Two New Species of Encotyllabe (Monogenea: Capsalidae) from Brazil: Morphological and Molecular Evidence
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Collection and Processing
2.2. Morphological and Statistical Analysis
2.3. Morphometric Analysis
2.4. Molecular Data and Phylogenetic Analyses
| GenBank ID | ||||||
|---|---|---|---|---|---|---|
| Species | Host | Family | Locality | cox1 | LSU rRNA | Reference |
| E. bifurcatum n. sp. | Pagrus pagrus | Sparidae | Brazil | MT968928 | This study | |
| E. parvum n. sp. | Orthopristis ruber | Haemulidae | Brazil | MT967362 | MT968927 | This study |
| E. cf. spari | O. ruber | Haemulidae | Brazil | KY553149 | [4] | |
| E. antofagastensis | Anisotremus scapularis | Haemulidae | Chile | JQ782836-40 | MT982166 | [3]/This study |
| E. caballeroi | Gymnocranius audleyi | Lethrinidae | Australia | AF026112 | [38] | |
| E. caranxi | Pseudocaranx dentex | Carangidae | Australia | FJ971990 | [39] | |
| E. cheilodactyli | Cheilodactylus variegatus | Cheilodactylidae | Chile | JQ782841-45 | MT982167 | [3]/This study |
| E. chironemi | Chironemus marmoratus | Chironemidae | Australia | AF382054 | [40] | |
| Encotyllabe sp.1 | Pagellus bogaraveo | Sparidae | Algeria | OL675214 | OL679678 | [12] |
| OL675215 | [12] | |||||
| OL675217 | [12] | |||||
| OL675220 | [12] | |||||
| OL675223 | [12] | |||||
| OL675214 | [12] | |||||
| Encotyllabe sp.2 | Diplodus vulgaris | Sparidae | Algeria | OL679688 | [12] | |
| Diplodus vulgaris | Sparidae | Algeria | OL675225 | [12] | ||
| Sparus aurata | Sparidae | Algeria | OL675222 | [12] | ||
| OL675226 | [12] | |||||
| Neobenedenia sp. | Cheilodactylus variegatus | Cheilodactylidae | Chile | JQ782846 | [3] | |
| Neobenedenia sp. | Paralabrax humeralis | Serranidae | Chile | MK202450 | [41] | |
3. Results
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- WoRMS Editorial Board. World Register of Marine Species. Available online: http://wwwmarinespeciesorg (accessed on 15 March 2023). [CrossRef]
- Tantalean, M. Nuevo Género y Nuevas Especies de Monogeneos Parásitos de Peces de Algunas Áreas de la Costa Peruana. Con redescripción de una Especie Conocida. Ph.D. Thesis, Universidad Nacional Mayor de San Marcos, Facultad de Ciencias, Lima, Peru, 1973. [Google Scholar]
- Sepúlveda, F.A.; González, M.T.; Oliva, M.E. Two new species of Encotyllabe (Monogenea: Capsalidae) based on morphometric and molecular evidence: Parasites of two inshore fish species of northern Chile. J. Parasitol. 2014, 100, 344–349. [Google Scholar] [CrossRef] [PubMed]
- Camargo, A.C.A.; Luque, J.L.; Santos, C.P. Mexicana rubra sp. nov. and Encotyllabe cf. spari off Rio de Janeiro. Helminthologia 2017, 54, 336–347. [Google Scholar] [CrossRef]
- Carvalho, A.R.; Luque, J.L. Three new species of monogeneans parasitic on Atlantic cutlassfish Trichiurus lepturus (Perciformes: Trichiuridae) from Southeastern Brazil. Acta Sci. Biol. Sci. 2012, 34, 359–365. [Google Scholar] [CrossRef]
- Diesing, K.M. Vierzhen Arten von Bdelliden. Denkschr. Acad. Wiss. Wien 1858, 14, 63–80. [Google Scholar]
- Whittington, I.D.; Cribb, B.W.; Hamwood, T.E.; Halliday, J.A. Host-specificity of monogenean (Platyhelminthes) parasites: A role for anterior adhesive areas? Int. J. Parasitol. 2000, 30, 305–320. [Google Scholar] [CrossRef] [PubMed]
- Rohde, K.; Watson, N. Morphology and geographical variation of Pseudokuhnia minor n. g, n. comb. (Monogenea: Polyopisthocotylea). Int. J. Parasitol. 1985, 15, 557–567. [Google Scholar] [CrossRef]
- Rohde, K.; Watson, N. Morphology, microhabitats and geographical variation of Kuhnia spp. (Monogenea: Polyopisthocotylea). Int. J. Parasitol. 1985, 15, 569–586. [Google Scholar] [CrossRef]
- Price, E.W. North American monogenetic trematodes III. The family Capsalidae (Capsaloidea). J. Wash. Acad. Sci. 1939, 29, 63–92. [Google Scholar]
- Fábio, S. Redescricao de Encotyllabe lintoni Monticelli, 1909 (Monogenea, Capsalidae) em Pagrus pagrus (Linnaeus, 1758). Bol. Mus. Nac. 1998, 385, 1–5. [Google Scholar]
- Lablack, L.; Rima, M.; Georgieva, S.; Marzoug, D.; Kostadinova, A. Novel molecular data for monogenean parasites of sparid fishes in the Mediterranean and a molecular phylogeny of the Microcotylidae Taschenberg, 1879. CRPVBD 2022, 2, 100069. [Google Scholar] [CrossRef]
- Egorova, T.P. Recent composition of the subfamily Encotyllabinae (Monogenea: Capsalidae). Parazitologiya 2000, 34, 295–301. (In Russian) [Google Scholar]
- Williams, A.; Beverley-Burton, M. Redescription of three species of the genus Encotyllabe (Capsalidae: Monogenea) from fishes of the east coast of Australia. Aust. J. Zool. 1989, 37, 45–53. [Google Scholar] [CrossRef]
- Ñacari, L.A.; Sepúlveda, F.A.; Escribano, R.; Oliva, M.E. Two new species of Acanthocotyle Monticelli, 1888 (Monogenea: Acanthocotylidae), parasites of two deep-sea skates (Elasmobranchii: Rajiformes) in the South-East Pacific. Parasit. Vectors 2019, 12, 512. [Google Scholar] [CrossRef] [PubMed]
- Chaves, N.D.; Luque, J.L. Ecology of metazoans parasites of Menticirrhus americanus (Osteichthyes: Sciaenidae), coast area from Rio de Janeiro State, Brazil. Rev. Bras. Parasitol. Vet. 1999, 8, 137–144. [Google Scholar]
- Cohen, S.C.; Justo, M.C.N.; Kohn, A. South American Monogenoidea Parasites of Fishes, Amphibians and Reptiles; Fundacion Oswaldo Cruz: Rio de Janeiro, Brazil, 2013; 663p. [Google Scholar]
- Cordeiro, A.; Luque, J.L. Metazoários parasitos do coió Dactylopterus volitans (Linnaeus, 1758) (Osteichthyes: Dactylopteridae) do litoral do Estado do Rio de Janeiro, Brasil. Acta Sci. Biol. Sci. 2005, 27, 119–123. [Google Scholar] [CrossRef]
- Domingues, S.A.; Alves, D. Metazoários parasitos de Thyrsitops lepidopoides (Osteichthyes: Gempylidae) do litoral do Estado do Rio de Janeiro, Brasil. Cadernos UniFOA 2015, 27, 95–103. [Google Scholar] [CrossRef]
- Kohn, A.; Abramson, B.; Macedo, B. Studies on some monogenean parasites of Haemulon sciurus (Shaw, 1803) (Pomadasyidae). J. Helminthol. 1984, 58, 213–218. [Google Scholar] [CrossRef]
- Kohn, A.; Cohen, S.C. South American Monogenea—List of species, hosts and geographical distribution. Int. J. Parasitol. 1998, 28, 1517–1554. [Google Scholar] [CrossRef]
- Luque, J.L.; Amato, J.F.R.; Takemoto, R.M. Comparative analysis of the communities of metazoan parasites of Orthopristis ruber and Haemulon steindachneri (Osteichthyes: Haemulidae) from the southeastern Brazilian littoral: I. Structure and influence of the size and sex of hosts. Rev. Brasil. Biol. 1996, 56, 279–292. [Google Scholar]
- Paraguassú, A.R.; Luque, J.L.; Alves, D.R. Community ecology of the metazoan parasites of red porgy, Pagrus pagrus (L. 1758) (Osteichthyes, Sparidae), from the coastal zone, state of Rio de Janeiro, Brazil. Acta Sci. Biol. Sci. 2002, 24, 461–467. [Google Scholar]
- Paschoal, F.; Cezar, A.D.; Luque, J.L. Checklist of metazoan associated with grunts (Perciformes, Haemulidae) from the Nearctic and Neotropical regions. Check List 2015, 11, 1–23. [Google Scholar] [CrossRef]
- Soares, I.A.; Luque, J.L. Seasonal variability of the composition and structure of parasite communities of red porgy, Pagrus pagrus (Perciformes: Sparidae) off Brazil. Helminthologia 2015, 52, 236–243. [Google Scholar] [CrossRef]
- Bush, A.O.; Lafferty, K.D.; Lotz, J.M.; Shostak, A.W. Parasitology meets ecology on its own terms: Margolis et al. Revisited. J. Parasitol. 1997, 83, 575–583. [Google Scholar] [CrossRef] [PubMed]
- Miller, S.A.; Dykes, D.D.; Polesky, H.F.R.N. A simple salting out procedure for extracting DNA from human nucleated cells. Nucleic Acids Res. 1998, 16, 1215. [Google Scholar] [CrossRef] [PubMed]
- Chisholm, L.A.; Morgan, J.A.T.; Adlard, R.D.; Whittington, I.D. Phylogenetic analysis of the Monocotylidae (Monogenea) inferred from 28S rDNA sequences. Int. J. Parasitol. 2001, 31, 1537–1547. [Google Scholar] [CrossRef]
- Littlewood, D.T.J.; Rohde, K.; Clough, K.A. Parasite speciation within or between host species? Phylogenetic evidence from site-specific polystome monogeneans. Int. J. Parasitol. 1997, 27, 1289–1297. [Google Scholar] [CrossRef]
- Verneau, O.; Du Preez, L.H.; Laurent, V.; Raharivololoniaina, L.; Glaw, F.; Vences, M. The double odyssey of Madagascan polystome flatworms leads to new insights on the origins of their amphibian hosts. Proc. Royal Soc. B 2009, 276, 1575–1583. [Google Scholar] [CrossRef] [PubMed]
- Filatov, D.A. Proseq: A software for preparation and evolutionary analysis of DNA sequence data sets. Mol. Ecol. Notes 2002, 2, 621–624. [Google Scholar] [CrossRef]
- Larkin, M.A.; Blackshields, G.; Brown, N.P.; Chenna, R.; McGettigan, P.A.; McWilliam, H.; Valentin, F.; Wallace, I.M.; Wilm, A.; Lopez, R.; et al. Clustal W and Clustal X version 2.0. Bioinformatics 2007, 23, 2947–2948. [Google Scholar] [CrossRef]
- Huelsenbeck, J.P.; Ronquist, F. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics 2001, 17, 754–755. [Google Scholar] [CrossRef]
- Trifinopoulos, J.; Nguyen, L.T.; von Haeseler, A.; Minh, B.Q. W-IQ-TREE: A fast online phylogenetic tool for maximum likelihood analysis. Nucleic Acids Res. 2016, 44, 232–235. [Google Scholar] [CrossRef]
- Posada, D. jModelTest: Phylogenetic Model Averaging. Mol. Biol. Evol. 2008, 25, 1256. [Google Scholar] [CrossRef]
- Rambaut, A.; Drummond, A.J.; Xie, D.; Baelee, G.; Suchard, M.A. Posterior Summarization in Bayesian Phylogenetics Using Tracer 1.7. Syst. Biol. 2018, 67, 901–904. [Google Scholar] [CrossRef]
- Tamura, K.; Peterson, D.; Peterson, N.; Stecher, G.; Nei, M.; Kumar, S. MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 2011, 28, 2731–2739. [Google Scholar] [CrossRef]
- Mollaret, I.; Jamieson, B.G.; Adlard, R.D.; Hugall, A.; Lecointre, G.; Chombard, C.; Justine, J.-L. Phylogenetic analysis of the Monogenea and their relationships with Digenea and Eucestoda inferred from 28S rDNA sequences. Mol. Biochem. Parasitol. 1997, 90, 433–438. [Google Scholar] [CrossRef] [PubMed]
- Perkins, E.M.; Donnellan, S.C.; Bertozzi, T.; Chisholm, L.A.; Whittington, I.D. Looks can deceive: Molecular phylogeny of a family of flatworm ectoparasites (Monogenea: Capsalidae) does not reflect current morphological classification. Mol. Phylogenet. Evol. 2009, 52, 705–714. [Google Scholar] [CrossRef]
- Olson, P.D.; Littlewood, D.T. Phylogenetics of the Monogenea—Evidence from a medley of Molecules. Int. J. Parasitol. 2002, 32, 233–244. [Google Scholar] [CrossRef] [PubMed]
- Sepúlveda, F.A.; González, M.T. DNA barcoding evidence for the first recorded transmission of Neobenedenia sp. from wild fish species to Seriola lalandi cultured in an open recirculating system on the Coast of Northern Chile. Aquaculture 2018, 501, 239–246. [Google Scholar] [CrossRef]
- Noble, E.R. The genus Encotyllabe (Class Trematoda) with a description of a new species. Trans. Am. Micr. Soc. 1966, 85, 144–151. [Google Scholar] [CrossRef]
- Whittington, I.D. The Capsalidae (Monogenea: Monopisthocotylea): A review of diversity, classification and phylogeny with a note about species complexes. Folia Parasitol. 2004, 51, 109–122. [Google Scholar] [CrossRef]
- Baeza, J.A.; Sepúlveda, F.A.; González, M.T. The complete mitochondrial genome and description of a new cryptic species of Benedenia Diesing, 1858 (Monogenea: Capsalidae), a major pathogen infecting the yellowtail kingfish Seriola lalandi Valenciennes in the South-East Pacific. Parasites Vectors 2019, 12, 490. [Google Scholar] [CrossRef] [PubMed]
- Sepulveda, F.A.; Gonzalez, M.T. Molecular and morphological analyses reveal that the pathogen Benedenia seriolae (Monogenea: Capsalidae) is a complex species: Implications for yellowtail Seriola spp. aquaculture. Aquaculture 2014, 418, 94–100. [Google Scholar] [CrossRef]
- Li, L.; Yan, J.; Wang, H. A new species of Encotyllabe from marine fishes in Minnan. Taiwan Bank, Fujian, China. J. Zhejiang Univ. Sci. 2003, 31, 207–210. [Google Scholar]
- Justine, J.L.; Beveridge, I.; Boxshall, G.A.; Bray, R.A.; Moravec, F.; Whittington, I.D. An annotated list of fish parasites (Copepoda, Monogenea, Digenea, Cestoda and Nematoda) collected from Emperors and Emperor Bream (Lethrinidae) in New Caledonia further highlights parasite biodiversity estimates on coral reef fish. Zootaxa 2010, 2691, 1–40. [Google Scholar] [CrossRef]
- Rohde, K.; Hayward, C.; Heap, M.; Gosper, D. A tropical assemblage of ectoparasites: Gill and head parasites of Lethrinus miniatus (Teleostei, Lethrinidae). Int. J. Parasitol. 1994, 24, 1031–1053. [Google Scholar] [CrossRef]
- Kardousha, M.M.; Al-Ansi, M.A.; Al-Khayat, J. Monogenea of the Arabian Gulf Fishes. 3. Encotyllabe spari and E. kuwaitensis (Capsalidae) from Qatari waters. Riv. Parassitol. 2002, 19, 227–235. [Google Scholar] [CrossRef]
- Khalil, L.F.; Abdul-Salam, J.B. The subfamily Encotyllabinae (Monogenea: Capsalidae) with the description of Alloencotyllabe caranxi n. g., n. sp. and Encotyllabe kuwaitensis n. sp. Syst. Parasitol. 1988, 11, 139–150. [Google Scholar] [CrossRef]
- Centeno, L.; Bashirullah, A.K.; Alvarez, M.E.; Álvarez, R. Análisis comparativo de las comunidades de parásitos metazoarios en dos especies de peces marinos del Golfo de Cariaco, Venezuela. Bioagro 2002, 14, 135–144. [Google Scholar]
- Faustino, K.D.; Martinez, R.R.; Tantaleán, M. Monogeneos parásitos en Caulolatilus affinis (Jenyns, 1840), (Malacanthidae) de Puerto Cabo Blanco, Piura, Perú. Peruv. J. Parasitol. 2015, 23, 38–53. [Google Scholar]
- Canel, D.; Levy, E.; Soares, I.A.; Braicovich, P.E.; Haimovicic, M.; Luque, J.L.; Timi, J.T. Stocks and migrations of the demersal fish Umbrina canosai (Sciaenidae) endemic from the subtropical and temperate Southwestern Atlantic revealed by its parasites. Fish. Res. 2019, 214, 214–218. [Google Scholar] [CrossRef]
- Mahmoud, N.E.; Mahmoud, A.M.; Fahmy, M.M. Parasitological and Comparative Pathological Studies on Monogenean Infestation of Cultured Sea Bream (Sparus aurata, Spariidae) in Egypt. Oceanography 2014, 2, 4. [Google Scholar] [CrossRef]
- Luque, J.L.; Cordeiro, A.S.; Oliva, M.E. Metazoan parasites as biological tags for stock discrimination of whitemouth croaker Micropogonias furnieri from south-western Atlantic Ocean waters. J. Fish Biol. 2010, 76, 591–600. [Google Scholar] [CrossRef] [PubMed]
- Robinson, E.S. Some Monogenetic Trematodes from Marine Fishes of the Pacific. Trans. Am. Micr. Soc. 1961, 80, 235–266. [Google Scholar] [CrossRef]
- Yamaguti, S. Two New Species of Encotyllabe (Monogenea: Capsalidae) from Brazil: Morphological and Molecular Evidence. Jpn. J. Zool. 1934, 5, 249–541. [Google Scholar]
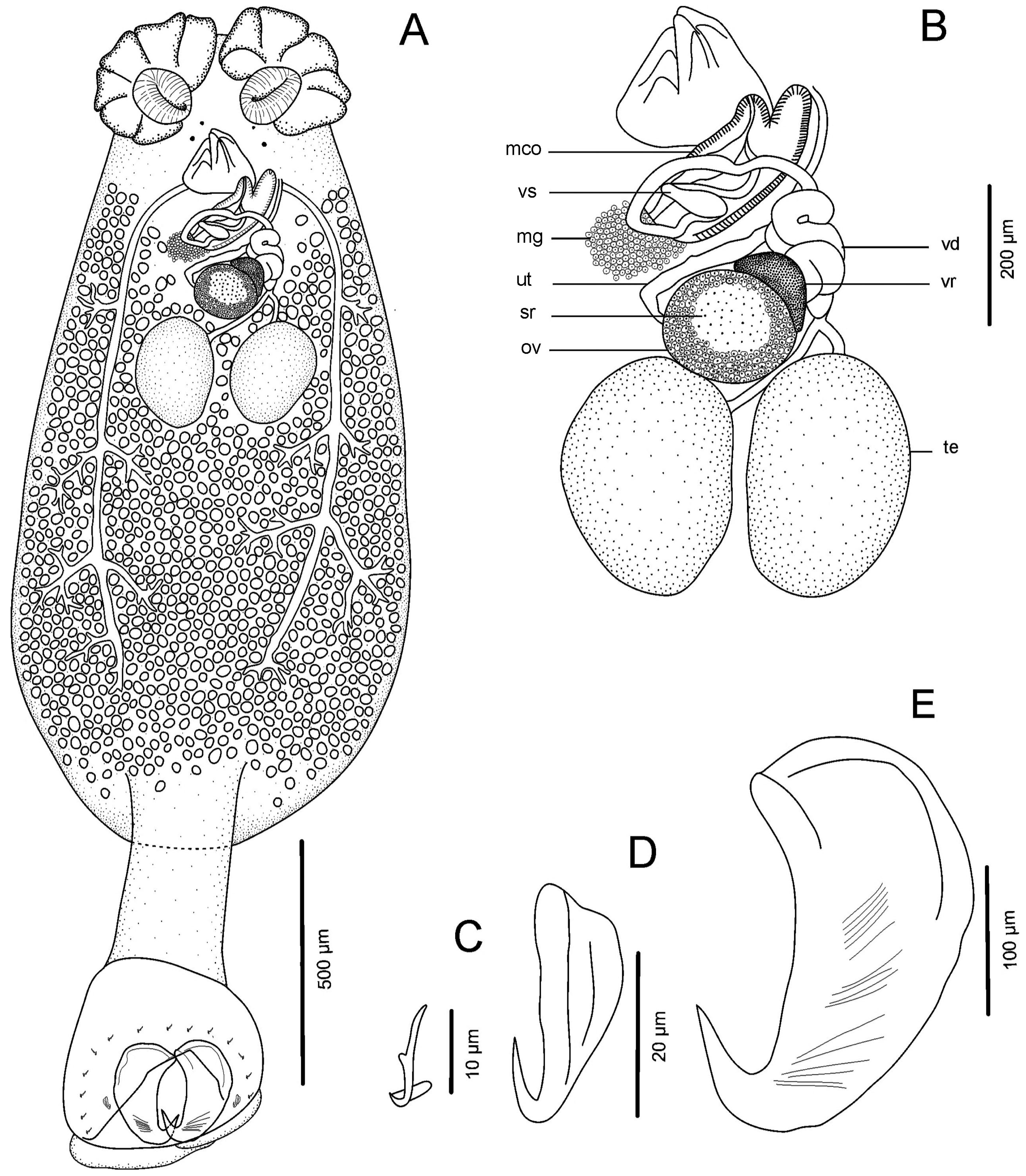
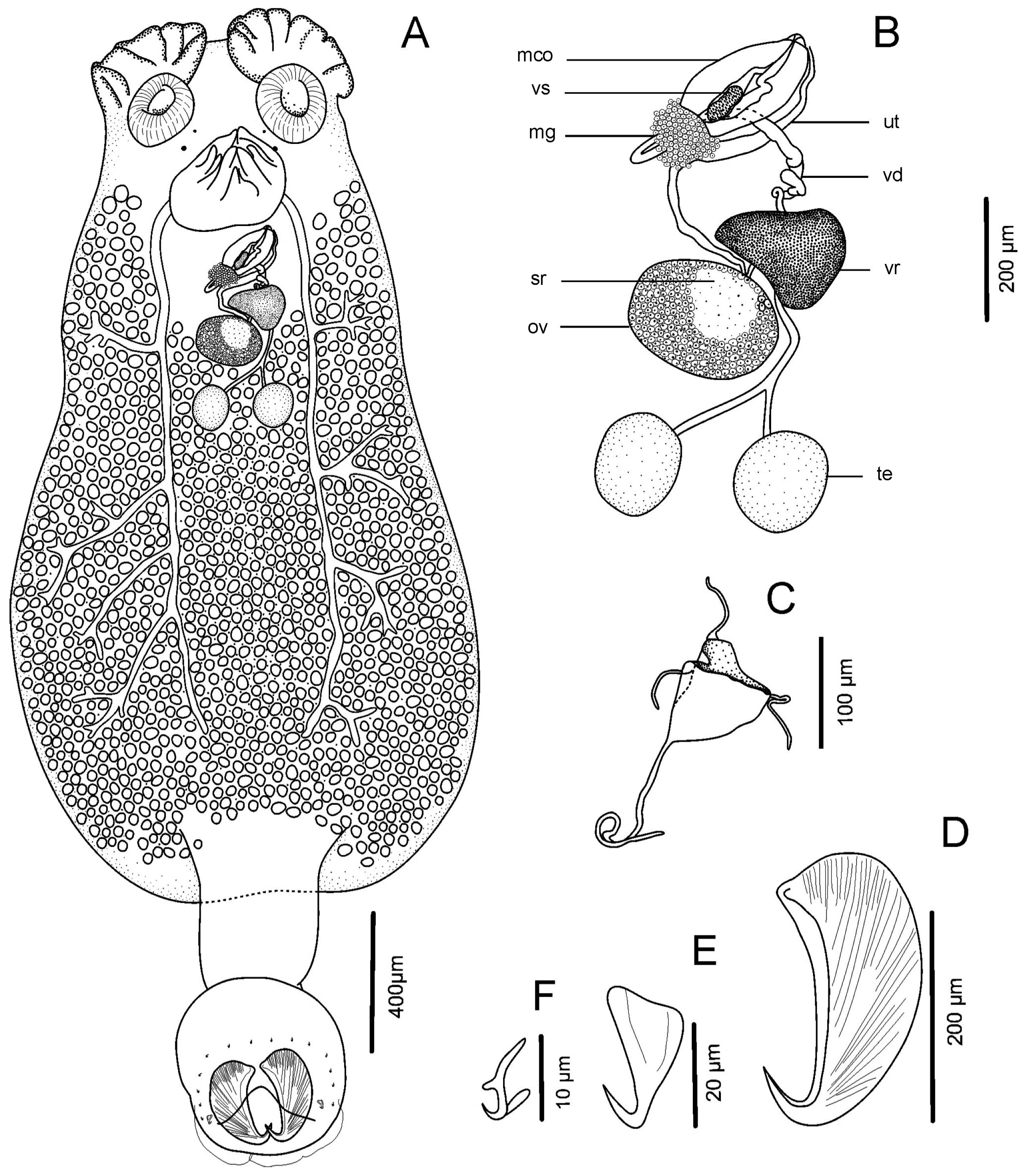


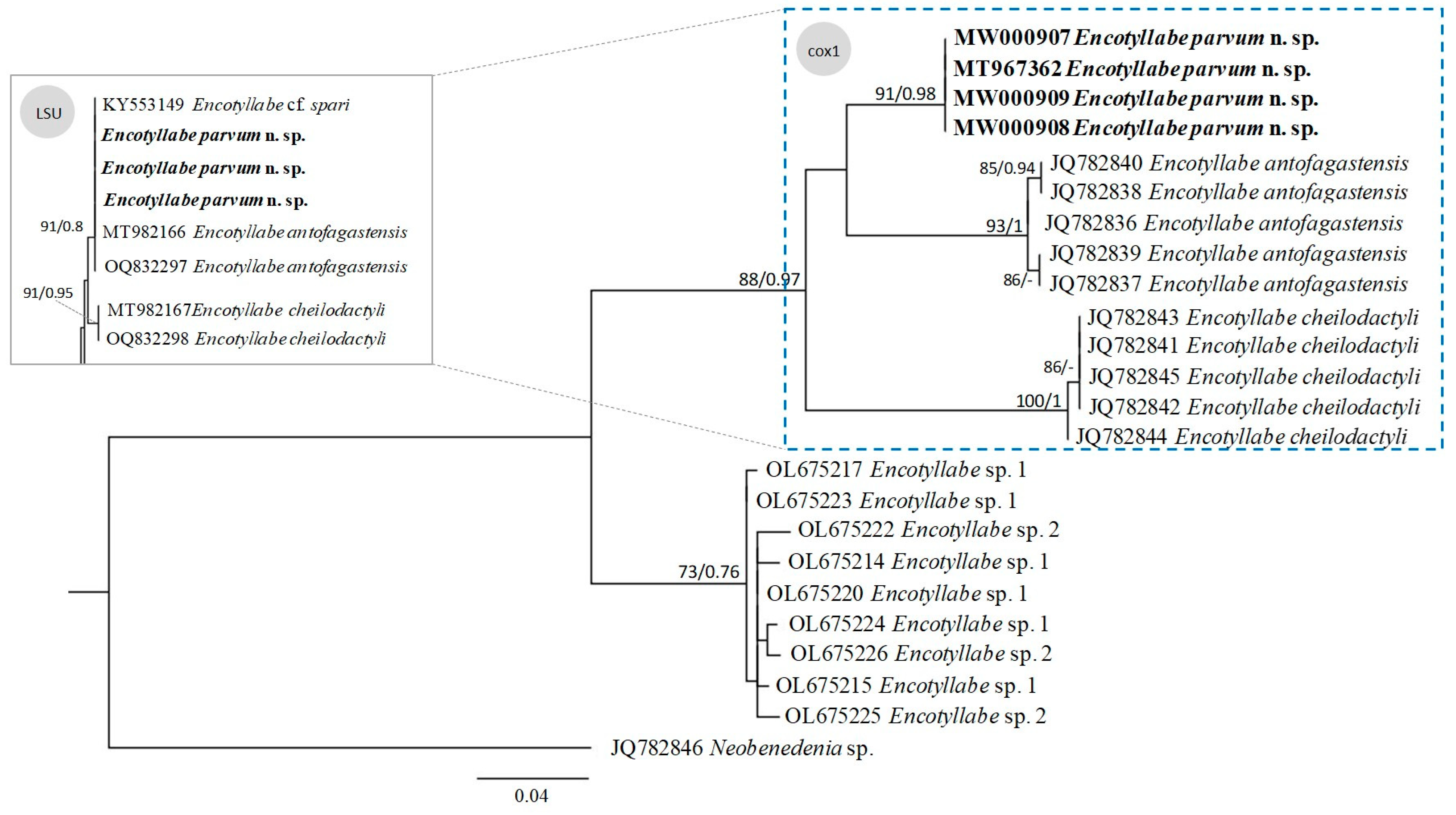
| ID | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | Encotyllabe bifurcatum (n = 2/0) | |||||||||
| 2 | Encotyllabe parvum (n = 3/4) | 0.8 (6) | 6.3–6.7 (17–18) | 8.6–8.9 (23–24) | 10.4–11.2 (28–30) | 11.2–11.5 (30–31) | ||||
| 3 | E. cf. spari (n = 1/0) | 0.8 (6) | 0 | - | ||||||
| 4 | E. antofagastensis (n = 2/5) | 0.8 (6) | 0 | 0 | 8.9–9.7 (25–26) | 10.0–11.5 (28–31) | 10.8–11.5 (29–31) | |||
| 5 | E. cheilodactyli (n = 2/5) | 1 (8) | 0.7 (5) | 0.7 (5) | 0.7 (5) | 11.5–12.3 (31–32) | 11.5–12.3 (31–32) | |||
| 6 | E. chironemi (n = 1/0) | 0.7 (5) | 0.4 (3) | 0.4 (3) | 0.4 (3) | 0.5 (4) | ||||
| 7 | E. caballeroi (n = 1/0) | 1.2 (9) | 1.2 (9) | 1.2 (9) | 1.2 (9) | 1.2 (9) | 1 (8) | |||
| 8 | Encotyllabe sp.1 (n = 1/6) | 1 (8) | 1 (8) | 1 (8) | 1 (8) | 1.2 (9) | 0.8 (6) | 1.6 (12) | 0.7–1.9 (2–4) | |
| 9 | Encotyllabe sp.2 (n = 1/3) | 1 (8) | 1.2 (9) | 1.2 (9) | 1.2 (9) | 1.4 (11) | 1 (8) | 1.6 (12) | 0.3 (2) | |
| 10 | E. caranxi (n = 1/0) | 1.4 (5) | 1.4 (5) | 1.4 (5) | 1.4 (5) | 1.4 (5) | 1.4 (5) | 2 (7) | 1.4 (5) | 1.4 (5) |
| Species | Recorded Hosts | Host Family | Locality | References |
|---|---|---|---|---|
| E. antofagastensis | Anisotremus scapularis | Haemulidae | Chile | [3] |
| E. bifurcatum | Pagrus pagrus | Sparidae | Brazil | This study |
| E. caballeroi | Gymnocranius audleyi | Lethrinidae | Australia | [13] |
| E. caballeroi | Lethrinus miniatus (*) | Lethrinidae | Australia; N. Caledonia | [13,17,18] |
| E. caballeroi | Lethrinus nebulosus | Lethrinidae | Philippines | [13] |
| E. caballeroi | Scolopsis monogramma | Nemipteridae | Australia | [13] |
| E. caballeroi | Scolopsis sp. | Nemipteridae | Vietnam | [13] |
| E. caranxi | Caranx lutescens | Carangidae | Australia | [13] |
| E. caranxi | Caranx sexfaciatus | Carangidae | Mediterranean Sea | [13] |
| E. caranxi | Caranx sp. | Carangidae | Australia | [13] |
| E. caranxi | Pseudocaranx dentex | Carangidae | Australia | [13] |
| E. cheilodactylid | Cheilodactylus variegatus | Cheilodactylidae | Chile | [3] |
| E. embiotocae | Amphistichus argenteus | Embiotocidae | California USA | [13] |
| E. embiotocae | Cymatogaster aggergata | Embiotocidae | California USA | [13] |
| E. kuwaitensis | Caranx sexfasciatus | Carangidae | Mediterranean Sea | [13] |
| E. kuwaitensis | Caranx sp. | Carangidae | Arabian Gulf | [13] |
| E. kuwaitensis | Plectorhinchus shotaf | Haemulidae | Arabian gulf | [18] |
| E. lutjanid | Lutjanus johni | Lutjanidae | India | [13] |
| E. pagrosomi | Caulolatilus princeps | Malacanthidae | Peru | [14] |
| E. pagrosomi | Caulolatilus sp. | Malacanthidae | Galapagos Islands | [13] |
| E. pagrosomi | Chrysophrys auratus | Sparidae | Australia | [13] |
| E. pagrosomi | Haemulon steindachneri | Haemulidae | Venezuela | [6] |
| E. pagrosomi | Orthopristis ruber | Haemulidae | Venezuela | [6] |
| E. pagrosomi | Pagrosomus auratus | Sparidae | Australia | [13] |
| E. pagrosomi | Pomadasys macracanthus | Haemulidae | Mexico | [13] |
| E. parvum | Orthorpistis ruber | Haemulidae | Brazil | This study |
| E. souzalimae | Thyrsitops lepidopoides | Gempylidae | Brazil | [11] |
| E. souzalimae | Trichiurus lepturus | Trichiuridae | Brazil | [5] |
| E. spari | Acanthopagrus bifasciatus | Sparidae | Arabian Gulf | [18] |
| E. spari | Anisotremus surinamensis | Haemulidae | Brazil | [35] |
| E. spari | Argyrops spimniofer | Sparidae | Arabian Gulf | [18] |
| E. spari | Carangoides bajad | Carangidae | Arabian Gulf | [18] |
| E. spari | Conodon nobilis | Haemulidae | Brazil | [35] |
| E. spari | Epinephelus akaara | Serranidae | Japan | [13] |
| E. spari | Gymnocranius griseus | Lethrinidae | Vietnam | [13] |
| E. spari | Haemulon Sciurus | Haemulidae | Brazil | [13] |
| E. spari | Lethrinus nebulosus | Lehtrinidae | Japan | [13] |
| E. spari | Menticirrhus americanus | Sciaenidae | Brazil | [7] |
| E. spari | Micropogonias furnieri | Sciaenidae | Brazil | [35] |
| E. spari | Nemipterus virgatus | Nemipteridae | Japan | [13] |
| E. spari | Orthopristis ruber | Haemulidae | Brazil | [9] |
| E. spari | Pagrus major | Sparidae | Japan | [13] |
| E. spari | Pagrus pagrus | Sparidae | Brazil | [23] |
| E. spari | Parapristipoma trilineatus | Haemulidae | Japan | [13] |
| E. spari | Plectorhinchus cinctus | Haemulidae | Arabian Gulf | [19] |
| E. spari | Plectorhinchus pictus | Haemulidae | Arabian Gulf | [13,19] |
| E. spari | Plectorhinchus schotaf | Haemulidae | Arabian Gulf | [19] |
| E. spari | Plectorhinchus sp. | Haemulidae | Vietnam | [13] |
| E. spari | Plectorhinchus spp. | Haemulidae | Arabian Gulf | [18] |
| E. spari | Sebastes inermis | Sebastidae | Japan | [13] |
| E. spari | Sparus aurata | Sparidae | Mediterranean Sea | [27] |
| E. spari | Sparus macrocephalus | Sparidae | Japan | [13] |
| E. spari | Umbrina canosai | Sciaenidae | Argentina | [4] |
| E. spari | Upeneus tragula | Mullidae | Japan | [13] |
| E. xiamenesis | Pagrosomus major | Sparidae | Taiwan | [23] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Taborda, N.; Sepulveda, F.A.; Luque, J.L.; Escribano, R.; Oliva, M.E. Two New Species of Encotyllabe (Monogenea: Capsalidae) from Brazil: Morphological and Molecular Evidence. Diversity 2023, 15, 706. https://doi.org/10.3390/d15060706
Taborda N, Sepulveda FA, Luque JL, Escribano R, Oliva ME. Two New Species of Encotyllabe (Monogenea: Capsalidae) from Brazil: Morphological and Molecular Evidence. Diversity. 2023; 15(6):706. https://doi.org/10.3390/d15060706
Chicago/Turabian StyleTaborda, Naraiana, Fabiola A. Sepulveda, Jose L. Luque, Rubén Escribano, and Marcelo E. Oliva. 2023. "Two New Species of Encotyllabe (Monogenea: Capsalidae) from Brazil: Morphological and Molecular Evidence" Diversity 15, no. 6: 706. https://doi.org/10.3390/d15060706
APA StyleTaborda, N., Sepulveda, F. A., Luque, J. L., Escribano, R., & Oliva, M. E. (2023). Two New Species of Encotyllabe (Monogenea: Capsalidae) from Brazil: Morphological and Molecular Evidence. Diversity, 15(6), 706. https://doi.org/10.3390/d15060706







