Molecular Phylogeny of the Genus Paracanthonchus (Nematoda: Chromadorida) with Description of P. yeongjongensis sp. nov. from Korea †
Abstract
1. Introduction
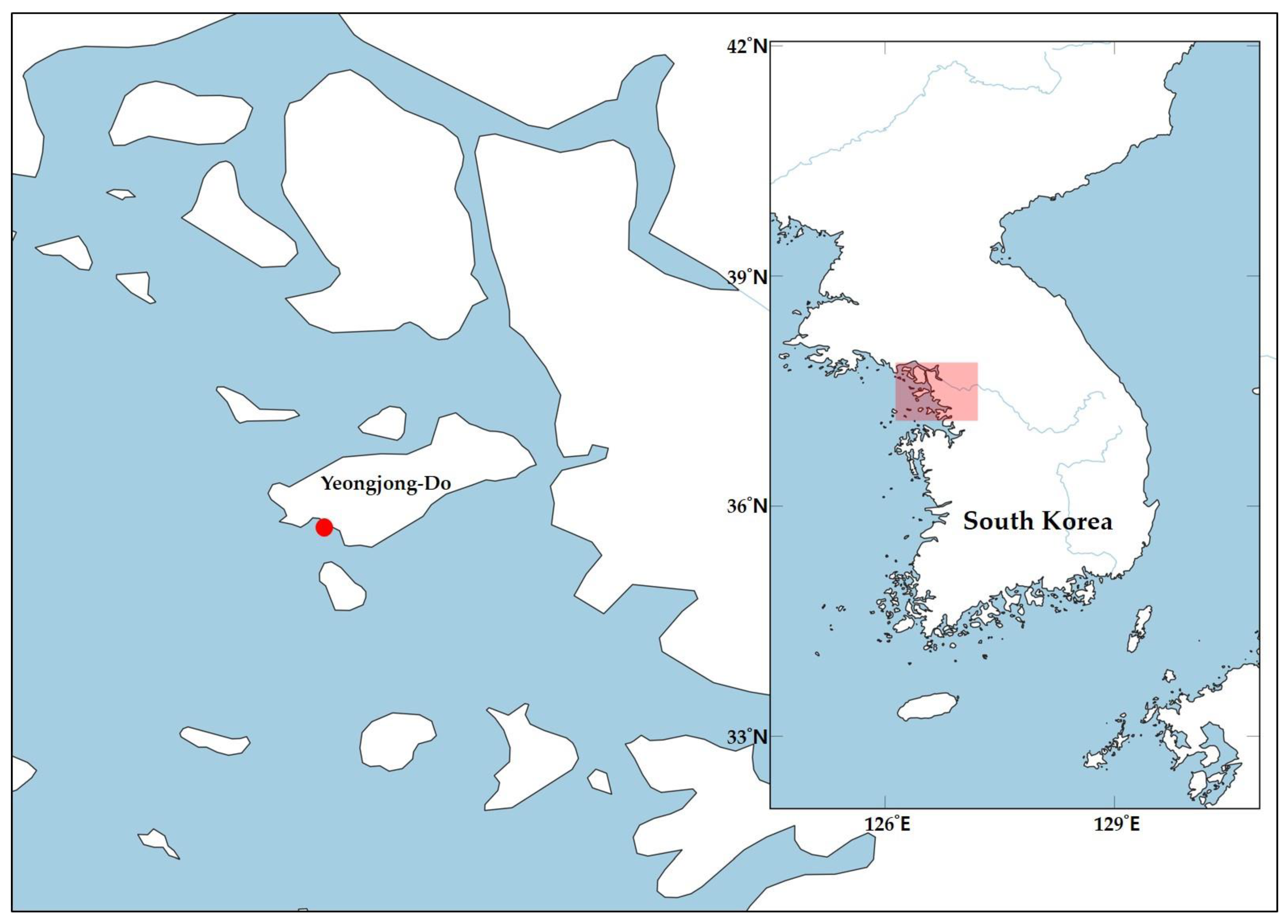
2. Materials and Methods
2.1. Morphological Analysis
2.2. gDNA Extraction and Amplification
2.3. Phylogenetic Analysis
3. Results
3.1. Systematics
3.2. Diagnosis (Followed Tchesunov, 2014)
3.3. List of Valid Species
3.4. Tabular Key to Valid Species (Table 2)
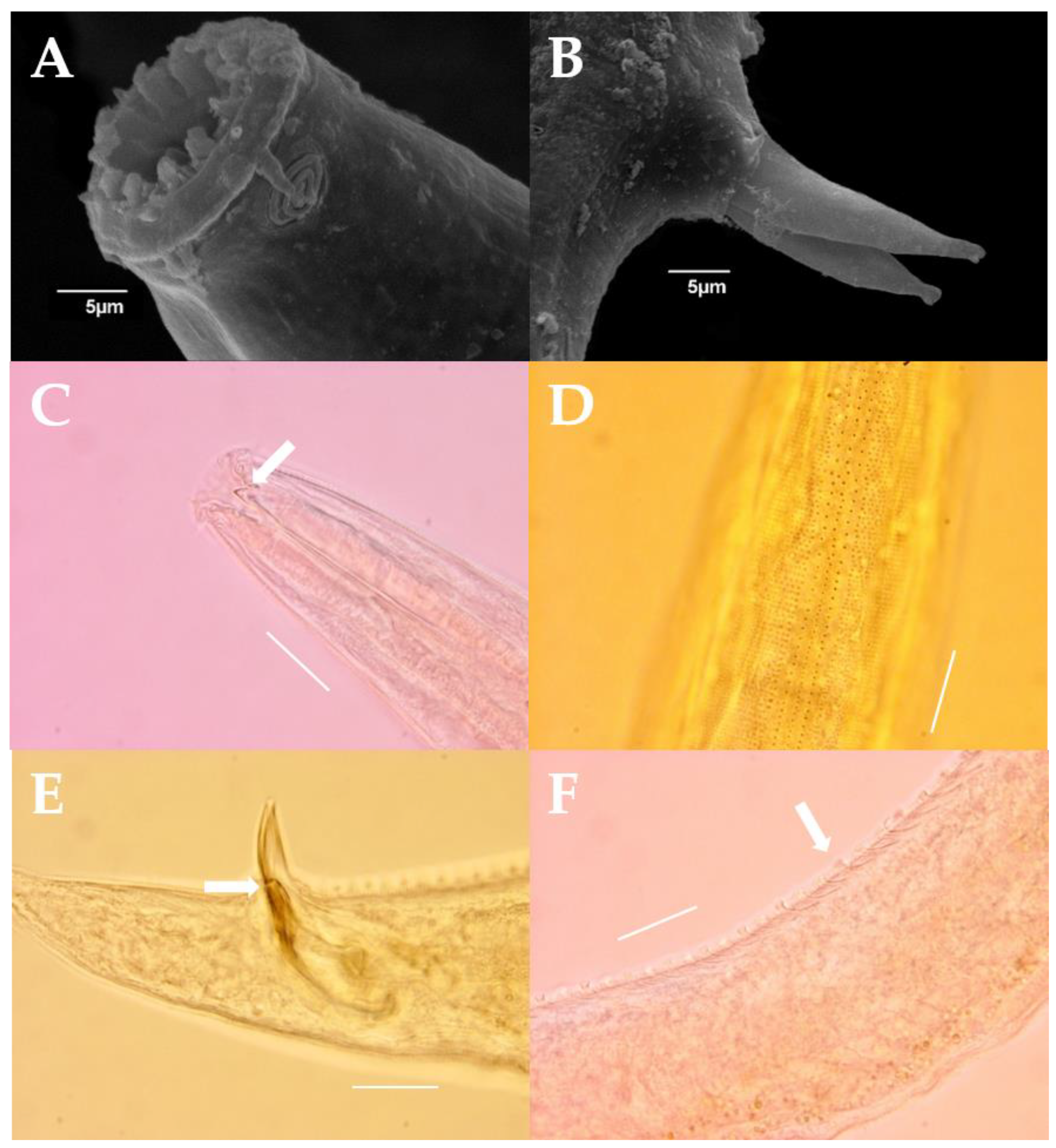
3.5. Taxonomic Description
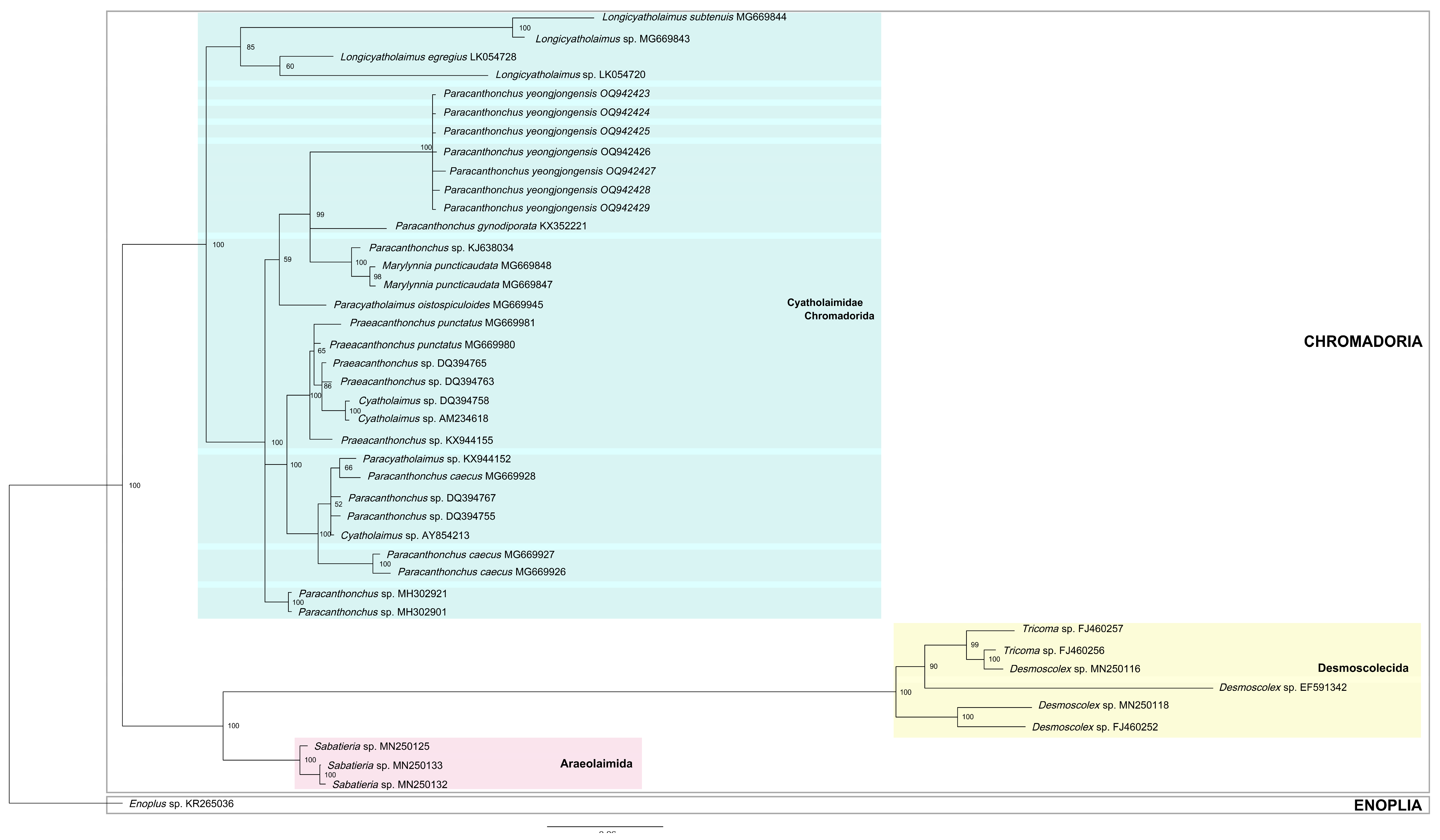
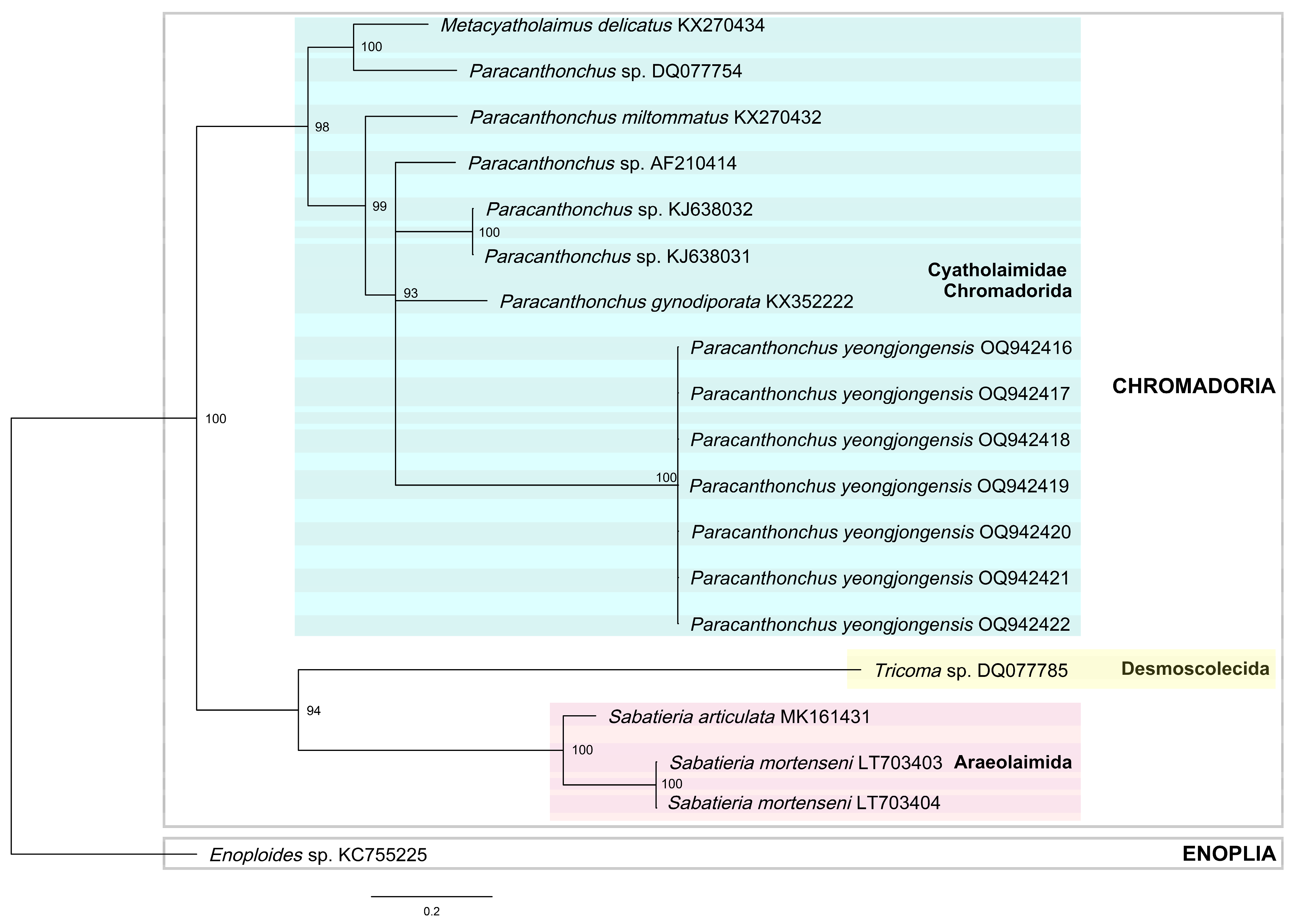
3.6. Molecular Analysis
4. Discussion
4.1. List of Valid Species
4.2. Differential Morphological Diagnoses
4.3. Phylogeny and Topology
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
References
- Leduc, D.; Zhao, Z. Phylogenetic relationships within the Cyatholaimidae (Nematoda: Chromadorida), the taxonomic significance of cuticle pore and pore-like structures, and a description of two new species. Mar. Biodivers. 2018, 48, 217–230. [Google Scholar] [CrossRef]
- Hodda, M. Phylum Nematoda: A classification, catalogue and index of valid genera, with a census of valid species. Zootaxa 2022, 5114, 1–289. [Google Scholar] [CrossRef] [PubMed]
- Miljutina, M.; Miljutin, D. A revision of the genus Paracanthonchus (Cyatholaimidae, Nematoda) with a tabularkey to species and a description of P. mamubiae sp. n. from the deep North-Western Pacific. Deep.-Sea Res. Pt. II 2015, 111, 104–118. [Google Scholar]
- Cunha, B.P.; Fonseca, G.; Amaral, A.C.Z. Diversity and Distribution of Cyatholaimidae (Chromadorida: Nematoda): A Taxonomic and Systematic Review of the World Records. Front. Mar. Sci. 2022, 9, 836670. [Google Scholar] [CrossRef]
- Vincx, M.; Sharma, J.; Smol, N. On the Identity and Nomenclature of Paracanthonchus caecus (Bastian, 1865), with a Redefinition of the Genus Paracanthonchus Micoletzky (Nematoda, Cyatholaimidae). Zool. Scr. 1982, 11, 243–263. [Google Scholar] [CrossRef]
- Filipjev, I.N. Free-living marine nematodes of the Sevastopol area. Trans. Zool. Lab. Sevastopol. Biol. Stat. Rus. Acad. Sci. 1918, 2, 1–255. [Google Scholar]
- De Coninck, L.A.; Schuurmans-Stekhoven, J.H. The freeliving marine nemas of the Belgian Coast. II With general remarks on the structure and the system of nemas. Mém. Mus. R. Hist. Nat. Belg. 1933, 58, 3–163. [Google Scholar]
- Bezerra, T.; Eisendle, U.; Hodda, M.; Holovachov, O.; Leduc, D.; Mokievsky, V. Nemys: World Database of Nematodes. 2022. Available online: http://nemys.ugent.be (accessed on 1 February 2023).
- Micoletzky, H. Letzter Bericht über freilebende Nematoden aus Suez. Sber. Akad. Wiss. Wien (I) 1924, 133, 137–179. [Google Scholar]
- Wieser, W. Free-Living Marine Nematodes II. Chromadoroidea; Gleerup: Lund, Sweden, 1954; Volume 50, pp. 1–148. [Google Scholar]
- Stekhoven, J.H. The free-living marine nemas of the Mediterranean. I. The Bay of Villefranche. Mem. Inst. R. Sci. Nat. Belg. Ser. 1950, 2, 1–220. [Google Scholar]
- Hope, W.D.; Murphy, D.G. A taxonomic hierarchy and checklist of the genera and higher taxa of marine nematodes. Smithson. Contrib. Zool. 1972, 137, 1–101. [Google Scholar] [CrossRef]
- Mawson, P.M. Some marine freeliving nematodes from the Australian coast. Trans. R. Soc. S. Aust. 1953, 76, 34–40. [Google Scholar]
- Wieser, W. Free-Living Nematodes and Other Small Invertebrates of Puget Sound Beaches; University of Washington Press: Seattle, WA, USA, 1959; pp. 1–179. [Google Scholar]
- Tchesunov, A. Order Chromadorida Chitwood, 1933. In Handbook of Zoology; Schmidt-Rhaesa, A., Ed.; De Gruyter: Berlin, Germany, 2014; Volume 2, pp. 373–398. [Google Scholar]
- Platt, H.; Warwick, R. Free-Living Marine Nematodes; Part 2: British Chromadorids; Brill, E.J., Backhuys, W., Eds.; Linnean Society of London: London, UK; Estuarine and Brackish-water Sciences Association: Leiden, The Netherlands, 1988; p. 502. [Google Scholar]
- Tchesunov, A. Free-living nematode species (Nematoda) dwelling in hydrothermal sites of the North Mid-Atlantic Ridge. Helgoland Mar. Res. 2015, 69, 343–384. [Google Scholar] [CrossRef]
- Lee, H.; Jung, J.; Rho, H. Two unrecorded marine nematode species of Paracanthonchus (Nematoda: Cyatholaimidae) from the East Sea of Korea. J. Species Res. 2016, 5, 503–513. [Google Scholar] [CrossRef]
- Kim, H.; Tchesunov, A.; Lee, W. A new species of the genus Marylynnia (Nematoda: Chromadorida: Cyatholaimidae) from Gwangyang Bay, Korea. Proc. Biol. Soc. Wash. 2015, 128, 227–238. [Google Scholar] [CrossRef]
- Jeong, R.; Tchesunov, A.; Lee, W. A new species of the genus Thalassironus (Nematoda: Enoplida: Ironidae) from the coasts of South Korea. Zootaxa 2019, 4563, 516–530. [Google Scholar] [CrossRef]
- Tchesunov, A.; Jeong, R.; Lee, W. Two New Marine Free-Living Nematodes from Jeju Island Together with a Review of the Genus Gammanema Cobb 1920 (Nematoda, Chromadorida, Selachinematidae). Diversity 2020, 12, 19. [Google Scholar] [CrossRef]
- Hong, J.; Tchesunov, A.; Lee, W. Revision of Cervonema Wieser, 1954 and Laimella Cobb, 1920 (Nematoda: Comesomatidae) with descriptions of two species from East Sea, Korea. Zootaxa 2016, 4098, 333–357. [Google Scholar] [CrossRef]
- Gagarin, V.; Nguyen Vu Thanh. Paracanthonchus brevicaudatus sp. n. (Nematoda, Chromadorida, Cyatholaimidae) from artificial reservoirs in Vietnam. Int. J. Nematol. 2016, 26, 9–14. [Google Scholar]
- Allgén, C. Pacific Freeliving Marine Nematodes. (Papers from Dr. Th. Mortensen’s Pacific Expedition 1914–16. LXXVI). Vidensk. Meddel. Naturhist. Foren. Kjobenhavn 1951, 113, 263–411. [Google Scholar]
- Huang, Y.; Xu, K. Two new free-living nematode species (Nematoda: Cyatholaimidae) from intertidal sediments of the Yellow Sea, China. Cah. Biol. Mar. 2013, 54, 1–10. [Google Scholar]
- Kito, K. Studies on the free-living marine nematodes from Hokkaido, IV. J. Fac. Sci. Hokkaido Univ. Ser. VI Zool. 1981, 22, 250–278. [Google Scholar]
- Gagarin, V. Paracanthonchus multisupplementatus sp.n. and Cyatholaimus minor sp.n. (Nematoda) from the coast of Vietnam. Zootaxa 2012, 3392, 60–68. [Google Scholar] [CrossRef]
- Nguyen Dinh Tu; Gagarin, V. Two new species of nematodes of the order chromadorida (Nematoda: Chromadorea) from Vietnam. Russ. J. Mar. Biol. 2018, 44, 86–95. [Google Scholar]
- Hopper, D. Drawing and measuring nematodes. In Laboratory Methods for Work with Plant and Soil Nematodes; Ministry of Agriculture, Fisheries and Food: Her Majesty’s Stationery Office: London, UK, 1970; pp. 59–65. [Google Scholar]
- Schindelin, J.; Arganda-Carreras, I.; Frise, E.; Kaynig, V.; Longair, M.; Pietzsch, T.; Preibisch, S.; Rueden, C.; Saalfeld, S.; Schmid, B.; et al. Fiji: An Open-Source Platform for Biological-Image Analysis. Nat. Methods 2012, 9, 676–682. [Google Scholar] [CrossRef] [PubMed]
- Derycke, S.; Vanaverbeke, J.; Rigaux, A.; Backeljau, T.; Moens, T. Exploring the Use of Cytochrome Oxidase c Subunit 1 (COI) for DNA Barcoding of Free-Living Marine Nematodes. PLoS ONE 2010, 5, e13716. [Google Scholar] [CrossRef] [PubMed]
- Bowles, J.; Bowles, J.; Blair, D.; McManus, D.P. Genetic variants within the genus Echinococcus identified by mitochondrial DNA sequencing. Mol. Biochem. Parasitol. 1992, 54, 165–173. [Google Scholar] [CrossRef]
- Carta, L.K.; Li, S. Improved 18S small subunit rDNA primers for problematic nematode amplification. J. Nematol. 2018, 50, 533–542. [Google Scholar] [CrossRef] [PubMed]
- Carta, L.K.; Li, S. PCR amplification of a long rDNA segment with one primer pair in agriculturally important nematodes. J. Nematol. 2019, 51, e2019–e2026. [Google Scholar] [CrossRef]
- De Ley, P.; De Ley, I.M.; Morris, K.; Abebe, E.; Mundo-Ocampo, M.; Yoder, M.; Heras, J.; Waumann, D.; Rocha-Olivares, A.; Jay Burr, A.H.; et al. An integrated approach to fast and informative morphological vouchering of nematodes for applications in molecular barcoding. Phil. Trans. R. Soc. B 2005, 360, 1945–1958. [Google Scholar] [CrossRef]
- Thompson, J.D.; Higgins, D.G.; Gibson, T.J. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994, 22, 4673–4680. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular evolutionary genetics analysis version 11. Mol. Boil. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Boil. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Kimura, M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 1980, 16, 111–120. [Google Scholar] [CrossRef]
- Minh, B.Q.; Schmidt, H.A.; Chernomor, O.; Schrempf, D.; Woodhams, M.D.; von Haeseler, A.; Lanfear, R. IQ-TREE 2: New models and efficient methods for phylogenetic inference in the genomic era. Mol Biol Evol. 2020, 37, 1530–1534. [Google Scholar] [CrossRef]
- Kalyaanamoorthy, S.; Minh, B.Q.; Wong, T.K.F.; von Haeseler, A.; Jermiin, L.S. ModelFinder: Fast model selection for accurate phylogenetic estimates. Nat. Methods 2017, 14, 587–589. [Google Scholar] [CrossRef]
- Minh, B.Q.; Nguyen, M.A.T.; von Haeseler, A. Ultrafast approximation for phylogenetic bootstrap. Mol. Biol. Evol. 2013, 30, 1188–1195. [Google Scholar] [CrossRef] [PubMed]
- Darriba, D.; Taboada, G.L.; Doallo, R.; Posada, D. jModelTest 2: More models, new heuristics and high-performance computing. Nat. Methods 2012, 9, 772. [Google Scholar] [CrossRef] [PubMed]
- Huelsenbeck, J.P.; Ronquist, F. Mrbayes: Bayesian inference of phylogenetic trees. Bioinformatics 2001, 17, 754–755. [Google Scholar] [CrossRef]
- Rambaut, A. FigTree Version 1.4.4. 2018. Available online: http://tree.bio.ed.ac.uk/software/figtree/ (accessed on 1 March 2023).
- Inglis, W. Marine nematodes from Banyuls-sur-mer: With a review of the genus Eurystomina. Bull. Br. Mus. Nat. Hist. 1962, 8, 209–287. [Google Scholar]
- Gerlach, S. Die Nematodenfauna des Sandstrandes an der Küste von Mittelbrasilien (BrasilianischeMeeres-NematodenIV). Mitt. Zool. Mus. Berl. 1957, 33, 411–459. [Google Scholar] [CrossRef]
- Kreis, H. Die freilebenden marinen Nematoden der Spitzbergen-Expedition von F. Roemer und F. Schaudinnim Jahre 1898. Mitt. Zool. Mus. Berl. 1928, 14, 131–197. [Google Scholar]
- Schiemer, F.; Jensen, P.; Riemann, F. Ecology and systematics of free-living nematodes from the Bothnian Bay, northern Baltic Sea. Ann. Zool. Fenn. 1983, 20, 277–291. [Google Scholar]
- Bussau, C. Taxonomische und ökologische Untersuchungen an Nematoden des Peru-Beckens. Ph.D. Thesis, University of Kiel, Kiel, Germany, 1993. [Google Scholar]
- Bastian, H.C. Monograph on the Anguillulidae, or free nematoids, marine, land, and fresh water; with descriptions of 100 new species. Trans. Linn. Soc. Lond. 1865, 25, 73–184. [Google Scholar] [CrossRef]
- Inglis, W.G. Cyatholaimidae (Nematoda) from the coast of Western Australia. Rec. S. Austr. Mus. 1970, 16, 1–13. [Google Scholar]
- De Man, J.G. Sur quelques espèces nouvelles ou peu connues de nématodes libres vivant sur les côtes de la Zélande. Tijdschr. Ned. Dierk. Vereen. 1907, 10, 227–244. [Google Scholar]
- Filipjev, I.N. Encore sur les Nématodes libres de la mer Noire. Tr. Stravrop. Skh. Inst. Zool. 1922, 1, 83–184. [Google Scholar]
- Oliveira, D.A.S.; de Decraemer, W.; Moens, T.; Dos Santos, G.A.P.; Derycke, S. Low genetic but high morphological variation over more than 1,000 km coastline refutes omnipresence of cryptic diversity in marine nematodes. BMC Evolution. Biol. 2017, 17, 71. [Google Scholar] [CrossRef]
- Schulz, E. Beitrage zur Kenntnis mariner Nematoden aus der Kieler Bucht. Zool. Jb. (Syst.) 1932, 62, 331–430. [Google Scholar]
- Gourbault, N. Nématodes abyssaux (Campagne Walda du navire océanographique Jean-Charcot). I. Espèces nouvelles de Cyatholaimidae. Cah. Biol. Mar. 1980, 21, 61–71. [Google Scholar]
- Gagarin, V.G.; Klerman, A.K. Two new species of free-living nematodes (Nematoda) from the Mediterranean Sea. Zool. Bespozvon. 2008, 5, 1–7. [Google Scholar]
- Warwick, R.M. The Cyatholaimidae (Nematoda, Chromadoroidea) off the coast of Northumberland. Cah. Biol. Mar. 1971, 12, 95–110. [Google Scholar]
- Pastor de Ward, C.T. Free-living nematodes of the Deseado River estuary (Chromadoroidea: Chromadoridae, Etholaimidae, Cyatholaimidae and Choniolaimidae) Santa Cruz, Argentina. 5. Cent. Nac. Patagon. Publ. Espec. 1985, 6, 1–83. [Google Scholar]
- Allgén, C. Über einige, meistens neue, freilebende marine Nematoden aus dem Feuerland-Archipel (Schwedische Südpolar-Expedition 1901–1903). Zool. Anz. 1959, 163, 222–243. [Google Scholar]
- Galtsova, V.V. Free-living marine nematodes as components of the meiofauna of the Chupa Bay, White Sea. Issled. Fauny Morej 1976, 15, 165–270. [Google Scholar]
- Stekhoven, J.H. Freilebende marine Nematoden des Mittelmeeres. IV. Freilebende marine Nematoden der Fischereigründe bei Alexandrien. Zool. Jb. (Syst.) 1943, 76, 323–378. [Google Scholar]
- Timm, R.W. The marine nematodes of the Bay of Bengal. Proc. Pak. Acad. Sci. 1961, 1, 25–88. [Google Scholar]
- Decraemer, W.; Coomans, A.C. Scientific report on the Belgian expedition to the Great Barrier Reefin 1967. Nematodes, XIII. A description of four new species and a redescription of four known species from in and around mangroves on LizardIsland. Aust. J. Mar. Freshw. Res. 1978, 29, 509–541. [Google Scholar] [CrossRef]
- Jayasree Vadhyar, K. A new species of Paracanthonchus Micoletzky, 1924 and a known species of Paracyatholaimus Micoletzky, 1922 (Nematoda, Cyatholaimidae, Paracanthonchinae) from polluted intertidal sand in Scotland. Cah. Bid. Mar. 1980, 21, 375–381. [Google Scholar]
- Wieser, W.; Hopper, B. Marine nematodes of the east coast of North America. I. Florida. Bull. Mus. Comp. Zool. Harv. 1967, 135, 239–344. [Google Scholar]
- Bouwman, L.A. A survey of nematodes from the Ems estuary. Part I: Systematics. Zool. Jb. (Syst.) 1981, 108, 335–385. [Google Scholar]
- Nasira, K.; Kamran, M.K.; Shanina, F. Two new species of free-living marine nematodes of the family Cytaholaimidae (Nematoda: Chromadorida) from the Arabian sea of Pakistan. Pak. J. Nematol. 2007, 25, 91–98. [Google Scholar]
- Allgén, C. Freilebende marine Nematoden von der Stateninsel (Feuerland-Archipel). II. Zool. Anz. 1930, 90, 27–38. [Google Scholar]
- Micoletzky, H. Neue freilebende Nematoden aus Suez. Sber. Akad. Wiss. Wien. 1922, 131, 77–103. [Google Scholar]
- Allgén, C. Die freilebenden Nematoden des Mittelmeers. Zool. Jb. (Syst.) 1942, 76, 1–102. [Google Scholar]
- Timm, R.W. New Marine Nematodes from St. Martin’s Island. Pak. J. Sci. 1957, 9, 133–138. [Google Scholar]
- Brunetti, B. Contributo alla conoscenza dei nematode del M. Tirreno. II. Alcune specie appartenenti alle famiglie: Enoplidae, Cyatholaimidae, Chromadoridae, Axonolaimidae. Monit. Zool. Ital. 1949, 57, 41–59. [Google Scholar]
- Gerlach, S.A. Die Nematodenbesiedlung des Sandstrandes und des Küstengrundwassers an der italienischen Küste. I. Systematischer Teil. Archo Zool. Ital. 1953, 37, 517–640. [Google Scholar]
- Leduc, D.; Zhao, Z.Q. The Marine Biota of Aotearoa New Zealand; Ngā toke o Parumoana: Common free-living Nematoda of Pāuatahanui Inlet, Te-Awarua-o-Porirua Harbour, Wellington; NIWA Biodiversity Memoir: Auckland, New Zealand, 2023; Volume 135, p. 212. [Google Scholar]
- Patterson, C.; Williams, D.M.; Humphries, C.J. Congruence between molecular and morphological phylogenies. Annu. Rev. Ecol. Syst. 1993, 24, 153–188. [Google Scholar] [CrossRef]
- Cohen, B.L. Match and mismatch of morphological and molecular phylogenies: Causes, implications, and new light on cladistics. Zool. J. Linn. Soc. 2018, 184, 516–527. [Google Scholar] [CrossRef]

| Marker | Primer (Direction) | Sequence 5′-3′ | Amplification Condition | Sequence Length (bp) | Reference |
|---|---|---|---|---|---|
| mtCOI | JB3 (f) | TTTTTTGGGCATCCTGAGGTTTAT | 94 °C for 5 min, 5 cycles of 94 °C for 30 s, 54 °C for 30 s and 72 °C for 30 s, followed by 35 cycles of denaturation at 94 °C for 30 s, annealing at 50 °C for 30 s, and extension at 72 °C for 30 s, followed by final step of 72 °C for 10 min | 330 | Derycke et al., 2010 [31]; Bowles et al., 1992 [32] |
| JB5 (r) | AGCACCTAAACTTAAAACATAATGAAAATG | ||||
| 18s | 18S-CL-F (f) | TCAAAGATTAAGCCATGCAT | 95 °C for 3 min followed by 36 cycles of denaturation at 95 °C for 30 s, annealing at 50 °C for 45 s, and extension at 72 °C for 3 min, followed by final step at 72 °C for 7 min | 494 | Carta and Li, 2018; Carta and Li, 2019 [33,34] |
| 530R ® | GCGGCTGCTGGCACCACACTT | ||||
| 28S | D2A (f) | ACAAGTACCGTGAGGGAAAGTTG | 95 °C for 5 min followed by 37 cycles of denaturation at 95 °C for 30 s, annealing at 56 °C for 1 min, and extension at 72 °C for 1 min 30 s min, followed by final step at 72 °C for 5 min | 731–741 bp | De Ley et al., 2005 [35] |
| D®(r) | TCGGAAGGAACCAGCTACTA |
| Species | Body Length (Male) | Body Length (Female) | Lateral Differentiation of Cuticle | Number of Subventral Teeth | Distance from Anterior to Amphid | Amphid Width | Turn Number of Amphid | Spicule Length as Arc | Gubernaculum Length | Number of Cusps on Distal End of Gubernaculum | Number of Supplements | Tail Shape |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| P. austrospectabilis | 1440–2090 | 1370–2280 | none/except on the tail | n/a | 9 | 13 | 4.5 | 65 | 65 | numerous | 6 | conical |
| P. barka | 860 | presence | n/a | 5 | 7–8 | 4.5–5.5 | 19 | 18 | 2 | 4 | conical | |
| P. batidus | 888 | none | n/a | 6 | 5.5 | 2.5 | 31 | 22 | n/a | 6 | conical | |
| P. bipapillatus | 2046 | none | none | 10 | 13 | 2.5 | 70.2 | 59.8 | 2 | 5 | conical | |
| P. bothnicus | 1568–1800 | 1715–1784 | presence | 4 | 9 | 10 | 6 | 50 | 45 | 4 | 5 | conical |
| P. brevicaudatus | 956–1230 | 992–1413 | none | 3 | 11 | 13–15 | 4.3–4.5 | 30 | 27 | 16–18 | 3 | conical |
| P. breviseta | 1092 | 1072 | none | n/a | 20 | 14 | 5.5 | 44 | n/a | 1 | 4 | conical |
| P. bulbicola | 1340 | 1230–1960 | presence | 4 | 11 | 15 | 6.25 | 38 | 23 | numerous | 5 | conical |
| P. caecus | 976–1470 | presence | 2–4 | 10 | 7–13 | 4.5–5.5 | 40–48 | 35–44 | 8–9 | 5 | conical | |
| P. canadensis | 960–1160 | 1089–1170 | none | 4 | 7 | 7 | 6 | 39.1–40.6 | 35.5–40.6 | numerous | 5 | conical |
| P. cheynei | 1240–1280 | 1090 | none | 4 | 14 | 9–10 | 4.25 | 46–48 | 39–42 | numerous | 6 | conical |
| P. cochlearis | 1123 | 1162 | presence | 2 | 10 | 17 | 6 | 36 | 30 | 2 | 5 | conical–cylindrical |
| P. cristatus | 1050–1360 | 1180–1300 | presence | n/a | 9–13 | 8 | 2.75–3 | 40 | 35 | n/a | 8–9 | conical |
| P. elongatus | 3025 | presence | none | 15 | 15 | 4–5 | 86 (as chord) | n/a | numerous | 5 | conical | |
| P. filipjevi | 1150 | 1300 | n/a | n/a | 7 | 9 | 4–5.5 | 46 | 41 | 2 | 3 | conical |
| P. gerlachi | 1194–1391 | 1045–1480 | none | 4 | 12 | 10 | 7.5 | 38–39 | 36 | 7 | 5 | conical |
| P. gynodiporata | 1001–1146 | 1075–1238 | presence | 4 | 11.3–12.9 | 7.4–10.7 | 4 | 38.9–42.1 | 34.5–40.8 | numerous | 4 | conical |
| P. hartogi | 1240–1420 | none | none | 15 | 12–13 | 3.25 | 44–49 | 39–44 | 4 | 6 | conical | |
| P. hawaiiensis | 1625 | presence | n/a | 13 | 10 | 3 | 40–44 | 39–44 | numerous | 4 | conical | |
| P. heterocaudatus | 1335–1570 | 1555–1750 | presence | 2 | 7 | 9–13 | 5–6 | 31–32 | 25–27 | 2 | 6 | conical–cylindrical |
| P. heterodontus | 1042–1668 | 1129–1842 | none | 4 | 13 | 13 | 4.5 | 56–67 | 46–54 | numerous | 5 | conical |
| P. kamui | 1658–1728 | 1761–1816 | presence | 4 | 6 | 10 | 4.25 | 53–60 | 45–55 | numerous | 6 | conical |
| P. kartanum | 870–1600 | 1450 | none | 4 | 15 | 9 | 3.5–4.5 | 23–38 | 26–36 | numerous | 6 | conical |
| P. latens | 1733–2458 | none | 4 | 8 | 10 | 6.5 | 84–87 | 43–50 | numerous | 4 | conical | |
| P. lissus | 1165–1252 | 1334 | none | n/a | 10 | 10–12 | 4–4.5 | 39–43 | 45–49 | 4 | 4 | conical |
| P. longicaudatus | 1330–1510 | 1570–1790 | none | 4 | 12 | 9–10 | 5.3–6.25 | 43–51 | 40–42 | numerous | 5 | conical–cylindrical |
| P. longispiculum | 1780–1800 | 1900 | n/a | 2 | 11 | 6–10 | 2.5–3 | 70–76 | 70 | none | 8 | conical |
| P. macrospiralis | 3000 | presence | n/a | 12 | 20 | 3 | 65 | 32 | n/a | n/a | conical | |
| P. major | 2608 | 2676 | none | n/a | n/a | 6.5–7.8 | 2.5 | 59.8 | 49.4 | none | 4 | conical |
| P. mamubiae | 1610–1831 | 1638–1971 | none | 4 | 3–7 | 7–14 | 4.15 | 60–65 | 46–51 | numerous | 3–5 | conical–cylindrical |
| P. margaretae | 1280–1480 | 1210 | presence | 4 | 10 | 10–11 | 4.75 | 47–48 | 44–45 | numerous | 6 | conical |
| P. medius | 913–1254 | 1232 | none | n/a | n/a | n/a | n/a | 48 | 44.4 | numerous | 4 | conical |
| P. micoletzkyi | 1020 | 1220 | presence | n/a | 12 | 12 | 4.5 | 41 | 25 | none | 4 | conical |
| P. micropapillatus | 960–1350 | 950–1400 | none/except on the tail | n/a | 5 | 8.5–10.5 | 3.25–2.5 | 34 | 25 | 2 | 7 | conical |
| P. miltommatus | 1827–2051 | 1885–2062 | none/except on the tail | 4 | 7–12 | 10–11 | 4.5–5 | 53–57 | 48–55 | 5–8 | 6 | conical |
| P. multisupplementatus | 1027–1308 | 1181–1365 | presence | 2(4?) | 3.5–6.5 | 8–13 | 4–4.5 | 56–61 | 36–43 | numerous | 57–62 | conical |
| P. multitubifer | 1100–1200 | 1100–1240 | none | n/a | 8 | 7.5 | 3.5 | 36 | 28 | 3 | 21–22 | conical–cylindrical |
| P. mutatus | 930 | 1100 | none/except anterior | 1 or 2 | 10 | 9–10 | 5 | 30 | 24 | none | 5 | conical |
| P. nannodontus | 1600 | 1430 | n/a | n/a | 17 | 9 | 3.5–4 | 60 | 57 | numerous | 3? | conical |
| P. olgae | 1300–1830 | 1445–2065 | none | 4 | 6–9.9 | 6–11 | 5 | 44.5–64 | 42–69 | numerous | 5 (rarely 2–4) | conical |
| P. parahartogi | 1350–1440 | 1410–1560 | presence | n/a | 8 | 8–12 | 3.75 | 49–52 | 50–53 | 6 | 6 | conical |
| P. perspicuus | 1269–1287 | presence | none | 15 | 11 | 4.25 | 31 | 22–24 | numerous | 5 | conical | |
| P. platti | 1500–1920 | 1820 | none | 2 | 12–14 | 12–13 | 5.5–6 | 40–45 | 35 | 9–11 | 5 | conical |
| P. platypus | 1180–1320 | none | n/a | 13–15 | 9–11 | 3.5 | 36 | 35 | none | 4 | conical | |
| P. quinquepapillatus | 1360 | presence | 2? | 13 | 13 | 4.5 | 38 | 28 | none | 5 | conical | |
| P. ruens | 1610 | 1450–1530 | none/except on the tail | n/a | 8 | 9 | 3 | 75 | 72 | none | 7 | conical |
| P. sabulicolus | 1700–1800 | 1500–1700 | none | 2 | 9 | 12 | 5 | 40–45 | 37 | 12 | 4 | conical |
| P. sandspitensis | 1200–1400 | 1000–1600 | none | 4 | 10 | 8–10 | 5–6 | 32–41 | 25–30 | numerous | 5 | conical |
| P. securus | 931–1112 | 944–1205 | none | 2 | 15 | 11–13 | 4.3–4.5 | 39–43 | 24–26 | 7–10 | 3 | conical–cylindrical |
| P. sonadiae | 1150–1270 | 1110–1820 | n/a | n/a | 5 | 9 | 5.5 | 32 | 25 | n/a | 6 | conical |
| P. stateni | 920–1720 | presence | n/a | 5 | 9 | 5.5 | 33 | 22 | 2 | 5 | conical | |
| P. stekhoveni | 1132 | presence | n/a | 12 | 10 | 4.5 | 45 (as chord) | 36 | numerous | 5 | conical | |
| P. steueri | 940 | 1070 | presence | 2–4? | 10 | 7 | 3.5 | 32 | 25 | n/a | 6 | conical |
| P. sunesoni | 800–1390 | presence | n/a | 16 | 7 | 2.5–3 | 32 | 26 | numerous | 5–7 | conical | |
| P. thaumasius | 1511–1902 | 1418–2001 | none | 4 | 14 | 10–15 | 5–6.5 | 43–65 | 36–49 | 9 | 5 | conical–cylindrical |
| P. tumepapillatus | 1200-1440 | none | 2 | 12 | 11 | 5 | 22 | 16 | n/a | 3 | conical–cylindrical | |
| P. tyrrhenicus | 1700 | 1700–1800 | n/a | 4? | 15 | 12–17 | 6 | 42 | 42 | 2 | 5 | conical |
| P. uniformis | 1272 | 1200 | n/a | none? | 11 | 7–12 | 4.5–5.5 | 21 | 18 | n/a | 4 | conical |
| P. wellsi | 1485–1512 | 1465–1595 | presence | 4 | 8–13 | 10 | 5 | 36–37 | 28–31 | numerous | 3 | conical–cylindrical |
| P. yeongjongensis | 1186–1295 | 1378–1401 | presence | 2 | 5 | 5.5–8 | 2.5 | 51–58 | 49–56 | None (little denticle) | 76 | conical–cylindrical |
| Characters | Holotype | Paratype (m1) | Paratype (m2) | Paratype (f1) | Paratype (f2) |
|---|---|---|---|---|---|
| L | 1186 | 1253 | 1295 | 1401 | 1378 |
| hd | 20 | 19 | 20 | 25 | 18.5 |
| LSL | 4.5 | 5 | 5.5 | 7 | 5 |
| CSL | 4.5 | 5 | 5.5 | 7 | 5 |
| bcl | 22 | 17 | 14 | 18 | 21 |
| amp | 6 | – | 5.5 | 8 | 7 |
| na | 2.5 | – | 2.5 | 2.5 | 2.5 |
| amp cbd | 25 | – | 21 | 27 | 21.3 |
| NL | 95 | 80 | 76 | 91 | 104 |
| ncbd | 45 | 40 | 41 | 53 | 52 |
| PL | 183 | 194 | 198 | 232 | 212 |
| pcbd | 57 | 47 | 52.5 | 70 | 60 |
| mbd | 62 | 54 | 65 | 84 | 64.5 |
| VL | n/a | n/a | n/a | 800 | 776 |
| ov | n/a | n/a | n/a | 768/987 | 662/884 |
| abd | 41 | 40 | 41 | 50 | 44.5 |
| spia | 58 | 51 | 54 | n/a | n/a |
| gub | 56 | 49 | 52 | n/a | n/a |
| ns | 76 | 76 | 76 | – | – |
| dps | 11 | 16 | 13 | – | – |
| das | 558 | 611 | 599 | – | – |
| TL | 98 | 106 | 103 | 121 | 117 |
| a | 19.13 | 23.20 | 19.92 | 16.68 | 21.36 |
| b | 6.48 | 6.46 | 6.54 | 6.04 | 6.50 |
| c | 12.10 | 11.82 | 12.57 | 11.58 | 11.78 |
| c’ | 2.39 | 2.65 | 2.51 | 2.42 | 2.63 |
| V | – | – | – | 0.57 | 0.56 |
| amp’ | 0.24 | – | 0.26 | 0.30 | 0.33 |
| s’ | 1.41 | 1.28 | 1.32 | – | – |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kim, H.; Lee, W.; Jeong, R. Molecular Phylogeny of the Genus Paracanthonchus (Nematoda: Chromadorida) with Description of P. yeongjongensis sp. nov. from Korea. Diversity 2023, 15, 664. https://doi.org/10.3390/d15050664
Kim H, Lee W, Jeong R. Molecular Phylogeny of the Genus Paracanthonchus (Nematoda: Chromadorida) with Description of P. yeongjongensis sp. nov. from Korea. Diversity. 2023; 15(5):664. https://doi.org/10.3390/d15050664
Chicago/Turabian StyleKim, Hyeonggeun, Wonchoel Lee, and Raehyuk Jeong. 2023. "Molecular Phylogeny of the Genus Paracanthonchus (Nematoda: Chromadorida) with Description of P. yeongjongensis sp. nov. from Korea" Diversity 15, no. 5: 664. https://doi.org/10.3390/d15050664
APA StyleKim, H., Lee, W., & Jeong, R. (2023). Molecular Phylogeny of the Genus Paracanthonchus (Nematoda: Chromadorida) with Description of P. yeongjongensis sp. nov. from Korea. Diversity, 15(5), 664. https://doi.org/10.3390/d15050664







