separation from wild boars, e.g., [10]. The significant FST value, together with the significant deviation of genotypes from overall Hardy–Weinberg expectations, as well as significant FIS values for several loci, indicate genetic substructuring of the wild boar in our relatively small study area. The individual patterns of genetic admixture, as revealed by our STRUCTURE models, and the remarkably high number of genetic clusters inferred for wild boar strongly suggest a fairly pronounced spatial gene pool partitioning, albeit at a low absolute and relative level, but consistent with previous data [43] from a smaller and geographically even more restricted sample. In line with that interpretation, a significant number of wild boars appeared to be very little genetically admixt, whereas others displayed quite a few of the ten inferred genetic clusters at varying frequencies. The Serbian and Hungarian Mangaliza samples were genetically distinct and, with a few exceptions, very little admixture, as were all commercial slaughter pigs. This pattern of distinct but almost not admixed gene pool characteristics for the three pig groupings (breeds) as obtained along with the wild boar clusters in the same STRUCTURE models, and in particular, the finding of many wild boar with almost no genetic admixture and many with extensive admixture patterns support our conclusion of a high number (10) of genetic clusters inherent to the currently studied wild boar. The few Turopolje pigs in our study that could not be distinguished from the commercial slaughter pigs with our marker system may be genetically too closely related to the commercial pigs, or the samples have been confused by the butcher with standard commercial pigs. Obviously, one of the Mangaliza samples appeared to be a hybrid between Mangaliza and commercial slaughter pigs according to its admixture pattern.
4.5. Spatial and Climate Patterns of Introgression
Contrary to our expectations, we found significantly varying levels of pig-typical gene pool characteristics introgressed in wild boar, but lower levels at peri-urban locations of Vienna, independent from climatic effects. This may result from rare historical pig-rearing in the forested areas of large parts of the environs of Vienna, particularly in the Vienna Forest and the riverine woodland along the Danube in Vienna that was frequently flooded before river regulations in the late 19th century and even later. However, that interpretation would mean that there has probably not been any significant immigration from areas further away from Vienna to the peri-urban habitats since the mid-1950s. A planned follow-up study aims to uncover gene flow patterns and the population genetic structure of wild boar in peri-urban Vienna as well as gene exchange in the wider area.
Ambient temperature and overall precipitation did not affect the level of pig-typical ancestry in individual wild boars, which, given the differential adaptation of wild boar and pigs to mid-latitude climate conditions [32], may indicate that pig-typical alleles of genes involved in thermoregulation and basal metabolism do not occur to any significant extent in the wild boar currently examined. Reference [32] pointed out a higher thermoneutral zone in pigs compared to wild boar, and in particular, the efficient adaptation of the latter to the cold season (or cold environments). On the other hand, heat stress is known to be a critical aspect in pig physiology [11] and in wild boar, but again, our model results did not indicate a significant accumulation of pig-typical gene pool characteristics at locations with lower temperatures. However, our models showed a statistically relevant minor precipitation effect on the individual level of pig-typical ancestry in wild boar, independent of geographical (spatial) differences. Specifically, wild boar exhibited increased introgression levels at locations with relatively low overall/summer precipitation and with relatively high winter precipitation. Without more detailed (ecological and physiological) information, we can only speculate about the possible underlying causes for this eco-genetic signal: sufficient winter precipitation may improve soil moisture, thereby stimulating plant growth in the following spring and warm season. This could lead to a better nutrition of the sows and improve milk production and consequently improve the growth of introgressed piglets, which are born in the study region in late winter/early spring.
Therefore, wild boar, which carry a relatively large number of pig-typical alleles with a low adaptive value, may survive better at locations with higher winter precipitation. On the other hand, increasing wetness in summer could increase the probability of infections (particularly coccidiosis) in growing young and, thus, their mortality, especially in wild boars with only low immunocompetent alleles typical of domestic pigs. However, even though the precipitation effect was statistically robust, the rather low loadings of the underlying factor in our PCA recommended the confirmation of the effect by a spatially more comprehensive sample array.
In conclusion, although all wild boar in our study displayed introgression by pig-typical gene pool characteristics, current hybridisation no longer seems to play a significant role in changes in gene pool composition. Rather, we interpret the constantly low levels of observed pig-typical signals in wild boar as gene pool characteristics that were common in various regional pig breeds in historical times that had the chance of introgressing into wild boars and that have formed a strong basis for the current widely reared commercial breeds for pork production in eastern central Europe and in parts on the Balkans. As in other regions of Europe, after the common medieval pannage of pigs, semi-free-ranging extensive pig farming practices in eastern central Europe and the Balkans have, until recently, i.e., around the mid-1950s or even later, allowed for gene exchange between pigs and wild boar over generations. This interpretation is in line with incomplete gene pool separation in the history of genetic admixture between wild boar and pigs, as concluded from SNP studies, e.g., [10]. It is also corroborated by the presence of various wild boar-typical genetic clusters in all our commercial pork samples from eastern central Europe and the Balkans, which are otherwise genetically very uniform and distinctly separate from wild boar gene pools. The wild boar-typical genetic partitions in pigs were, on average greater than the pig-typical genetic partitions in wild boar, even though there was basically no chance of introgression of wild boar into pigs, given the current breeding and rearing system in agro-industrial units rather than in sites on small family farms. This latter observation is consistent with the asymmetric gene flow from wild boar to pigs during domestication, as suggested by SNP data [10]. Nowadays, hybridisation may occasionally occur in the study region, but would not essentially affect the gene pool architecture of wild boar, in line with many regions of Europe; see, e.g., [6,13]. Moreover, contrary to our initial hypothesis, habitats of peri-urban Vienna, with probably more anthropogenic disturbances than in rural habitats, do not seem to specifically favour wild boar with higher levels of pig-typical ancestry. However, this does not rule out the possibility of various selective forces acting differently on certain (pig-typical) alleles [4] in peri-urban and rural habitats.
Author Contributions
Conceptualisation, D.B. and F.S.; methodology, D.B., F.S., F.K., M.S. and S.S.; software, D.B., F.S., F.K., M.S. and S.S.; validation, F.S., S.S. and M.S.; formal analysis, D.B., F.K., S.S. and F.S.; investigation, D.B. and F.S.; resources, R.Z., A.K.-H., A.P., C.B., A.D., V.S., H.D., M.D., N.V. and C.D.Z.; data curation, R.Z., A.K.-H., A.P., C.B., A.D., V.S., H.D., M.D., N.V. and C.D.Z.; writing—original draft preparation, D.B. and F.S.; writing—review and editing, D.B. and F.S.; visualisation, D.B. and F.S.; supervision, F.S.; project administration, F.S.; funding acquisition, F.S. All authors have read and agreed to the published version of the manuscript.
Funding
Financial support was provided by the Kulturabteilung of the City of Vienna, Austria, and of the Government of the Province of Lower Austria, as well as by Wildlife Research—Franz Suchentrunk.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Acknowledgments
We thank Anita Haiden (Vienna) for her excellent assistance in the laboratory. Open Access Funding by the University of Veterinary Medicine Vienna.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Scandura, M.; Iacolina, L.; Apollonio, M. Genetic diversity in the European wild boar Sus scrofa: Phylogeography, population structure and wild x domestic hybridization. Mamm. Rev. 2011, 41, 125–137. [Google Scholar] [CrossRef]
- Stillfried, M.; Fickel, J.; Borner, K.; Wittstatt, U.; Heddergott, M.; Ortmann, S.; Kramer-Schadt, S.; Frantz, A.C. Do cities represent sources, sinks or isolated islands for urban wild boar population structure? J. Appl. Ecol. 2017, 54, 272–281. [Google Scholar] [CrossRef]
- Hagemann, J.; Conejero, C.; Stillfried, M.; Mentaberre, G.; Castillo-Contreras, R.; Fickel, J.; López-Olvera, J.R. Genetic population structure defines wild boar as an urban exploiter species in Barcelona, Spain. Sci. Total Environm. 2022, 833, 155126. [Google Scholar] [CrossRef] [PubMed]
- Zsolnai, A.; Csókas, A.; Szabó, L.; Patkó, L.; Csány, S.; Márton, M.; Lakatos, E.A.; Anton, I.; Deutsch, F.; Heltai, M. Genetic adaptation to urban living: Molecular DNA analyses of wild boar populations in Budapest and surrounding area. Mamm. Biol. 2022, 102, 221–234. [Google Scholar] [CrossRef]
- Nikolov, I.S.; Stoeckle, B.C.; Markov, G.; Kuehn, R. Substantial hybridisation between wild boars (Sus scrofa scrofa) and East Balkan Pigs (Sus scrofa f. domestica) in natural environment as a result of semi-wild rearing in Bulgaria. Czech J. Anim. Sci. 2017, 62, 1–8. [Google Scholar] [CrossRef]
- Iacolina, L.; Pertoldi, C.; Amills, M.; Kusza, S.; Megens, H.-J.; Bâlteanu, V.L.; Bakan, J.; Cubric-Curik, V.; Oja, R.; Saarma, U.; et al. Hotspots of recent hybridization between pigs and wild boars in Europe. Sci. Rep. 2018, 8, 17372. [Google Scholar] [CrossRef]
- Vetter, S.G.; Ruf, T.; Bieber, C.; Arnold, W. What Is a Mild Winter? Regional Differences in Within-Species Responses to Climate Change. PLoS ONE 2015, 10, e0132178. [Google Scholar] [CrossRef]
- Fulgione, D.; Rippa, D.; Buglione, M.; Trapanese, M.; Petrelli, S.; Maselli, V. Unexpected but welcome. Artificially selected traits may increase fitness in wild boar. Evol. Appl. 2016, 9, 769–776. [Google Scholar] [CrossRef]
- Vetter, S.; Brandstätter, C.; Macheiner, M.; Suchentrunk, F.; Gerritsmann, H.; Bieber, C. Shy is sometimes better: Personality and juvenile body mass affect adult reproductive success in wild boars, Sus scrofa. Behaviour 2016, 115, 193–205. [Google Scholar] [CrossRef]
- Frantz, L.A.F.; Schraiber, J.G.; Madsen, O.; Megens, H.-J.; Cagan, A.; Bosse, M.; Paudel, Y.; Crooijmans, R.P.M.A.; Larson, G.; Groenen, M.A.M. Evidence of long-term gene flow and selection during domestication from analyses of Eurasian wild and domestic pig geomes. Nat. Genet. 2015, 47, 1141–1148. [Google Scholar] [CrossRef]
- Gourdine, J.-L.; Rauw, W.M.; Gilbert, H.; Poullet, N. The Genetics of Thermoregulation in Pigs: A Review. Front. Vet. Sci. 2021, 8, 1–14. [Google Scholar] [CrossRef]
- Rotschild, M.F.; Ruvinsky, A. The Genetics of the Pig, 2nd ed.; CAB International: Wallingford, UK; Cambridge, MA, USA, 2011; 507p. [Google Scholar]
- Scandura, M.; Iacolina, L.; Crestanello, B.; Pecchioli, E.; Di Benedetto, M.F.; Russo, V.; Davoli, R.; Apollonio, M.; Bertorelle, G. Ancient vs. recent processes as factors shaping the genetic variation of the European wild boar: Are the effects of the last glaciation still detectable? Mol. Ecol. 2008, 17, 1745–1762. [Google Scholar] [CrossRef]
- Costa, V.; Pérez-González, P.; Santos, P.; Fernández-Llario, P.; Carranza, J.; Zsolnai, A.; Anton, I.; Buzgó, J.; Varga, G.; Monteiro, N.; et al. Microsatellite markers for identification and parentage analysis in the European wild boar (Sus scrofa). BMC Res. Notes 2012, 5, 479. [Google Scholar] [CrossRef] [PubMed]
- Frantz, A.C.; Zachos, F.E.; Kirschning, J.; Cillina, S.; Bertouille, S.; Mamuris, Z.; Koutsogiannouli, E.A.; Burke, T. Genetic evidence for introgression between domestic pigs and wild boars (Sus scrofa) in Belgium and Luxembourg: A comparative approach with multiple marker systems. Biol. J. Linn Soc. 2013, 110, 104–115. [Google Scholar] [CrossRef]
- Anderson, D.; Negishi, Y.; Tomaa, R.; Nagatab, J.; Tamatec, H.; Kaneko, S. Robust microsatellite markers for hybrid analysis between domesticated pigs and wild boar. Markers for pig and wild boar hybridization. Genet. Resour. 2020, 1, 29–41. [Google Scholar] [CrossRef]
- Vetter, S.G.; Bieber, C.; Suchentrunk, F. Evaluating commonly used microsatellites for parenthood analysis and population assignment in wild boar (Sus scrofa). In Proceedings of the 10th Int. Symp. on Wild Boar and other Suids, Velenje, Slovenia, 6–9 September 2014; book of abstracts. p. 118. [Google Scholar]
- Belkhir, K.; Castric, V.; Bonhomme, F. IDENTIX, a software to test for relatedness in a population using permutation methods. Mol. Ecol. Notes 2002, 2, 611–614. [Google Scholar] [CrossRef]
- Belkhir, K. GENETIX V. 4.0, Logiciel sous WindowsTM pour la Génétique des Populations. Laboratoire Génome, Populations, Interactions CNRS UMR 5000; Université de Montpellier II: Montpellier, France, 2004. [Google Scholar]
- Queller, D.C.; Goodnight, K.F. Estimating relatedness using genetic markers. Evolution 1989, 43, 258–275. [Google Scholar] [CrossRef] [PubMed]
- Goudet, J. FSTAT (version 1.2): A computer program to calculate F-statistics. J. Hered. 1995, 86, 485–486. [Google Scholar] [CrossRef]
- Piry, S.; Alapetite, A.; Cornuet, J.-M.; Paetkau, D.; Baudouin, L.; Estoup, A. GeneClass2: A software for genetic assignment and first generation migrant detection. J. Hered. 2004, 95, 536–539. [Google Scholar] [CrossRef]
- Cornuet, J.M.; Piry, S.; Luikart, G.; Estoup, A.; Solignac, M. New methods employing multilocus genotypes to select or exclude populations as origins of individuals. Genetics 1999, 153, 1989–2000. [Google Scholar] [CrossRef]
- Rannala, B.; Mountain, J.L. Detecting immigration by using multilocus genotypes. Proc. Natl. Acad. Sci. USA 1997, 94, 9197–9201. [Google Scholar] [CrossRef] [PubMed]
- Paetkau, D.; Slade, R.; Burden, M.; Estoup, A. Direct real-time estimation of migration rate using assignment methods: A simulation-based exploration of accuracy and power. Mol. Ecol. 2004, 13, 55–65. [Google Scholar] [CrossRef]
- Schneider, S.; Roessli, D.; Excoffier, L. Arlequin: A software for Population Genetics Data Analysis, 2000. Vers. 2.000. Available online: http://cmpg.unibe.ch/software/arlequin3/.
- Falush, D.; Stephens, M.; Pritchard, J.K. Inference of population structure: Extensions to linked loci and correlated allele frequencies. Genetics 2003, 164, 1567–1587. [Google Scholar] [CrossRef] [PubMed]
- Pritchard, J.K.; Stephens, M.; Donnelly, P. Inference of population structure using multilocus genotype data. Genetics 2000, 155, 945–959. [Google Scholar] [CrossRef]
- Pritchard, J.K.; Wen, W.; Falush, D. Available from 2010. Documentation for Structure Software: Version 2.3. Available online: http://pritch.bsd.uchicago.edu.
- Evanno, G.; Regnaut, S.; Goudet, J. Detecting the number of clusters of individuals using the software STRUCTURE: A simulation study. Mol. Ecol. 2005, 14, 2611–2620. [Google Scholar] [CrossRef]
- Earl, D.A.; von Holdt, B.M. STRUCTURE HARVESTER: A website and program for visualizing STRUCTURE output and implementing the Evanno method. Conserv. Gen. Res. 2012, 4, 35. [Google Scholar] [CrossRef]
- Ruf, T.; Vetter, S.G.; Painer, J.; Stalder, G.; Bieber, C. Thermogregulation in the wild boar (Sus scrofa). Subm. Ms.
- Barton, K. Multi-Model Inference. R Package Version 1.9.13. 2013. Available online: http://CRAN.R-project.org/package=MuMIn.
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2016; Available online: https://www.R-project.org/.
- Burnham, K.P.; Anderson, D.R. Model Selection and Multimodel Inference: A Practical Information-Theoretic Approach, 22nd ed.; Springer: New York, NY, USA, 2002; 488p. [Google Scholar]
- ENETWILD-Consortium; Illanas, S.; Fernández-López, J.; Acevedo, P.; Apollonio, M.; Blanco-Aguiar, J.A.; Brivio, F.; Croft, S.; Ferroglio, E.; Keuling, O.; et al. Analysis of wild boar-domestic pig interface in Europe: Spatial overlapping and fine resolution approach in several countries. EFSA Support. Publ. 2021, 18, 23. [Google Scholar] [CrossRef]
- Györffy, I. Alföldi Népélet; Gondolat: Budapest, Hungary, 1983; 510p, ISBN 9632812794. (In Hungarian) [Google Scholar]
- Manunza, A.; Amills, M.; Noce, A.; Cabrera, B.; Zidi, A.; Eghbalsaied, S.; Carillo de Albornoz, E.; Portell, M.; Mercadé, A.; Sánchez, A.; et al. Romanian wild boars and Mangalitza pigs have a European ancestry and harbour genetic signatures compatible with past population bottlenecks. Scient. Rep. 2016, 6, 29913. [Google Scholar] [CrossRef]
- von Matlekovits, A. Das Königreich Ungarn. Volkswirtschaftlich und Statistisch Dargestellt; Duncker & Humblot: Berlin, Germany, 1900; Volume 1, 616p, Reprint ISBN 978-3-428-16611-4. [Google Scholar]
- Falkenberg, H.; Hammer, H. Zur Geschichte und Kultur der Schweinezucht und -haltung. 3. Mitt.: Schweinezucht und -haltung in Deutschland von 1650 bis 1900. Züchtungsk 2007, 79, 92–110. [Google Scholar]
- Miorini Edler von Sebtenberg, A. Der Schlachtviehmarkt St. Marx. In Zweiund-Zwanzigster Jahres-Bericht der Landwirtschaftlichen Lehranstalt “Francisco-Josephinum“ in Mödling; Verlag der Landwirtschaftlichen Lehranstalt: Vienna, Austria, 1891. [Google Scholar]
- VÖS—Verband Österreichischer Schweinebauern, Vienna. Available online: https://voes-online.at/ (accessed on 17 January 2023).
- Suchentrunk, F.; Himmler, E.; Habe, M.; Zink, R.; Bieber, C.; Vetter, S.G. Spatial genetics for the managment of wild boar (Sus scrofa) in peri-urban Vienna. In Proceedings of the 10th International Symposium on Wild Boar and other Suids, Velenje, Slovenia, 1–5 September 2014; SEP 1-4; book of abstracts. p. 117. [Google Scholar]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).