Isolating Greater Cane Rat Populations (Thryonomys swinderianus) from Eastern Arc Mountains, Tanzania: Linking Diversity to Morphometric and Molecular Characteristics
Abstract
1. Introduction
2. Materials and Methods
2.1. Sampling Sites and Sampling Procedure
2.2. Data Analysis
2.2.1. Geometric Morphometric Assessment
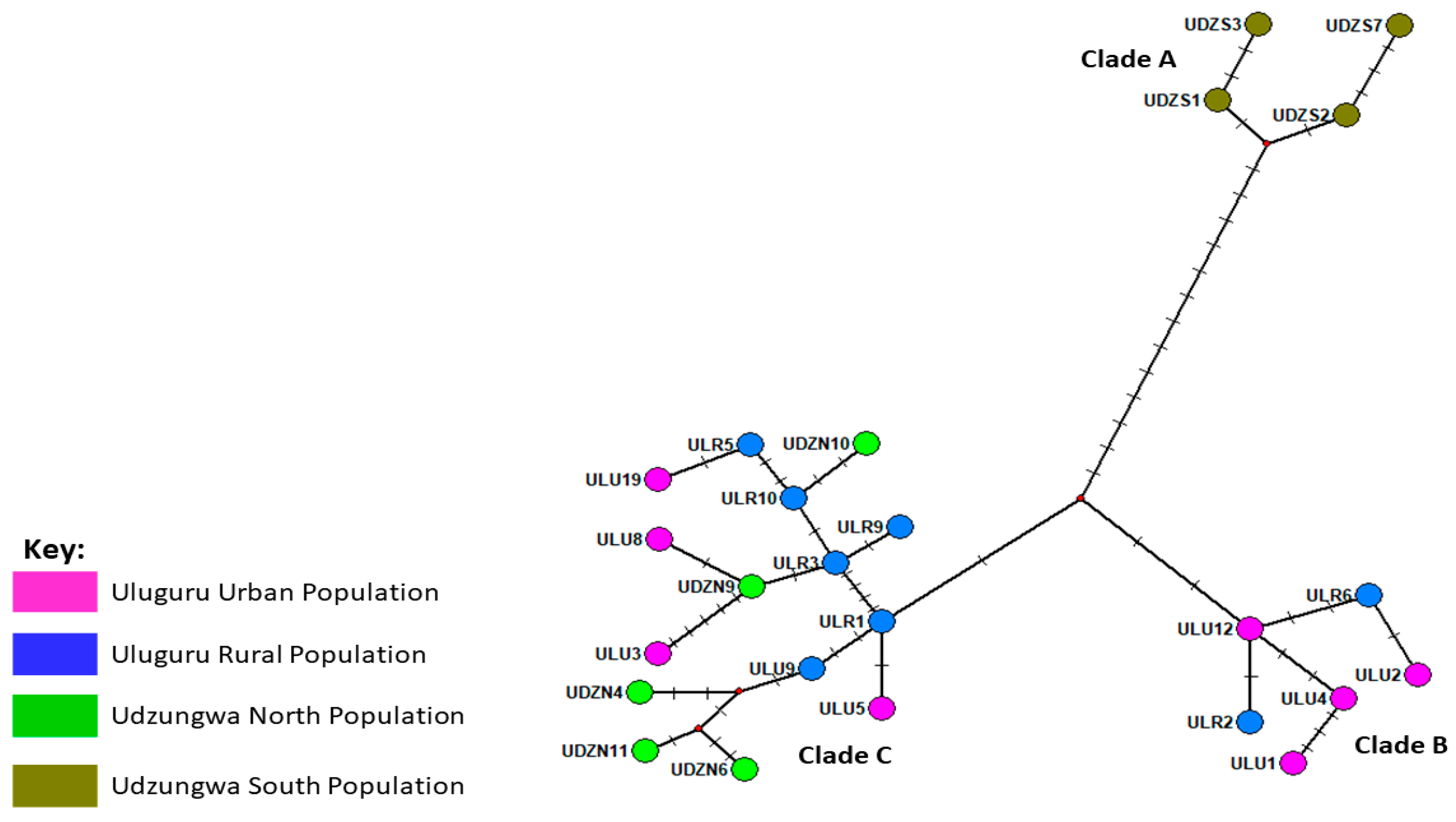
2.2.2. Molecular Analysis
3. Results
3.1. Matching Geometric and Genetic Variation Evidence in T. swinderianus Populations
Differences between Populations
3.2. Variations in Skull Size
3.3. Variations in Skull Shape
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Olude, M.A.; Mustapha, O.A.; Sonubi, A.C.; Falade, T.E.; Ogunbunmi, T.K.; Adebayo, A.O.; Akinloye, A.K. Morphometric study of the skull of the greater cane rat (Thryonomys swinderianus, Temmnick). Niger. Vet. J. 2014, 35, 1026–1037. [Google Scholar]
- Padial, J.M.; Miralles, A.; De La Riva, I.; Vences, M. The integrative future of taxonomy. Front. Zool. 2010, 7, 16. [Google Scholar] [CrossRef] [PubMed]
- Gurgis, G.P.; Daza, J.D.; Brennan, I.G.; Hutchinson, M.; Bauer, A.M.; Stocker, M.R.; Olori, J.C. Ecomor-phometric analysis of diversity in cranial shape of pygopodid geckos. Integr. Org. Biol. 2021, 3, obab013. [Google Scholar] [CrossRef] [PubMed]
- Bennett, C.V.; Goswami, A. Morphometric Analysis of Cranial Shape in Fossil and Recent Euprimates. Anat. Res. Int. 2012, 2012, 478903. [Google Scholar] [CrossRef] [PubMed]
- Evin, A.; Baylac, M.; Ruedi, M.; Mucedda, M.; Pons, J.-M. Taxonomy, skull diversity and evolution in a species complex of Myotis (Chiroptera: Vespertilionidae): A geometric morphometric appraisal. Biol. J. Linn. Soc. 2008, 95, 529–538. [Google Scholar] [CrossRef]
- Voss, R.S.; Marcus, L.F. Morphological evolution in muroid rodents II. Craniometric factor divergence in seven Neotropical genera, with experimental results from Zygodontomys. Evolution 1992, 46, 1918–1934. [Google Scholar] [CrossRef]
- Thomassen, H.A.; Buermann, W.; Milá, B.; Graham, C.H.; Cameron, S.E.; Schneider, C.J.; Pollinger, J.P.; Saatchi, S.; Wayne, R.K.; Smith, T.B. Modeling environmentally associated morphological and genetic variation in a rainforest bird, and its application to conservation prioritization. Evol. Appl. 2009, 3, 1–16. [Google Scholar] [CrossRef]
- Ordóñez-Garza, N.; Matson, J.O.; Strauss, R.E.; Bradley, R.D.; Salazar-Bravo, J. Patterns of phenotypic and genetic variation in three species of endemic Mesoamerican Peromyscus (Rodentia: Cricetidae). J. Mammal. 2010, 91, 848–859. [Google Scholar] [CrossRef]
- Bittner, N.K.J.; Mack, K.L.; Nachman, M.W. Gene expression plasticity and desert adaptation in house mice *. Evolution 2021, 75, 1477–1491. [Google Scholar] [CrossRef]
- Rohlf, F.J.; Marcus, L.F. A revolution morphometrics. Trends Ecol. Evol. 1993, 8, 129–132. [Google Scholar] [CrossRef]
- Czarnomska, S.D.; Jędrzejewska, B.; Borowik, T.; Niedziałkowska, M.; Stronen, A.V.; Nowak, S.; Mysłajek, R.W.; Okarma, H.; Konopiński, M.; Pilot, M.; et al. Concordant mitochondrial and microsatellite DNA structuring between Polish lowland and Carpathian Mountain wolves. Conserv. Genet. 2013, 14, 573–588. [Google Scholar] [CrossRef]
- Kawakami, M.; Yamamura, K.-I. Cranial bone morphometric study among mouse strains. BMC Evol. Biol. 2008, 8, 73. [Google Scholar] [CrossRef] [PubMed]
- Kilwanila, S.I.; Msalya, G.M.; Lyimo, C.M.; Rija, A.A. Geographic biases in cane rat (Thryonomyds) research may impede broader wildlife utilization and conservation in Africa: A systematic review. Sci. Afr. 2021, 12, e00785. [Google Scholar] [CrossRef]
- Morgan, K.; Mboumba, J.F.; Ntie, S.; Mickala, P.; Miller, C.A.; Zhen, Y.; Harrigan, R.J.; Le Underwood, V.; Ruegg, K.; Fokam, E.B.; et al. Precipitation and vegetation shape patterns of genomic and craniometric variation in the central African rodent Praomys misonnei. Proc. R. Soc. B 2020, 287, 20200449. [Google Scholar] [CrossRef] [PubMed]
- Nichols, V.; Akpatou, B.; Wendelen, W.I.M.; Kerbis Peterhans, J.U.L.I.A.N.; Olayemi, A.; Decher, J.A.N.; Colyn, M. Molecular and morphometric variation in two sibling species of the genus Praomys (Rodentia: Muridae): Implications for biogeography. Zool. J. Linn. Soc. 2010, 160, 397–419. [Google Scholar] [CrossRef]
- Blackford, C.; Germain, R.M.; Gilbert, B. Species Differences in Phenology Shape Coexistence. Am. Nat. 2020, 195, E168–E180. [Google Scholar] [CrossRef]
- Monteiro-Filho, E.L.; Monteiro, L.R.; dos Reis, S.F. Skull Shape and Size Divergence in Dolphins of the Genus Sotalia: A Tridimensional Morphometric Analysis. J. Mammal. 2002, 83, 125–134. [Google Scholar] [CrossRef]
- Roberts, R.B.; Hu, Y.; Albertson, R.C.; Kocher, T.D. Craniofacial divergence and ongoing adaptation via the hedgehog pathway. Proc. Natl. Acad. Sci. USA 2011, 108, 13194–13199. [Google Scholar] [CrossRef]
- Lalis, A.; Baylac, M.; Cosson, J.F.; Makundi, R.H.; Machangu, R.S.; Denys, C. Cranial morphometric and fine scale genetic variability of two adjacent Mastomys natalensis (Rodentia: Muridae) populations. Acta Theriol. 2009, 54, 171–181. [Google Scholar] [CrossRef]
- Auffray, C.; Imbeaud, S.; Roux-Rouquié, M.; Hood, L. From functional genomics to systems biology: Concepts and practices Molecular biology and genetics. Comptes Rendus Biol. 2003, 326, 879–892. [Google Scholar] [CrossRef]
- Eckert, C.G.; Samis, K.E.; Lougheed, S.C. Genetic variation across species’ geographical ranges: The central–marginal hypothesis and beyond. Mol. Ecol. 2008, 17, 1170–1188. [Google Scholar] [CrossRef] [PubMed]
- Voss, R.S.; Hubbard, C.; Jansa, S.A. Phylogenetic relationships of New World porcupines (Rodentia, Erethizontidae): Implications for taxonomy, morphological evolution, and biogeography. Am. Mus. Novit. 2013, 3769, 1–36. [Google Scholar] [CrossRef]
- Martínez-Abadías, N.; Esparza, M.; Sjøvold, T.; González-José, R.; Santos, M.; Hernández, M. Heritability of human cranial dimensions: Comparing the evolvability of different cranial regions. J. Anat. 2014, 214, 19–35. [Google Scholar] [CrossRef] [PubMed]
- Kawakami, K.; Ege, K.; Yamada, T. Method of producing flat skin and skeleton specimens of laboratory mice for exhibition at a university museum. Shimane J. Med. Sci. 2015, 32, 37–43. [Google Scholar]
- Mustapha, O.A.; Aderounmu, O.A.; Olude, M.A.; Okandeji, M.E.; Akinloye, A.K.; Oke, B.O.; Olopade, J.O. Anatomical studies on the spinal cord of the greater cane rat (Thryonomys swinderianus, Temminck) I: Gross morphometry. Niger. Vet. J. 2015, 36, 1192–1202. [Google Scholar]
- Mustapha, O.; Ezekiel, O.; Olaolorun, F.; Awala-Ajakaiye, M.; Popoola, E.; Olude, M.; Olopade, J. Morpho-logical Characterization of the Developing Greater Cane Rat (Thryonomys swinderianus) Brain. Dev. Neurosci. 2020, 42, 114–123. [Google Scholar] [CrossRef]
- Olude, M.A.; Mustapha, O.A.; Olopade, J.O. Morphological characterization of the African Giant Rat (Cricetomys gambianus, Waterhouse) brain across age groups: Gross features of cortices. Niger. J. Physiol. Sci. 2016, 31, 133–138. [Google Scholar]
- Igado, O.O.; Adebayo, A.O.; Oriji, C.C.; Oke, B.O. Cranio-facial and ocular morphometrics of the male greater cane rat (Thryonomys swinderianus). Niger. Vet. J. 2016, 37, 54–63. [Google Scholar]
- Biseko, B.B.; Rija, A.A. Correlates of flight response in some common birds of a rapidly expanding African city. J. East Afr. Ornithol. 2022, 42, 10–22. [Google Scholar]
- Rija, A.A.; Critchlow, R.; Thomas, C.D.; Beale, C.M. Global extent and drivers of mammal population declines in protected areas under illegal hunting pressure. PLoS ONE 2020, 15, e0227163. [Google Scholar] [CrossRef]
- Adenyo, C.; Hayano, A.; Kayang, B.B.; Owusu, E.H.; Inoue-Murayama, M. Mitochondrial D-loop diversity of grasscutter (Thryonomys swinderianus Rodentia: Hystricomorpha) in Ghana. Open J. Anim. Sci. 2013, 3, 145–153. [Google Scholar] [CrossRef]
- Carleton, M.D.; Musser, G.G. Systematic studies of oryzomyine rodents (Muridae, Sigmodontinae): A Synopsis of Microryzomys. Bulletin of the AMNH; no. 191. 1989. Available online: http://hdl.handle.net/2246/953 (accessed on 15 October 2022).
- Olivera-Angel, M.; Betancur, J. Body shape variation between farms of tilapia (Oreochromis sp.) in Colombian Andes using landmark-based geometric morphometrics Geometric morphometrics tilapia. Lat. Am. J. Aquat. Res. 2019, 47, 194–200. [Google Scholar]
- Rohlf, F.J. The tps series of software. Hystrix 2015, 26, 9–12. [Google Scholar] [CrossRef]
- Bookstein, F.L.; Grayson, B.; Cutting, C.B.; Kim, H.-C.; McCarthy, J.G. Landmarks in three dimensions: Reconstruction from cephalograms versus direct observation. Am. J. Orthod. Dentofac. Orthop. 1991, 100, 133–140. [Google Scholar] [CrossRef] [PubMed]
- Klingenberg, C.P. MorphoJ: An integrated software package for geometric morphometrics. Mol. Ecol. Resour. 2011, 11, 353–357. [Google Scholar] [CrossRef]
- Adams, D.C.; Rohlf, F.J.; Slice, D.E. Geometric morphometrics: Ten years of progress following the ‘revo-lution’. Ital. J. Zool. 2004, 71, 5–16. [Google Scholar] [CrossRef]
- Huson, D.H.; Bryant, D. Application of Phylogenetic Networks in Evolutionary Studies. Mol. Biol. Evol. 2005, 23, 254–267. [Google Scholar] [CrossRef]
- Adams, D.C.; Collyer, M.L. On the comparison of the strength of morphological integration across morphometric datasets. Evolution 2016, 70, 2623–2631. [Google Scholar] [CrossRef]
- Adams, D.C.; Otárola-Castillo, E. Geomorph: An R package for the collection and analysis of geometric morphometric shape data. Methods Ecol. Evol. 2013, 4, 393–399. [Google Scholar] [CrossRef]
- Oksanen, J.; Kindt, R.; Legendre, P.; O’Hara, B.; Stevens, M.H.H.; Oksanen, M.J.; Suggests, M.A.S.S. The vegan package. Community Ecol. Package 2007, 10, 719. [Google Scholar]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef] [PubMed]
- Kilwanila, S.I.; Lyimo, C.M.; Rija, A.A. Mitochondrial genetic diversity of the Greater Cane rat (Thryonomys swinderianus) populations from the Eastern Arc Mountains ecosystem, Tanzania. Mol. Biol. Rep. 2022, 49, 10431–10442. [Google Scholar] [CrossRef] [PubMed]
- Hall, T. BioEdit version 7.0. 0. Distributed by the Author. 2004. Available online: www.mbio.ncsu.edu/BioEdit/bioedit.html (accessed on 12 October 2022).
- Excoffier, L.; Lischer, H.E.L. Arlequin suite ver 3.5: A new series of programs to perform population genetics analyses under Linux and Windows. Mol. Ecol. Resour. 2010, 10, 564–567. [Google Scholar] [CrossRef] [PubMed]
- Wright, S. Isolation by distance. Genetics 1943, 28, 114. [Google Scholar] [CrossRef] [PubMed]
- Lieberman, D.E.; Krovitz, G.E.; Yates, F.W.; Devlin, M.; Claire, M.S. Effects of food processing on masticatory strain and craniofacial growth in a retrognathic face. J. Hum. Evol. 2004, 46, 655–677. [Google Scholar] [CrossRef]
- Kilawe, C.J.; Kaaya, O.E.; Kolonel, C.P.; Macrice, S.A.; Mshama, C.P.; Lyimo, P.J.; Emily, C.J. Wildfires in the Eastern Arc Mountains of Tanzania: Burned areas, underlying causes and management challenges. Afr. J. Ecol. 2020, 59, 204–215. [Google Scholar] [CrossRef]
- Stevens, K.; Harrisson, K.A.; Hogan, F.E.; Cooke, R.; Clarke, R.H. Reduced gene flow in a vulnerable species reflects two centuries of habitat loss and fragmentation. Ecosphere 2018, 9, e02114. [Google Scholar] [CrossRef]
- Hanski, I. Metapopulation dynamics. Nature 1998, 396, 41–49. [Google Scholar] [CrossRef]
- Frankham, R.; Bradshaw, C.J.; Brook, B.W. Genetics in conservation management: Revised recommendations for the 50/500 rules, Red List criteria and population viability analyses. Biol. Conserv. 2014, 170, 56–63. [Google Scholar] [CrossRef]
- Freudiger, A.; Josi, D.; Thünken, T.; Herder, F.; Flury, J.M.; Marques, D.A.; Taborsky, M.; Frommen, J.G. Ecological variation drives morphological differentiation in a highly social vertebrate. Funct. Ecol. 2021, 35, 2266–2281. [Google Scholar] [CrossRef]
- Fornel, R.; Cordeiro-Estrela, P.; Freitas, T.R.O. Skull shape and size variation in Ctenomys minutus (Rodentia: Ctenomyidae) in geographical, chromosomal polymorphism, and environmental contexts. Biol. J. Linn. Soc. 2010, 101, 705–720. [Google Scholar] [CrossRef]
- Fadda, C.; Corti, M. Three dimensional geometric morphometric study of the Ethiopian Myomys-Stenocephalemys complex (Murinae, Rodentia). Hystrix Ital. J. Mammal. 2000, 11, 131–143. [Google Scholar]
- Kerschbaumer, M.; Mitteroecker, P.; Sturmbauer, C. Evolution of body shape in sympatric versus non-sympatric Tropheus populations of Lake Tanganyika. Heredity 2013, 112, 89–98. [Google Scholar] [CrossRef]
- Bradburd, G.S.; Ralph, P.L.; Coop, G.M. Disentangling the effects of geographic and ecological isolation on genetic differentiation. Evolution 2013, 67, 3258–3273. [Google Scholar] [CrossRef]
- Abiadh, A.; Colangelo, P.; Capanna, E.; Lamine-Cheniti, T.; Chetoui, M. Morphometric analysis of six Gerbillus species (Rodentia, Gerbillinae) from Tunisia. Comptes Rendus Biol. 2010, 333, 680–687. [Google Scholar] [CrossRef]
- Dobigny, G.; Baylac, M.; Denys, C. Geometric morphometrics, neural networks and diagnosis of sibling Taterillus species (Rodentia, Gerbillinae). Biol. J. Linn. Soc. 2002, 77, 319–327. [Google Scholar] [CrossRef]
- Wilson, V.J. Mammalian Vestibular Physiology; Springer Science & Business Media: Berlin/Heidelberg, Germany, 2013. [Google Scholar]

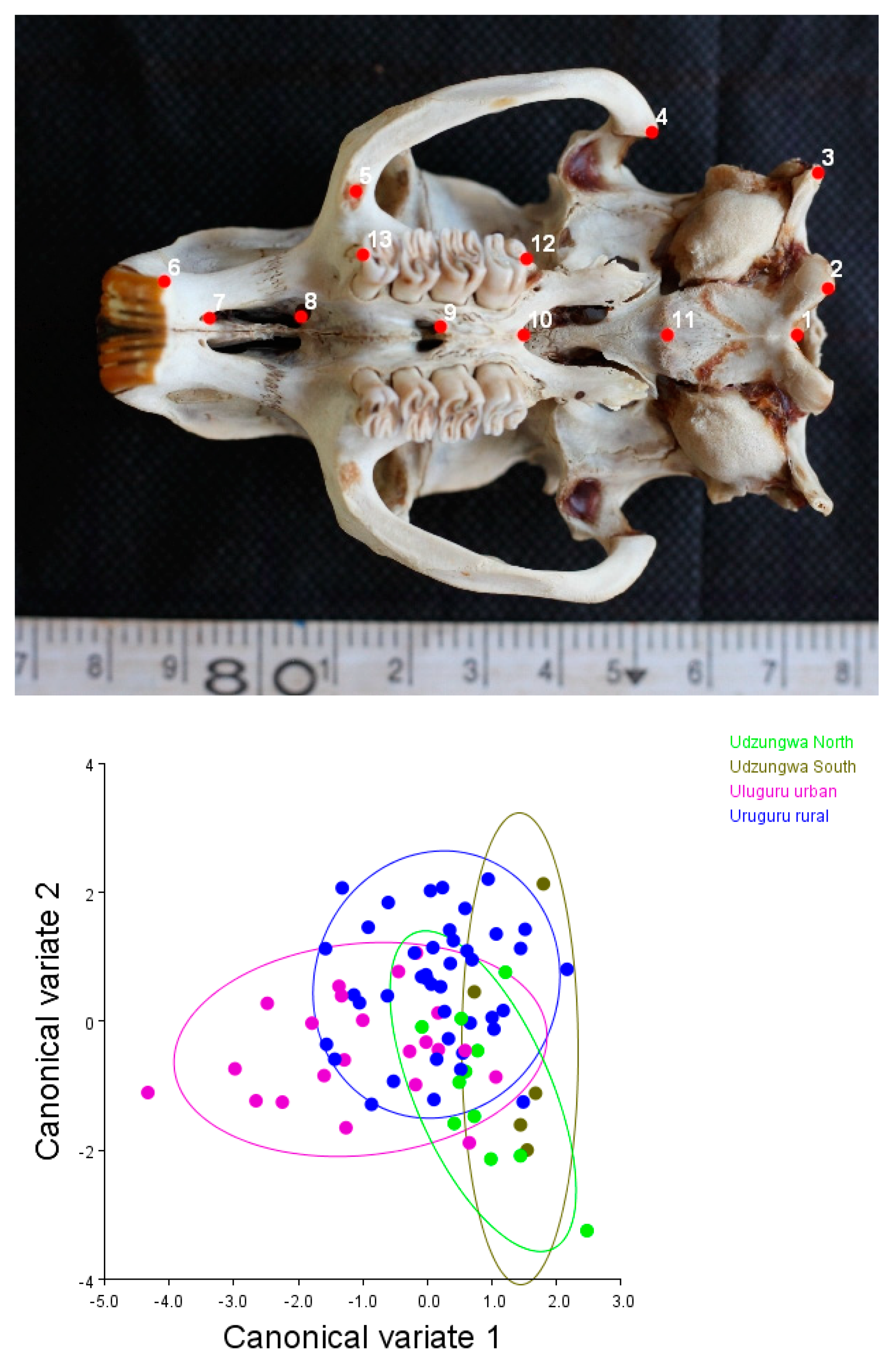
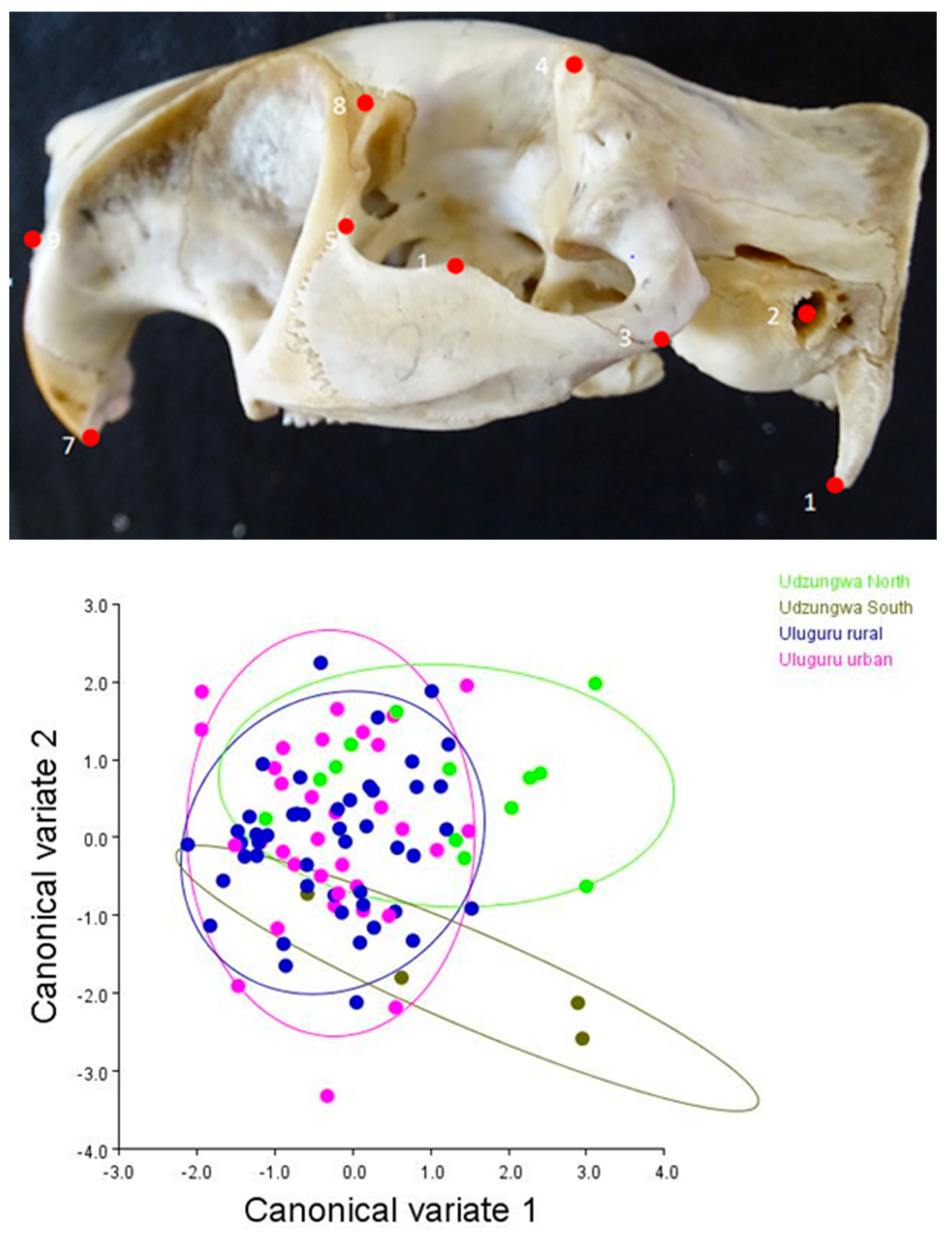
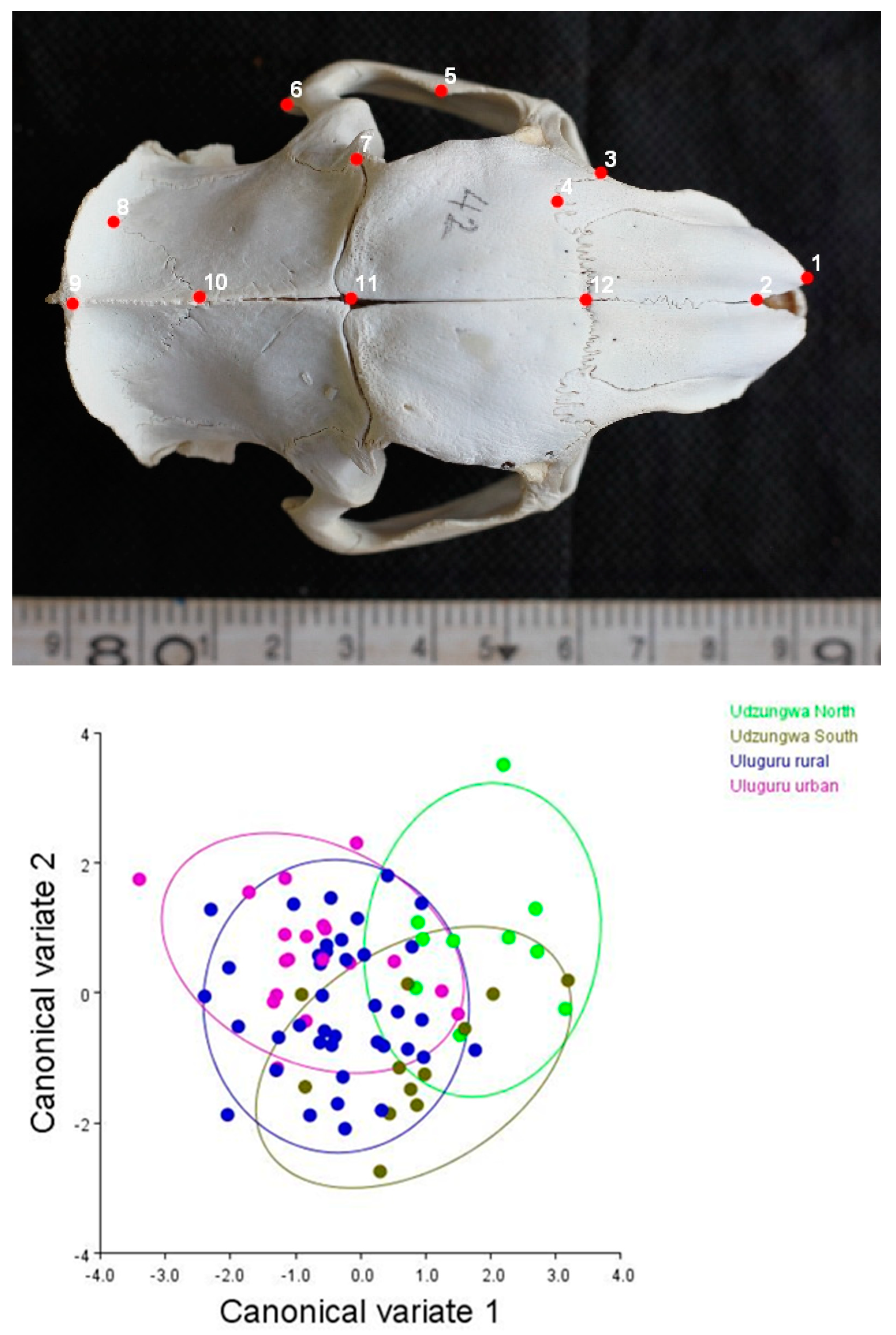


| Mahalanobis Distance | |||
| Population | Udzungwa North | Udzungwa South | Uluguru Urban |
| Udzungwa south | 2.2472 (0.7864) | ||
| Uluguru urban | 2.1609 (0.0548) | 2.7865 (0.1180) | |
| Uruguru rural | 1.8783 (0.0582) | 2.2598 (0.3564) | 1.5776 (0.0111) |
| Procrustes Distance | |||
| Population | Udzungwa North | Udzungwa South | Uluguru Urban |
| Udzungwa south | 0.0202 (0.7139) | ||
| Uluguru urban | 0.0572 (0.3530) | 0.0575 (0.4616) | |
| Uruguru rural | 0.0244 (0.2347) | 0.0218 (0.5781) | 0.0462 (0.0895) |
| Mahalanobis Distance | |||
| Populations | Udzungwa North | Udzungwa South | Uluguru Rural |
| Udzungwa south | 2.5012 (0.2837) | ||
| Uluguru rural | 1.6627 (0.0109 | 2.5284 (0.0418) | |
| Uluguru urban | 1.7111 (0.0142) | 2.5772 (0.0473) | 0.9281(0.2344) |
| Procrustes Distance | |||
| Populations | Udzungwa North | Udzungwa South | Uluguru Rural |
| Udzungwa south | 0.0501 (0.3014) | ||
| Uluguru rural | 0.0285(0.2059) | 0.0628 (0.0394) | |
| Uluguru urban | 0.0292 (0.1532) | 0.0630 (0.0124) | 0.0117 (0.7840) |
| Mahalanobis Distance | |||
| Population | Udzungwa North | Udzungwa South | Uluguru Rural |
| Udzungwa South | 2.2643 (0.1584) | ||
| Uluguru rural | 2.4629 (0.0003) | 1.7541(0.0725) | |
| Uluguru urban | 2.7200 (0.0005) | 2.2279 (0.0040) | 1.2913 (0.3434) |
| Procrustes Distance | |||
| Population | Udzungwa North | Udzungwa South | Uluguru Rural |
| Udzungwa South | 0.0341 (0.0182) | ||
| Uluguru rural | 0.0279 (0.0154) | 0.0150 (0.3154) | |
| Uluguru urban | 0.0239 (0.0828) | 0.0196 (0.0927) | 0.0109 (0.5324) |
| Source of Variation | df | Sum of Squares | Variance Components | Percentage of Variations | FST | p-Value |
|---|---|---|---|---|---|---|
| Between Populations | 3 | 71.452 | 2.14201 | 40.30 | 0.40297 | 0.00001 |
| Within Population | 42 | 133.287 | 3.17349 | 59.70 | ||
| Total | 45 | 204.739 | 4.86248 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kilwanila, S.I.; Lyimo, C.M.; Makundi, R.H.; Rija, A.A. Isolating Greater Cane Rat Populations (Thryonomys swinderianus) from Eastern Arc Mountains, Tanzania: Linking Diversity to Morphometric and Molecular Characteristics. Diversity 2023, 15, 626. https://doi.org/10.3390/d15050626
Kilwanila SI, Lyimo CM, Makundi RH, Rija AA. Isolating Greater Cane Rat Populations (Thryonomys swinderianus) from Eastern Arc Mountains, Tanzania: Linking Diversity to Morphometric and Molecular Characteristics. Diversity. 2023; 15(5):626. https://doi.org/10.3390/d15050626
Chicago/Turabian StyleKilwanila, Shadia I., Charles M. Lyimo, Rhodes H. Makundi, and Alfan A. Rija. 2023. "Isolating Greater Cane Rat Populations (Thryonomys swinderianus) from Eastern Arc Mountains, Tanzania: Linking Diversity to Morphometric and Molecular Characteristics" Diversity 15, no. 5: 626. https://doi.org/10.3390/d15050626
APA StyleKilwanila, S. I., Lyimo, C. M., Makundi, R. H., & Rija, A. A. (2023). Isolating Greater Cane Rat Populations (Thryonomys swinderianus) from Eastern Arc Mountains, Tanzania: Linking Diversity to Morphometric and Molecular Characteristics. Diversity, 15(5), 626. https://doi.org/10.3390/d15050626





