Insights into the Gut Microbiota of the Freshwater Crab Sinopotamon planum across Three Seasons and Its Associations with the Surrounding Aquatic Microbiota
Abstract
1. Introduction
2. Materials and Methods
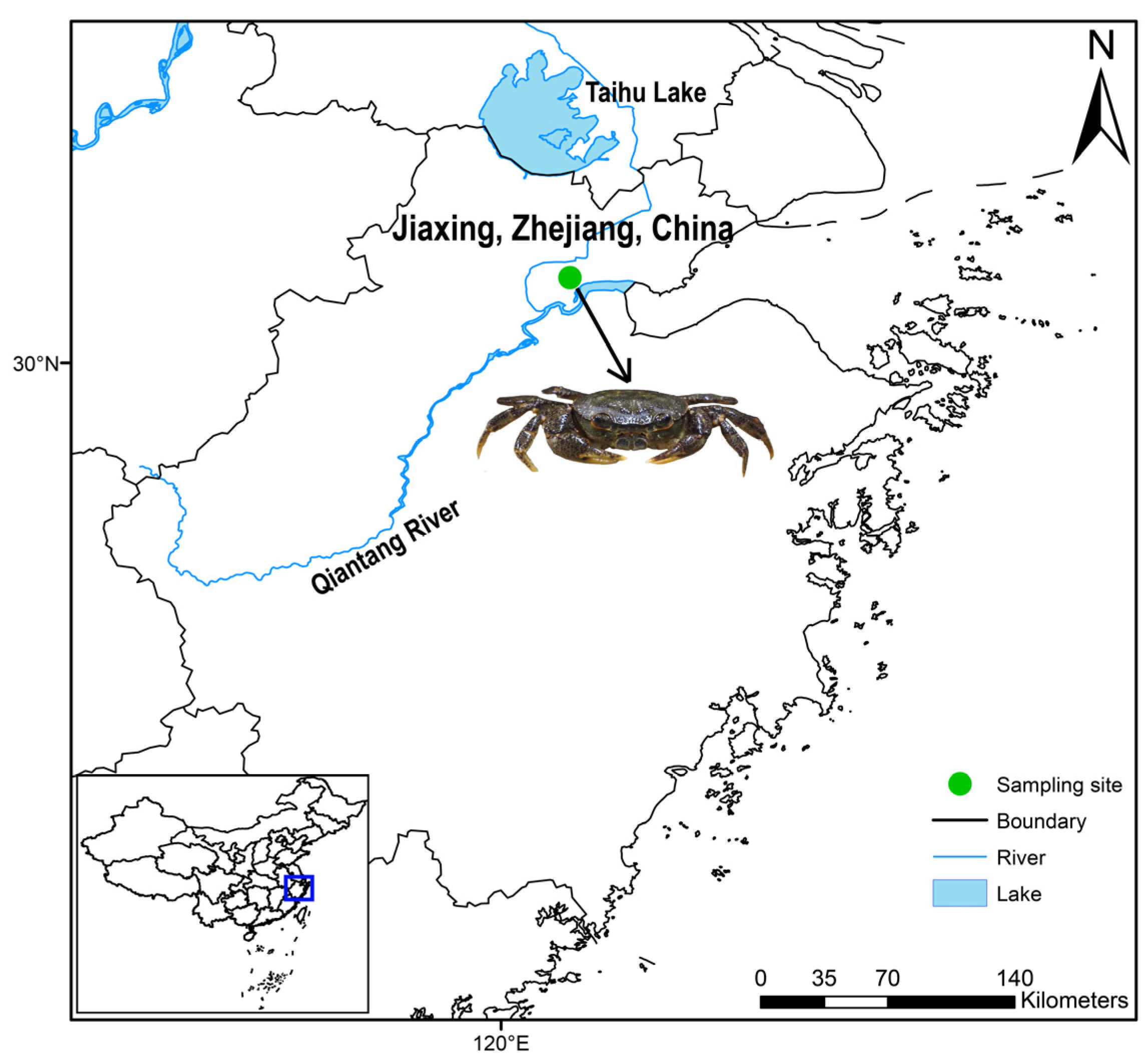
2.1. Samples Collection
2.2. DNA Extraction and Sequencing
2.3. Statistical Analysis
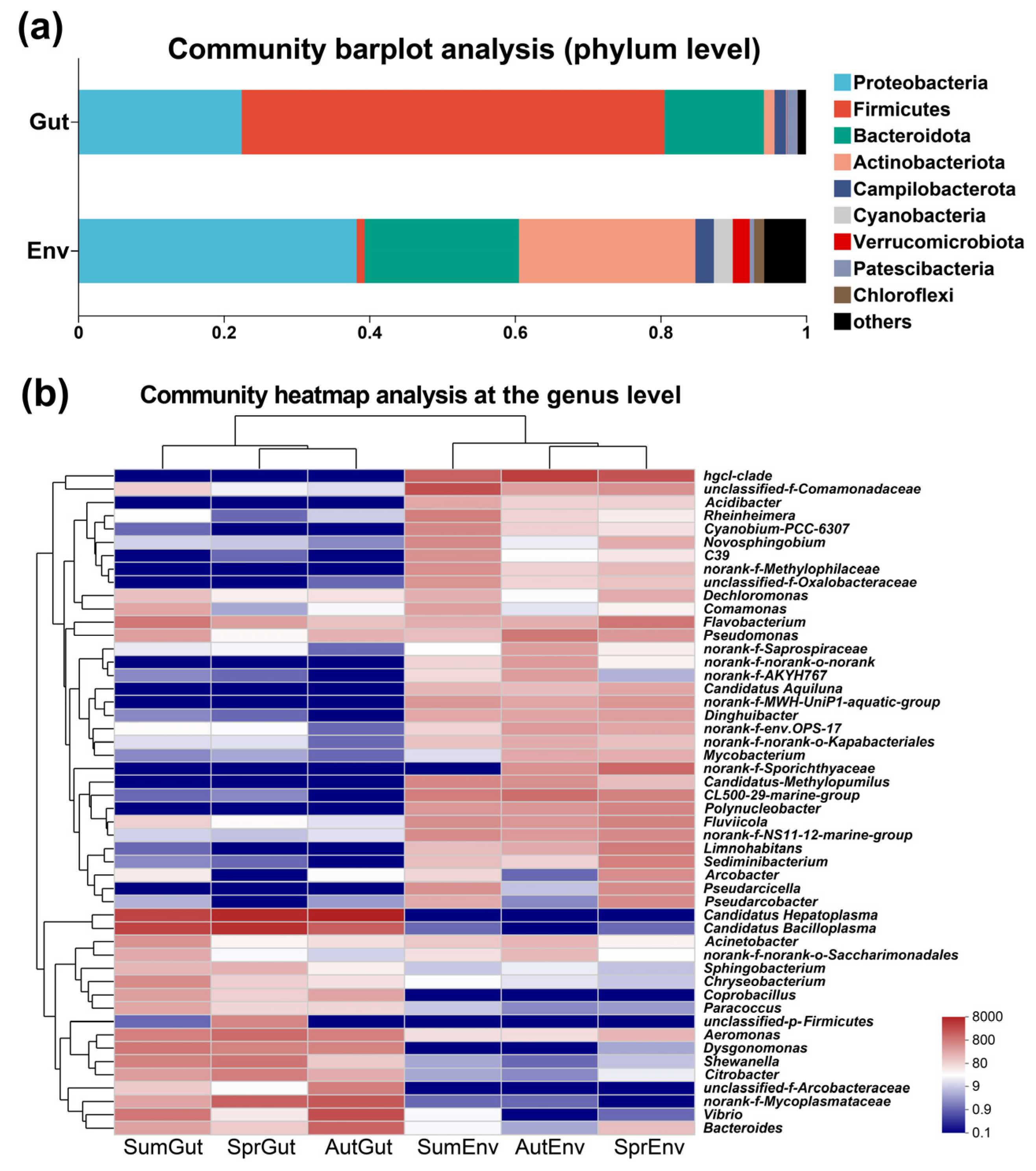
3. Results
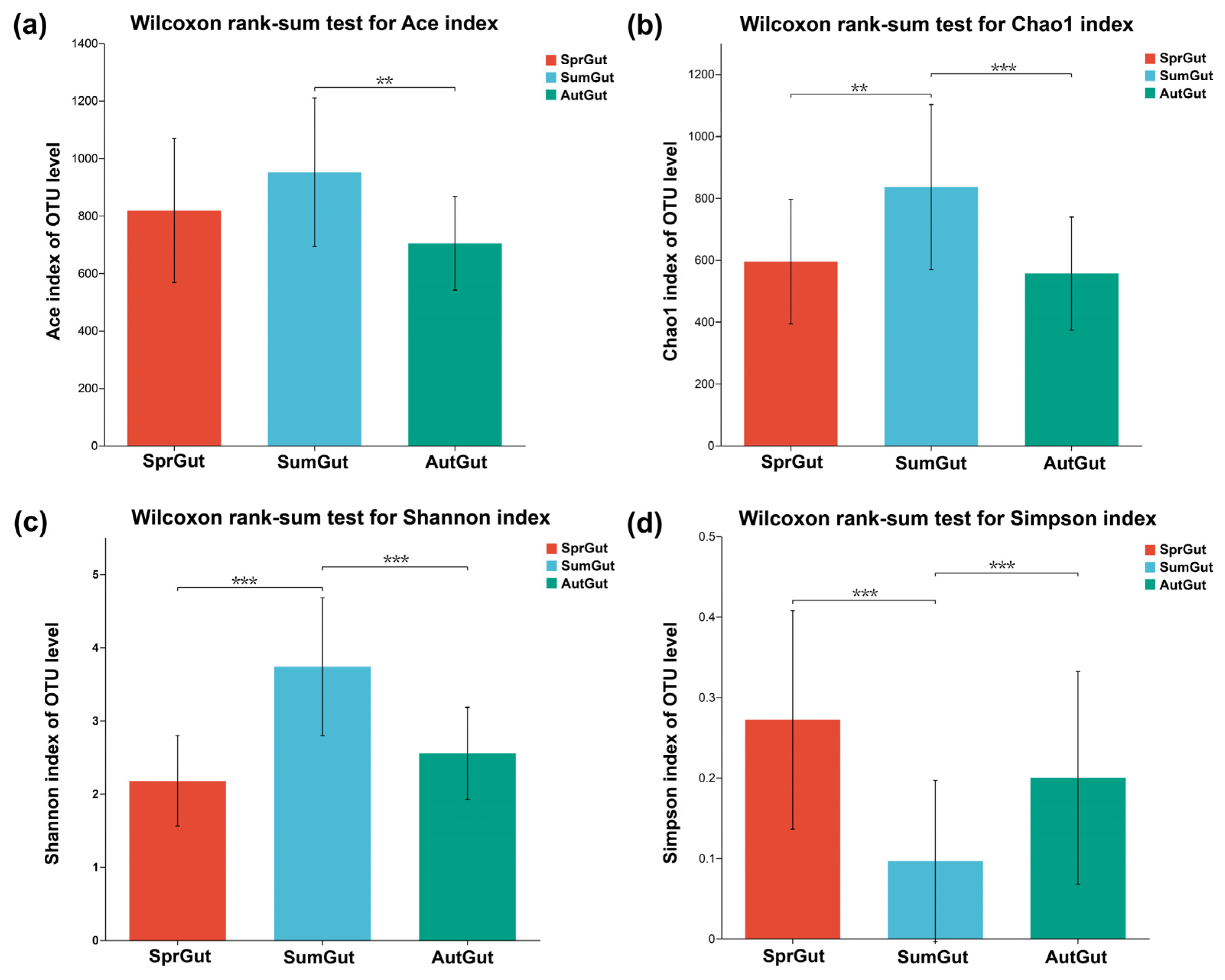
3.1. Sequencing Data and Alpha Diversity Indexes
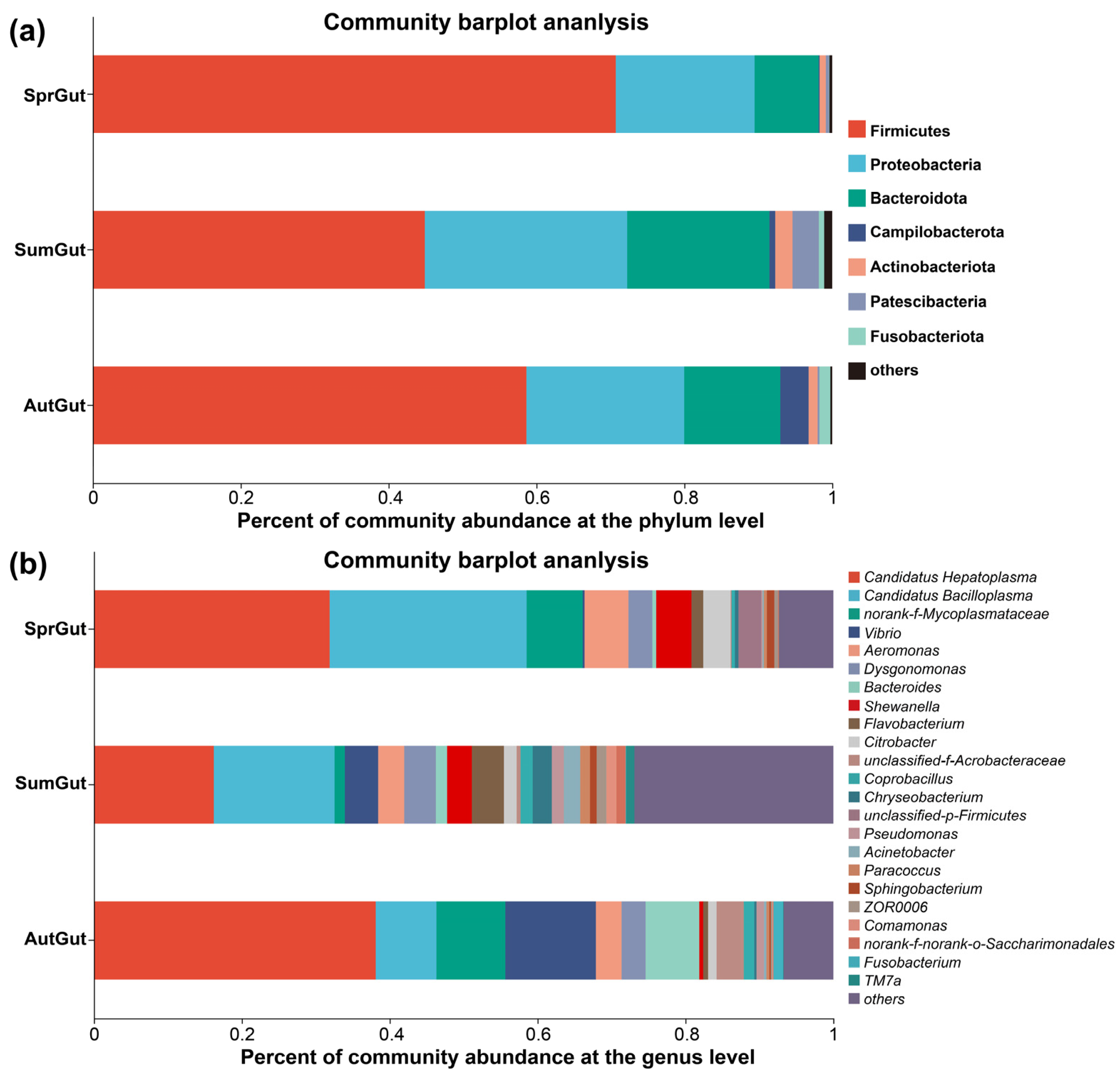
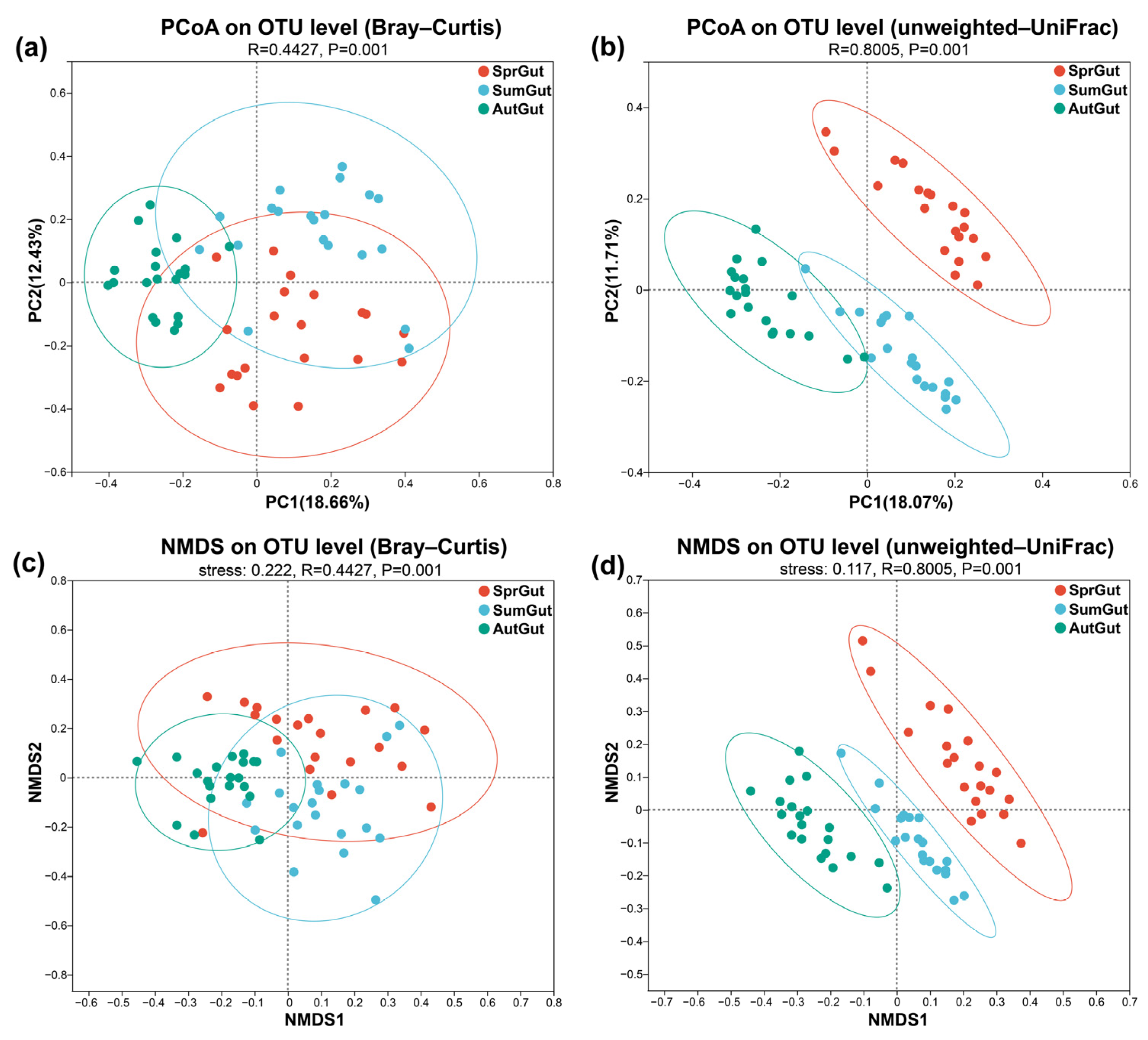
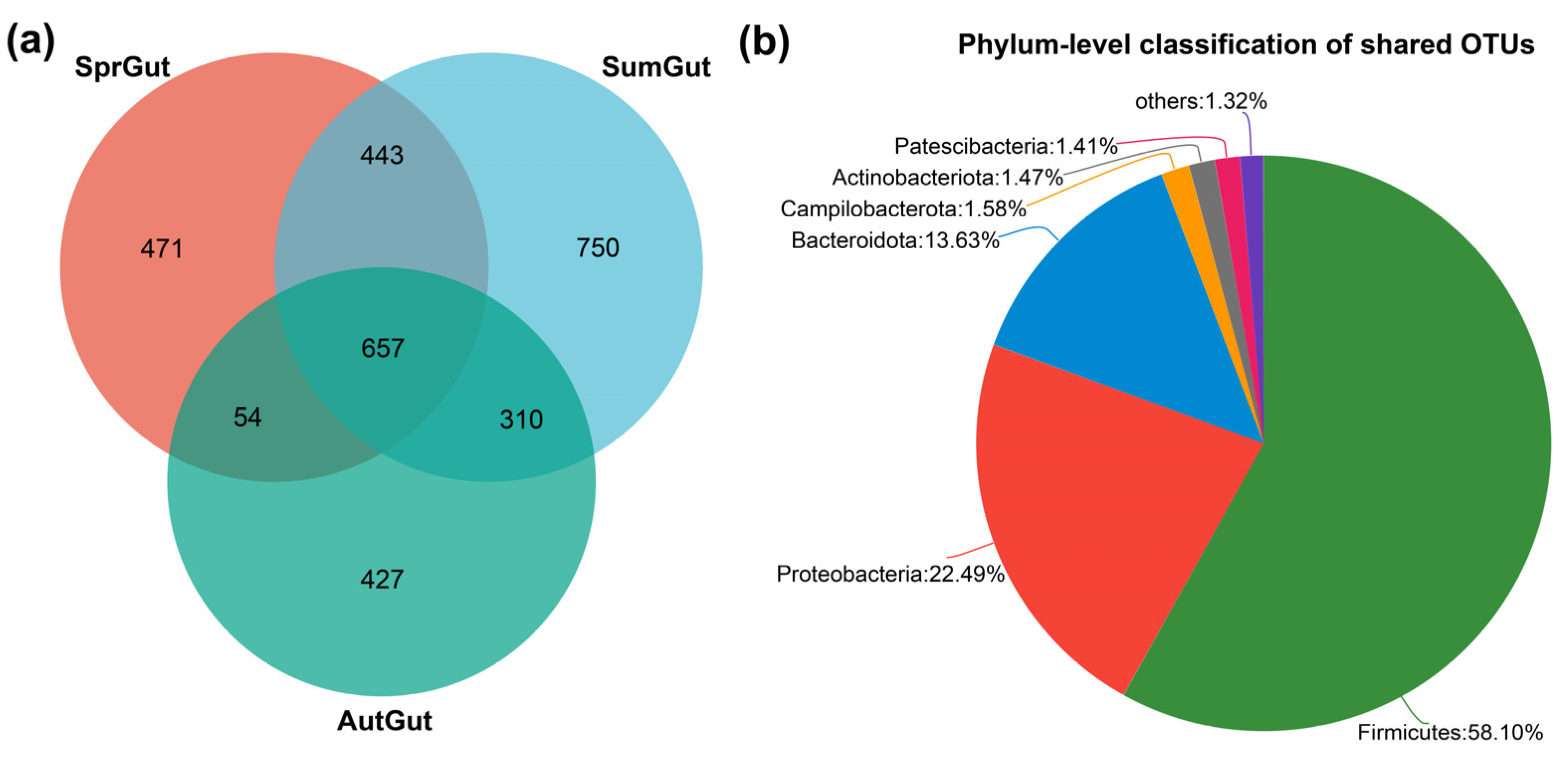
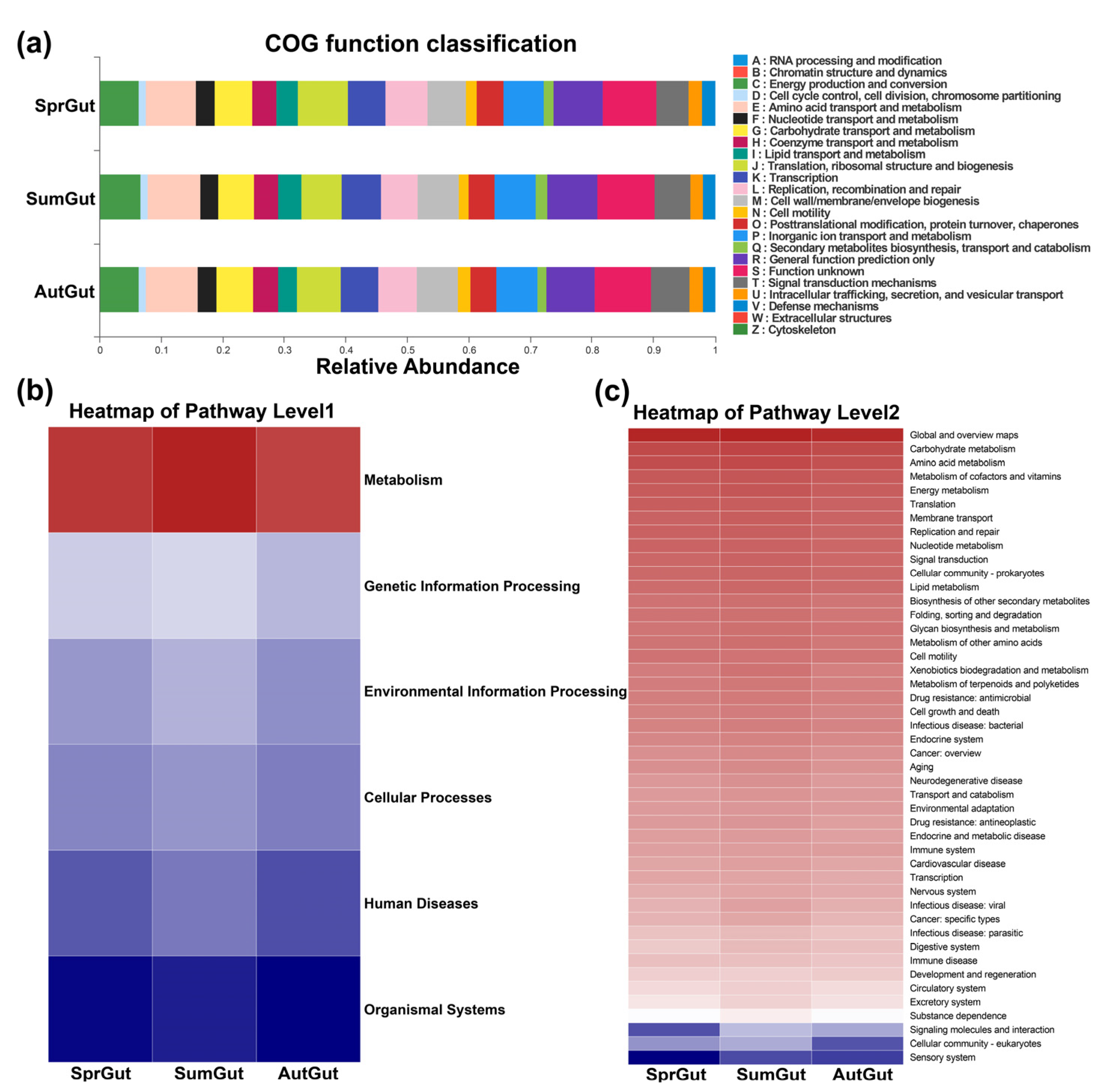
3.2. Composition and Beta Diversity Analysis of Gut Microbiota
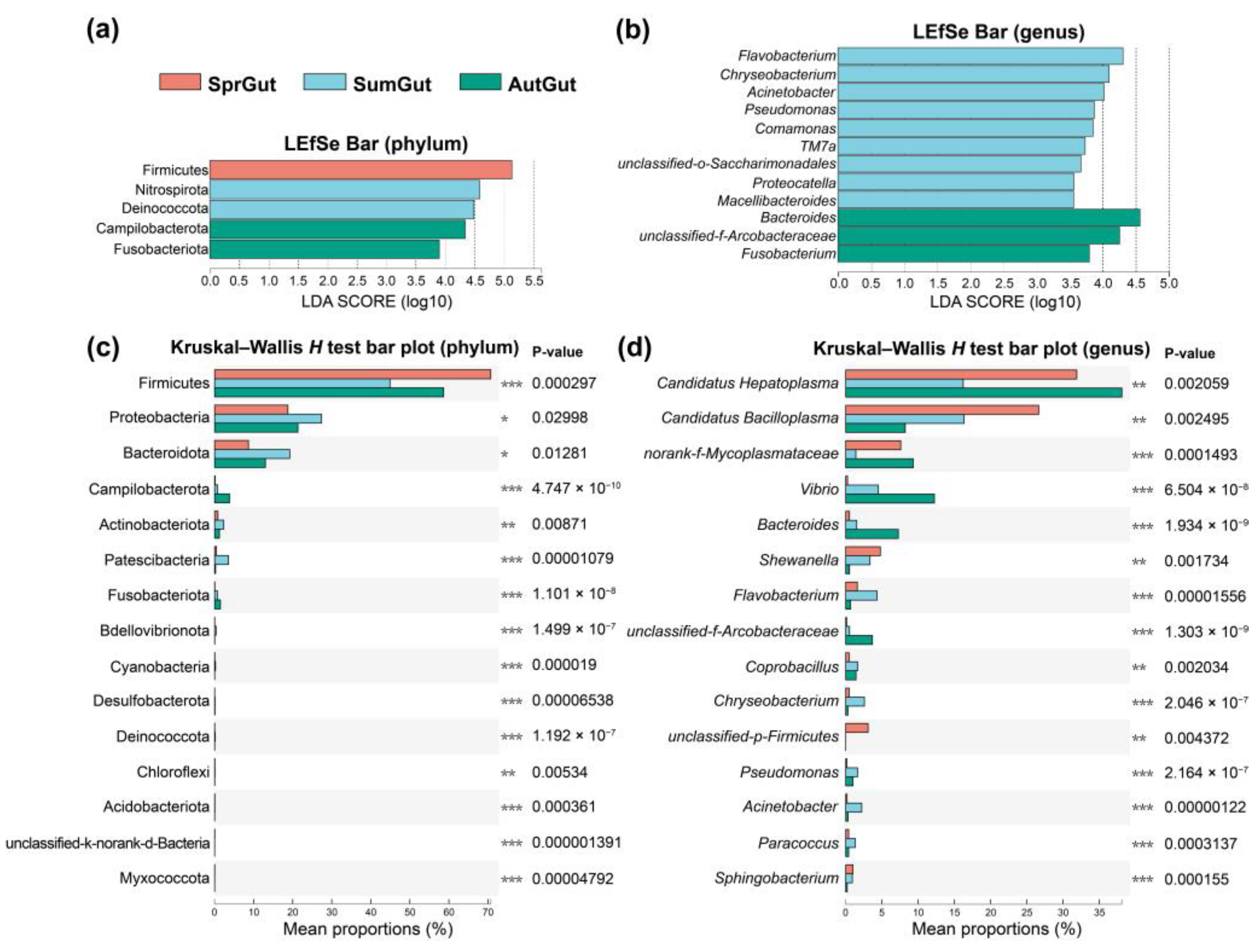
3.3. Differences in Gut Microbes in the Three Studied Seasons
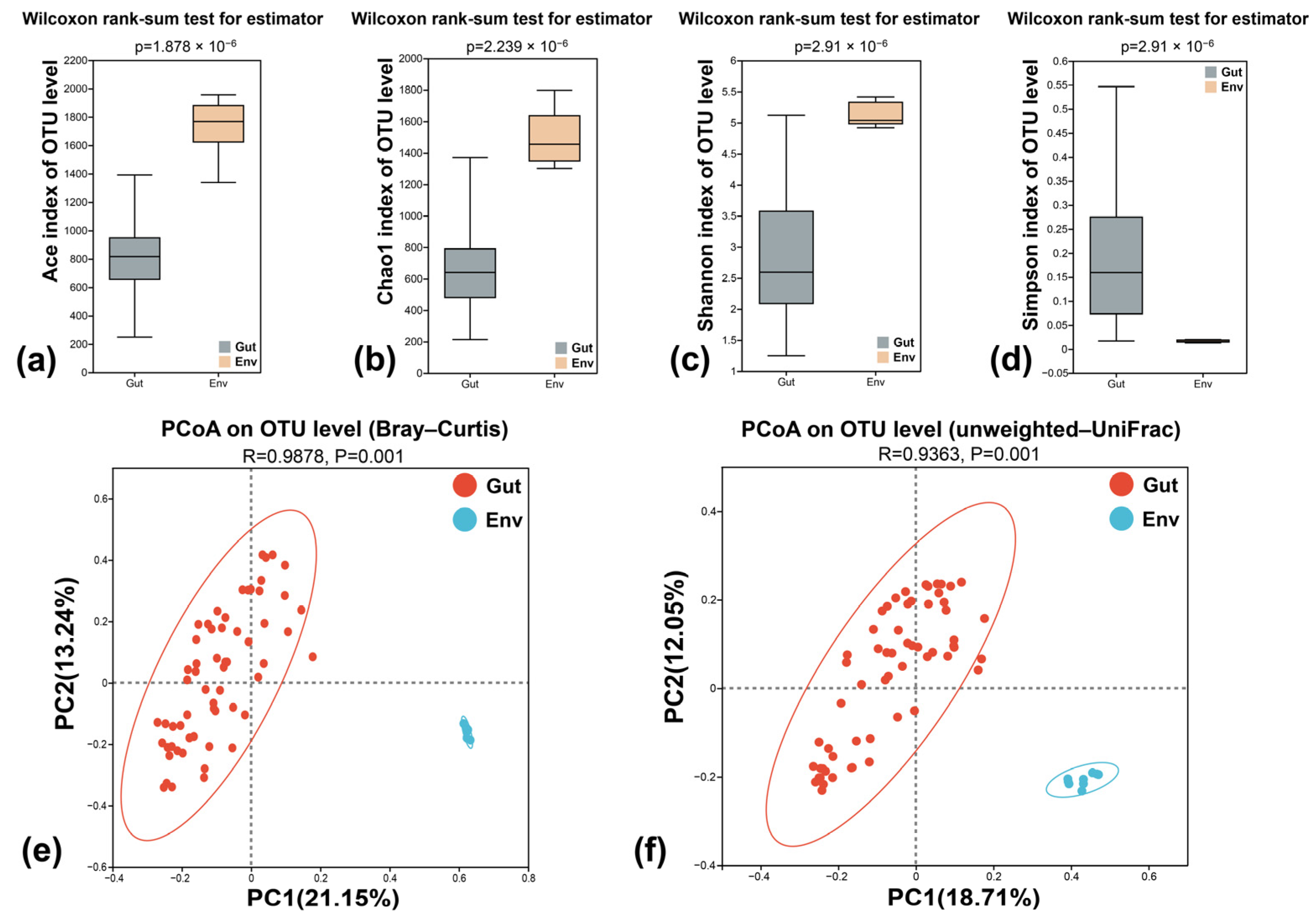
3.4. Microbial Diversity of Surrounding Water
3.5. Analysis of the Microbiota Variance between S. planum and Surrounding Water
4. Discussion
4.1. Structure and Function of Gut Microbes of S. planum in Three Seasons
4.2. Relationships between Crab Gut Microbes and Surrounding Water Microbes
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Lee, W.-J.; Hase, K. Gut microbiota–generated metabolites in animal health and disease. Nat. Chem. Biol. 2014, 10, 416–424. [Google Scholar] [CrossRef] [PubMed]
- Moszak, M.; Szulińska, M.; Bogdański, P. You are what you eat—The relationship between diet, microbiota, and metabolic disorders—A review. Nutrients 2020, 12, 1096. [Google Scholar] [CrossRef]
- Quigley, E.M.M. Prebiotics and probiotics; modifying and mining the microbiota. Pharmacol. Res. 2010, 61, 213–218. [Google Scholar] [CrossRef] [PubMed]
- Nicholson, J.K.; Holmes, E.; Kinross, J.; Burcelin, R.; Gibson, G.; Jia, W.; Pettersson, S. Host-gut microbiota metabolic interactions. Science 2012, 336, 1262–1267. [Google Scholar] [CrossRef] [PubMed]
- Maslowski, K.M.; Mackay, C.R. Diet, gut microbiota and immune responses. Nat. Immunol. 2011, 12, 5–9. [Google Scholar] [CrossRef]
- Sommer, F.; Bäckhed, F. The gut microbiota—Masters of host development and physiology. Nat. Rev. Microbiol. 2013, 11, 227–238. [Google Scholar] [CrossRef]
- Tremaroli, V.; Bäckhed, F. Functional interactions between the gut microbiota and host metabolism. Nature 2012, 489, 242–249. [Google Scholar] [CrossRef]
- Amato, K.R.; Leigh, S.R.; Kent, A.; Mackie, R.I.; Yeoman, C.J.; Stumpf, R.M.; Wilson, B.A.; Nelson, K.E.; White, B.A.; Garber, P.A. The role of gut microbes in satisfying the nutritional demands of adult and juvenile wild, black howler monkeys (Alouatta pigra). Am. J. Phys. Anthropol. 2014, 155, 652–664. [Google Scholar] [CrossRef] [PubMed]
- Ley, R.E.; Hamady, M.; Lozupone, C.; Turnbaugh, P.J.; Ramey, R.R.; Bircher, J.S.; Schlegel, M.L.; Tucker, T.A.; Schrenzel, M.D.; Knight, R.; et al. Evolution of mammals and their gut microbes. Science 2008, 320, 1647–1651. [Google Scholar] [CrossRef] [PubMed]
- David, L.A.; Maurice, C.F.; Carmody, R.N.; Gootenberg, D.B.; Button, J.E.; Wolfe, B.E.; Ling, A.V.; Devlin, A.S.; Varma, Y.; Fischbach, M.A.; et al. Diet rapidly and reproducibly alters the human gut microbiome. Nature 2014, 505, 559–563. [Google Scholar] [CrossRef] [PubMed]
- Sepulveda, J.; Moeller, A.H. The effects of temperature on animal gut microbiomes. Front. Microbiol. 2020, 11, 384. [Google Scholar] [CrossRef]
- Sullam, K.E.; Essinger, S.D.; Lozupone, C.A.; O’Connor, M.P.; Rosen, G.L.; Knight, R.; Kilham, S.S.; Russell, J.A. Environmental and ecological factors that shape the gut bacterial communities of fish: A meta-analysis. Mol. Ecol. 2012, 21, 3363–3378. [Google Scholar] [CrossRef]
- Shang, S.; Li, L.-Y.; Liu, X.-X.; Wang, J.; Tang, X.-X. Bacterial Community in the Gut of Neanthes japonica and Its Association with Surrounding Environment. Diversity 2022, 14, 514. [Google Scholar] [CrossRef]
- Koliada, A.; Moseiko, V.; Romanenko, M.; Piven, L.; Lushchak, O.; Kryzhanovska, N.; Guryanov, V.; Vaiserman, A. Seasonal variation in gut microbiota composition: Cross-sectional evidence from Ukrainian population. BMC Microbiol. 2020, 20, 100. [Google Scholar] [CrossRef] [PubMed]
- Li, S.-X.; Qian, Z.-J.; Yang, J.-N.; Lin, Y.-F.; Li, H.; Chen, L. Seasonal variation in structure and function of gut microbiota in Pomacea canaliculata. Ecol. Evol. 2022, 12, e9162. [Google Scholar] [CrossRef] [PubMed]
- Hu, X.-L.; Liu, G.; Li, Y.-M.; Wei, Y.-T.; Lin, S.-B.; Liu, S.-Q.; Zheng, Y.-L.; Hu, D.-F. High-throughput analysis reveals the seasonal variation of the gut microbiota composition within forest musk deer (Moschus berezovskii). Front. Microbiol. 2018, 9, 1674. [Google Scholar] [CrossRef]
- Day, A.S.; Keenan, J.I.; Tannock, G.W. The intestinal microbiota in health and disease. J. R. Soc. N. Z. 2020, 50, 367–370. [Google Scholar] [CrossRef]
- O’Sullivan, D.J. Methods for analysis of the intestinal microflora. Curr. Issues Intest. Microbiol. 2000, 1, 39–50. [Google Scholar]
- Eckburg, P.B.; Bik, E.M.; Bernstein, C.N.; Purdom, E.; Dethlefsen, L.; Sargent, M.; Gill, S.T.; Nelson, K.E.; Relman, D.A. Diversity of the human intestinal microbial flora. Science 2005, 308, 1635–1638. [Google Scholar] [CrossRef]
- Dethlefsen, L.; Huse, S.; Sogin, M.L.; Relman, D.A. The pervasive effects of an antibiotic on the human gut microbiota, as revealed by deep 16S rRNA sequencing. PLoS Biol. 2008, 6, e280. [Google Scholar] [CrossRef]
- Zhernakova, A.; Kurilshikov, A.; Bonder, M.J.; Tigchelaar, E.F.; Schirmer, M.; Vatanen, T.; Mujagic, Z.; Vila, A.V.; Falony, G.; Vieira-Sliva, S.; et al. Population-based metagenomics analysis reveals markers for gut microbiome composition and diversity. Science 2016, 352, 565–569. [Google Scholar] [CrossRef]
- Qin, J.-J.; Li, R.-Q.; Raes, J.; Arumugam, M.; Burgdorf, K.S.; Manichanh, C.; Nielsen, T.; Pons, N.; Levenez, F.; Yamada, T.; et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature 2010, 464, 59–65. [Google Scholar] [CrossRef]
- Liu, Z.-Q.; Yu, P.; Cai, M.-Q.; Wu, D.-L.; Zhang, M.; Chen, M.-H.; Zhao, Y.-L. Effects of microplastics on the innate immunity and intestinal microflora of juvenile Eriocheir sinensis. Sci. Total Environ. 2019, 685, 836–846. [Google Scholar] [CrossRef] [PubMed]
- Li, E.-C.; Xu, C.; Wang, X.-D.; Wang, S.-F.; Zhao, Q.; Zhang, M.-L.; Qin, J.G.; Chen, L.-Q. Gut microbiota and its modulation for healthy farming of Pacific white shrimp Litopenaeus vannamei. Rev. Fish. Sci. Aquac. 2018, 26, 381–399. [Google Scholar] [CrossRef]
- Dai, A.-Y. A new species of genus Sinopotamon (Decapoda, Brachyura). Acta Zootaxonomica Sin. 1992, 17, 283–285. (In Chinese) [Google Scholar]
- Dai, A.-Y. Fauna Sinica (Arthropoda: Crustacea: Malacostraca: Decapoda: Parathelphusidae, Potamidae); Science Press: Beijing, China, 1999; pp. 37–38. (In Chinese) [Google Scholar]
- Xie, R.-L.; Zhao, G.-F.; Yang, J.-H.; Wang, Z.-H.; Xu, Y.-P.; Zhang, X.-W.; Wang, Z.-J. eDNA metabarcoding revealed differential structures of aquatic communities in a dynamic freshwater ecosystem shaped by habitat heterogeneity. Environ. Res. 2021, 201, 111602. [Google Scholar] [CrossRef] [PubMed]
- Wong, S.; Rawls, J.F. Intestinal microbiota composition in fishes is influenced by host ecology and environment. Mol. Ecol. 2012, 21, 3100–3102. [Google Scholar] [CrossRef]
- Ji, Y.-K.; Sun, Y.-F.; Gao, W.; Chu, K.-L.; Wang, R.-C.; Zhao, Q.; Sun, H.-Y. Out of the Sichuan Basin: Rapid species diversification of the freshwater crabs in Sinopotamon (Decapoda: Brachyura: Potamidae) endemic to China. Mol. Phylogenet. Evol. 2016, 100, 80–94. [Google Scholar] [CrossRef]
- Covich, A.P.; Palmer, M.A.; Crowl, T.A. The role of benthic invertebrate species in freshwater ecosystems: Zoobenthic species influence energy flows and nutrient cycling. BioScience 1999, 49, 119–127. [Google Scholar] [CrossRef]
- Pinto, I.; Rodrigues, S.; Antunes, S.C. Assessment of the benthic macroinvertebrate communities in the evaluation of the water quality of Portuguese reservoirs: An experimental approach. Water 2021, 13, 3391. [Google Scholar] [CrossRef]
- Chen, S.-F.; Zhou, Y.Q.; Chen, Y.R.; Gu, J. fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 2018, 34, i884–i890. [Google Scholar] [CrossRef] [PubMed]
- Magoč, T.; Salzberg, S.L. FLASH: Fast length adjustment of short reads to improve genome assemblies. Bioinformatics 2011, 27, 2957–2963. [Google Scholar] [CrossRef] [PubMed]
- Edgar, R.C. UPARSE: Highly accurate OTU sequences from microbial amplicon reads. Nat. Methods 2013, 10, 996–998. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Garrity, G.M.; Tiedje, J.M.; Cole, J.R. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 2007, 73, 5261–5267. [Google Scholar] [CrossRef]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glöckner, F.O. The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 2012, 41, D590–D596. [Google Scholar] [CrossRef]
- Schloss, P.D.; Westcott, S.L.; Ryabin, T.; Hall, J.R.; Hartmann, M.; Hollister, E.B.; Lesniewski, R.A.; Oakley, B.B.; Parks, D.H.; Robinson, C.J.; et al. Introducing mothur: Open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 2009, 75, 7537–7541. [Google Scholar] [CrossRef]
- Caporaso, J.G.; Kuczynski, J.; Stombaugh, J.; Bittinger, K.; Bushman, F.D.; Costello, E.K.; Fierer, N.; Peña, A.G.; Goodrich, J.K.; Gordon, J.I.; et al. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 2010, 7, 335–336. [Google Scholar] [CrossRef]
- Oksanen, J.; Blanchet, F.G.; Friendly, M.; Kindt, R.; Legendre, P.; McGlinn, D.; Minchin, P.R.; O’Hara, R.B.; Simpson, G.L.; Solymos, P.; et al. Package ‘Vegan’. Community Ecology Package. Version 2.5-6. 2019. Available online: https://CRAN.R-project.org/package=vegan (accessed on 23 June 2020).
- Wickham, H. ggplot2: Elegant Graphics for Data Analysis, 2nd ed.; Springer Nature: Berlin/Heidelberg, Germany, 2016; pp. 189–201. [Google Scholar]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2013. [Google Scholar]
- Segata, N.; Izard, J.; Waldron, L.; Gevers, D.; Miropolsky, L.; Garrett, W.S.; Huttenhower, C. Metagenomic biomarker discovery and explanation. Genome Biol. 2011, 12, R60. [Google Scholar] [CrossRef]
- Langille, M.G.I.; Zaneveld, J.; Caporaso, J.G.; McDonald, D.; Knights, D.; Reyes, J.A.; Clemente, J.C.; Burkepile, D.E.; Thurber, R.L.V.; Knight, R.; et al. Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat. Biotechnol. 2013, 31, 814–821. [Google Scholar] [CrossRef]
- Tong, Q.; Hu, Z.-F.; Du, X.-P.; Bie, J.; Wang, H.-B. Effects of seasonal hibernation on the similarities between the skin microbiota and gut microbiota of an Amphibian (Rana dybowskii). Microb. Ecol. 2020, 79, 898–909. [Google Scholar] [CrossRef]
- Stoffel, M.A.; Acevedo-Whitehouse, K.; Morales-Durán, N.; Grosser, S.; Chakarov, N.; Krüger, O.; Nichols, H.J.; Elorriaga-Verplancken, F.R.; Hoffman, J.I. Early sexual dimorphism in the developing gut microbiome of northern elephant seals. Mol. Ecol. 2020, 29, 2109–2122. [Google Scholar] [CrossRef]
- Pierce, M.L.; Ward, J.E.; Holohan, B.A.; Zhao, X.-W.; Hicks, R.E. The influence of site and season on the gut and pallial fluid microbial communities of the eastern oyster, Crassostrea virginica (Bivalvia, Ostreidae): Community-level physiological profiling and genetic structure. Hydrobiologia 2016, 765, 97–113. [Google Scholar] [CrossRef]
- Fan, C.; Zhang, L.-Z.; Jia, S.-G.; Tang, X.-J.; Fu, H.-B.; Li, W.-J.; Liu, W.-J.; Liu, C.-F.; Zhang, H.; Cheng, Q.; et al. Seasonal variations in the composition and functional profiles of gut microbiota reflect dietary changes in plateau pikas. Integr. Zool. 2022, 17, 379–395. [Google Scholar] [CrossRef] [PubMed]
- Dulski, T.; Kozłowski, K.; Ciesielski, S. Habitat and seasonality shape the structure of tench (Tinca tinca L.) gut microbiome. Sci. Rep. 2020, 10, 4460. [Google Scholar] [CrossRef] [PubMed]
- Sun, P.; Jin, M.; Ding, L.-Y.; Lu, Y.; Ma, H.-N.; Yuan, Y.; Zhou, Q.-C. Dietary lipid levels could improve growth and intestinal microbiota of juvenile swimming crab, Portunus trituberculatus. Aquaculture 2018, 490, 208–216. [Google Scholar] [CrossRef]
- Hao, Y.-W.; Guan, W.; Wu, H.-N.; Li, L.-H.; Abe, E.M.; Xue, J.-B.; Qin, Z.-Q.; Wang, Q.; Lv, S.; Xu, J.; et al. Intestinal microbiome profiles in Oncomelania hupensis in mainland China. Acta Trop. 2020, 201, 105202. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.-T.; Liu, J.-L.; Jin, X.-X.; Liu, C.; Fan, C.-W.; Guo, L.; Liang, Y.-X.; Zheng, J.-S.; Peng, N. Developmental, dietary, and geographical impacts on gut microbiota of red swamp crayfish (Procambarus clarkii). Microorganisms 2020, 8, 1376. [Google Scholar] [CrossRef]
- Chen, X.-W.; Lu, D.-Y.; Li, Z.-H.; Yue, W.-C.; Wang, J.; Jiang, X.-Y.; Han, H.; Wang, C.-H. Plant and animal-type feedstuff shape the gut microbiota and metabolic processes of the Chinese mitten crab Eriocheir sinensis. Front. Vet. Sci. 2021, 8, 589624. [Google Scholar] [CrossRef]
- Yang, J.-C.; Zhang, Q.-Q.; Zhang, T.-L.; Wang, S.-Y.; Hao, J.-W.; Wu, Z.-B.; Li, A.-H. Comparative analusis of the symbiotic microbiota in the Chinese mitten crab (Eriocheir sinensis): Microbial structure, co-occurrence patterns, and predictive functions. Microorganisms 2023, 11, 544. [Google Scholar] [CrossRef]
- Wei, H.-L.; Wang, H.; Tang, L.; Mu, C.-K.; Ye, C.-Y.; Chen, L.-Z.; Wang, C.-L. High-throughput sequencing reveals the core gut microbiota of the mud crab (Scylla paramamosain) in different coastal regions of southern China. BMC Genom. 2019, 20, 829. [Google Scholar] [CrossRef]
- Wei, H.-L.; Li, X.; Tang, L.; Yao, H.-Z.; Ren, Z.-M.; Wang, C.-L.; Mu, C.-K.; Shi, C.; Wang, H. 16S rRNA gene sequencing reveals the relationship between gut microbiota and ovarian development in the swimming crab Portunus trituberculatus. Chemosphere 2020, 254, 126891. [Google Scholar] [CrossRef] [PubMed]
- Turnbaugh, P.J.; Bäckhed, F.; Fulton, L.; Gordon, J.I. Diet-induced obesity is linked to marked but reversible alterations in the mouse distal gut microbiome. Cell Host Microbe 2008, 3, 213–223. [Google Scholar] [CrossRef] [PubMed]
- Becker, A.A.; Hesta, M.; Hollants, J.; Janssens, G.P.; Huys, G. Phylogenetic analysis of faecal microbiota from captive cheetahs reveals underrepresentation of Bacteroidetes and Bifidobacteriaceae. BMC Microbiol. 2014, 14, 43. [Google Scholar] [CrossRef]
- Wu, X.-Y.; Zhang, H.-H.; Chen, J.; Shang, S.; Wei, Q.-G.; Yan, J.-K.; Tu, X.-Y. Comparison of the fecal microbiota of dholes high-throughput Illumina sequencing of the V3–V4 region of the 16S rRNA gene. Appl. Microbiol. Biotechnol. 2016, 100, 3577–3586. [Google Scholar] [CrossRef] [PubMed]
- Wexler, A.G.; Goodman, A.L. An insider’s perspective: Bacteroides as a window into the microbiome. Nat. Microbiol. 2017, 2, 17026. [Google Scholar] [CrossRef]
- Murphy, E.F.; Cotter, P.D.; Healy, S.; Marques, T.M.; O’Sullivan, O.; Fouhy, F.; Clarke, S.F.; O’Toole, P.W.; Quigley, E.M.; Stanton, C.; et al. Composition and energy harvesting capacity of the gut microbiota: Relationship to diet, obesity and time in mouse models. Gut 2010, 59, 1635–1642. [Google Scholar] [CrossRef]
- Holt, C.C.; Bass, D.; Stentiford, G.D.; van der Giezen, M. Understanding the role of the shrimp gut microbiome in health and disease. J. Invertebr. Pathol. 2021, 186, 107387. [Google Scholar] [CrossRef]
- You, Z.-Q.; Deng, J.; Liu, J.-L.; Fu, J.-H.; Xiong, H.; Luo, W.; Xiong, J.-L. Seasonal variations in the composition and diversity of gut microbiota in white-lipped deer (Cervus albirostris). PeerJ 2022, 10, e13753. [Google Scholar] [CrossRef] [PubMed]
- Huang, G.-P.; Qu, Q.-Y.; Wang, M.; Huang, M.-P.; Zhou, W.-L.; Wei, F.-W. Global landscape of gut microbiome diversity and antibiotic resistomes across vertebrates. Sci. Total Environ. 2022, 838, 156178. [Google Scholar] [CrossRef]
- Sun, F.-L.; Xu, Z.-T. Significant differences in intestinal microbial communities in aquatic animals from an aquaculture area. J. Mar. Sci. Eng. 2021, 9, 104. [Google Scholar] [CrossRef]
- Sun, Y.-F.; Han, W.-F.; Liu, J.; Liu, F.; Cheng, Y.-X. Microbiota comparison in the intestine of juvenile Chinese mitten crab Eriocheir sinensis fed different diets. Aquaculture 2020, 515, 734518. [Google Scholar] [CrossRef]
- Fraune, S.; Zimmer, M. Host-specificity of environmentally transmitted Mycoplasma-like isopod symbionts. Environ. Microbiol. 2008, 10, 2497–2504. [Google Scholar] [CrossRef] [PubMed]
- Kostanjšek, R.; Štrus, J.; Avguštin, G. “Candidatus Bacilloplasma”, a novel lineage of Mollicutes associated with the hindgut wall of the terrestrial isopod Porcellio scaber (Crustacea: Isopoda). Appl. Environ. Microbiol. 2007, 73, 5566–5573. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.-H.; Zhou, Y.-F.; Lv, D.-W.; Ge, Y.; Li, H.; You, Y. Change in the intestinal bacterial community structure associated with environmental microorganisms during the growth of Eriocheir sinensis. MicrobiologyOpen 2019, 8, e00727. [Google Scholar] [CrossRef]
- Rameshkumar, G.; Ravichandran, S.; Chandrasekar; Ajithkumar, T. Gut microbiota of crabs from the Vellar estuary, southeast coast of India. Microb. Ecol. Health Dis. 2009, 21, 178–182. [Google Scholar]
- Zhang, Z.; Li, B.; Wang, Y.-G.; Liao, M.-J.; Wang, L.; Rong, X.-J. The microflora structure in digestive tract of half-smooth tongue sole (Cynoglossus semilaevis Günther) cultured in outdoor pond basing on high-through sequencing technique. Acta Hydrobiol. Sin. 2015, 39, 38–45. [Google Scholar]
- Wang, X.-F.; Zhao, Y.-F.; Song, Z.-F.; Zhong, S.-P.; Huang, G.-Q.; Tong, T.; Nie, Z.-P.; Su, Q.; Yang, J.-L. Application of high-throughput sequencing techniques for analyzing bacterial communities in pond-raised mud crab (Scylla paramamosain) intestine and its aquaculture environment. J. Fish. Sci. China 2017, 24, 1245–1253. [Google Scholar] [CrossRef]
- Sun, Y.-G.; Zhang, S.-S.; Nie, Q.-X.; He, H.-J.; Tan, H.-Z.; Geng, F.; Ji, H.-H.; Hu, J.-L.; Nie, S.P. Gut firmicutes: Relationship with dietary fiber and role in host homeostasis. Crit. Rev. Food Sci. Nutr. 2022, 1–16. [Google Scholar] [CrossRef]
- Ghai, R.; Rodŕíguez-Valera, F.; McMahon, K.D.; Toyama, D.; Rinke, R.; Cristina Souza de Oliveira, T.; Garcia, J.T.; de Miranda, F.P.; Henrique-Silva, F. Metagenomics of the water column in the pristine upper course of the Amazon river. PLoS ONE 2011, 6, e23785. [Google Scholar] [CrossRef]
- Zhang, M.-L.; Sun, Y.-H.; Chen, L.-Q.; Cai, C.-F.; Qiao, F.; Du, Z.-Y.; Li, E.-C. Symbiotic bacteria in gills and guts of Chinese mitten crab (Eriocheir sinensis) differ from the free-living bacteria in water. PLoS ONE 2016, 11, e0148135. [Google Scholar] [CrossRef]
- Xavier, R.; Soares, M.C.; Silva, S.M.; Banha, F.; Gama, M.; Ribeiro, L.; Anastácio, P.; Cardoso, S.C. Environment and host-related factors modulate gut and carapace bacterial diversity of the invasive red swamp crayfish (Procambarus clarkii). Hydrobiologia 2021, 848, 4045–4057. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, C.; Liu, M.; Wang, Y.; Shi, B.; Pan, D. Insights into the Gut Microbiota of the Freshwater Crab Sinopotamon planum across Three Seasons and Its Associations with the Surrounding Aquatic Microbiota. Diversity 2023, 15, 519. https://doi.org/10.3390/d15040519
Liu C, Liu M, Wang Y, Shi B, Pan D. Insights into the Gut Microbiota of the Freshwater Crab Sinopotamon planum across Three Seasons and Its Associations with the Surrounding Aquatic Microbiota. Diversity. 2023; 15(4):519. https://doi.org/10.3390/d15040519
Chicago/Turabian StyleLiu, Caixin, Meijun Liu, Yifan Wang, Boyang Shi, and Da Pan. 2023. "Insights into the Gut Microbiota of the Freshwater Crab Sinopotamon planum across Three Seasons and Its Associations with the Surrounding Aquatic Microbiota" Diversity 15, no. 4: 519. https://doi.org/10.3390/d15040519
APA StyleLiu, C., Liu, M., Wang, Y., Shi, B., & Pan, D. (2023). Insights into the Gut Microbiota of the Freshwater Crab Sinopotamon planum across Three Seasons and Its Associations with the Surrounding Aquatic Microbiota. Diversity, 15(4), 519. https://doi.org/10.3390/d15040519








