Abstract
The aim of the present study was to investigate the genetic diversity and structure of eight Trifolium fragiferum populations in Latvia in the context of the Baltic Sea region. In addition, one wild population from Estonia and one from Denmark were analyzed, as well as the commercial cultivar ‘Palestine’. The genetic diversity of wild populations was low, compared to the higher diversity in the cultivar ‘Palestine’. The wild populations were differentiated into four clusters, separate from the cultivar ‘Palestine’. Three of the genetically similar Latvian populations were also geographically proximal, but the four populations geographically located near the capital, Riga, were genetically differentiated from each other. The genetic results were compared to previously published physiological studies of the same populations. The results from this study can be used as a basis for further studies of T. fragiferum populations in the Baltic Sea region, and to develop in situ and ex situ conservation strategies for this crop wild relative species.
1. Introduction
Crop wild relative (CWR) species represent a valuable resource for the breeding of resilient and stress-tolerant crops, enhancing the sustainability of agroecosystems facing increasing threats from anthropogenic pressures and global climate change [1]. CWR species are an important source of genetic diversity, and contain alleles and adaptive traits that can be utilized in crop breeding programs. The incorporation of these diverse sources of variation can assist the agricultural system to adjust to increasing disease pressures, changing farming practices and consumer demands, and unstable climatic and environmental conditions [2]. However, given the anthropogenic and climatic pressures mentioned previously, CWR species are increasingly threatened, and therefore the development of both in situ and ex situ conservation strategies is urgently required to ensure that these valuable resources are not lost, and can provide a long-term base for sustainable agriculture and food security [3].
Among CWR species, legumes are especially important in ecosystem sustainability due to their unique ability to establish symbioses with N2-fixing bacteria [4]. In the Baltic Sea region, CWR species are mainly represented by forage crop species, perennial grasses and legumes. Trifolium fragiferum (strawberry clover) is an herbaceous perennial semi-rosette plant with proliferating creeping basal shoots, which has a high potential for clonal reproduction due to the ability of monopodially branching shoots (stolons) to form roots at the nodes in high moisture conditions [5]. The main distribution area in Europe is centered around the Mediterranean, and is widely distributed in Western and Central Europe. It is also native to North Africa and Southwest Asia, and has been introduced to North and South America, Australia and New Zealand [6]. In contrast to other clover species, T. fragiferum is tolerant to a number of adverse soil conditions, including salinity, alkalinity and flooding [7]. Due to efficient N2-fixing ability of the rhizobial symbionts, T. fragiferum can maintain optimum or close to optimum N concentrations in plant tissues, despite growing in soils with low plant-available N concentrations [8]. Therefore, these wild populations not only represent valuable pools of adaptive T. fragiferum alleles, but also sources of stress-tolerant symbiotic N2-fixing bacteria that are required to ensure sustainable agriculture [9].
Studies of the diversity of wild clover species in the Baltic region have been performed with respect to their use as forage crops [10,11,12,13,14]. Trifolium fragiferum L. is a rare clover species in Northern Europe, characteristic of the endangered habitat ‘Boreal Baltic coastal meadows’ (1630*) [15]. T. fragiferum has not been included in the European list of CWR species [16], and is not in a priority list of the taxa of Nordic CWR species [17]. However, due to the high resilience and abiotic stress tolerance of this species, T. fragiferum has been commercially grown in the temperate zone of Australia and in several other countries [18,19,20].
Wild populations of T. fragiferum represent a valuable resource in forage crop breeding, due to the high diversity in abiotic stress tolerance found within this species [20]. Recently, we characterized the physiological diversity in responses to abiotic and biotic factors among Latvian accessions of wild T. fragiferum in controlled conditions [9,21,22,23]. These studies indicated that all analyzed accessions were tolerant to increased soil moisture, trampling and cutting; however, significant accession-specific differences in tolerance to individual factors were found. In addition, while Latvian T. fragiferum accessions were tolerant to cadmium and lead in soils, differences in morphological responses to heavy metals were identified between populations. As a result, we concluded that geographically isolated populations of Latvia represent various physiotypes, and possibly belong to several different ecotypes. Therefore, further studies are necessary to determine the genetic diversity and structure of these populations in order to develop effective in situ and ex situ conservation strategies for this crop wild relative species.
In situ and ex situ conservation are complementary approaches, and both are needed to effective conserve and utilize CWR species, and the actions needed to establish CWR species conservation strategies have been outlined [24]. After the prioritization of species, CWR species populations need to be surveyed, and conservation sites identified and established. These sites require long-term monitoring to identify threats and changes in genetic diversity in a timely manner. The conserved CWR species populations should be placed into ex situ collections to facilitate the distribution of the material to breeders and other potential users. Detailed knowledge about the reproductive biology and population structure of CWR species populations is required to identify the most diverse and valuable wild populations, as well as to develop protocols for the collection of propagating material for inclusion into ex situ collections [25].
The use of DNA markers provides valuable information about the genetic diversity and differentiation of plant populations, which is required for efficient management of CWR species populations [1]. DNA markers can also provide insights into the provenance and connectivity of populations. In contrast to phenotypic markers, DNA markers are not influenced by external or environmental conditions, and can be assessed at any developmental stage [26]. Microsatellites or simple sequence repeat (SSR) markers are often used in population genetic studies, due to their high polymorphism and codominance [27]. However, specific PCR primers need to be designed for each locus, requiring substantial investments for the development of these markers [28]. Microsatellite markers are developed for specific species, but often can be transferred to related species. This can enable the use of previously developed SSR markers in related species, but can result in a lower amplification success, and the transferred markers can have lower polymorphism levels compared to the species for which they were developed [29].
Molecular studies of T. fragiferum genetic diversity are scarce. One study used isozymes to assess the genetic diversity and differentiation of 22 Polish T. fragiferum populations [30]. Microsatellite or SSR markers have not been specifically developed for T. fragiferum; however, the cross-species amplification of SSR markers developed for Trifolium pratense has been assessed in a number of Trifolium species, including T. fragiferum [31]. These SSR markers were used to investigate the genetic diversity and differentiation of eight T. fragiferum populations in Iran [32]. The aim of the present study was to analyze the genetic diversity and structure of Latvian T. fragiferum populations, as well as other populations in the Baltic Sea region.
2. Materials and Methods
2.1. Sample Collection and Plant Establishment
Seeds of T. fragiferum from eight previously identified geographically isolated micropopulations (populations) in Latvia were collected (Figure 1; Table S1) [8]. In addition, seeds from coastal populations of T. fragiferum in Denmark and Estonia were collected for comparison. T. fragiferum cv. ‘Palestine’ seeds obtained from Sheffields Seeds Company (Locke, NY, USA), was used as a reference genotype. As T. fragiferum occupied relatively small areas in all sampling sites, 24 fruit capsules from individual plants spatially separated in the population were collected. From each capsule, three seeds were randomly selected and used for the establishment of seedlings, as described previously [21]. For each population, 24 two-week-old seedlings, each derived from a separate seed capsule, were selected for genetic analysis.
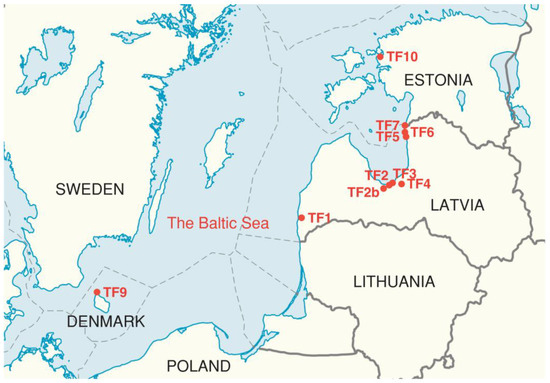
Figure 1.
Map of the Baltic Sea region indicating the locations of the analyzed wild Trifolium fragiferum populations. TF1 to TF10 indicate different populations.
2.2. Genetic Analyses
DNA was extracted using a modified CTAB method [33]. From each population, 24 individuals were analyzed. Ten SSR markers developed for red clover were tested on a subset of samples (TPSSR09, TPSSR46, TPSSR13, TPSSR17, TPSSR34, TPSSR16, TPSSR44, TPSSR50, TPSSR29, and TPSSR40) [34], as well as six SSR markers developed for red clover, but tested for cross-species transferability, including T. fragiferum (RCS0883, RCS2667, RCS1928, RCS1225, RCS3666, and RCS1897) [31,35]. The six RCS markers were previously used to analyze T. fragiferum populations in Iran [32].
PCR reactions were performed in a volume of 10 µL containing approximately 50 ng DNA, 2 µL HOT FIREPol® Blend Master Mix (Solis BioDyne, Tartu, Estonia) (containing 10 mM MgCl2), 0.3 µM forward, and reverse primers. PCR was carried out in a thermocycler (Eppendorf Mastercycler Epgradient; Eppendorf, Hamburg, Germany) using the following protocol: initial pre-denaturation step at 95 °C for 15 min, followed by 40 cycles of 95 °C for 30 s, annealing for 30 s, and 72 °C for 45 s, and a final extension step of 72 °C for 10 min. Annealing temperatures were 60 °C for the TPSSR markers, and 55 °C for the RCS markers. Forward primers were labeled with 6-FAM, HEX, or TMR dyes. All PCR reaction products were diluted 1:10 with deionized water, and visualized on an Applied Biosystems ABI Prism 3100xl Genetic Analyzer. Genotyping was performed using GeneMapper 4.0. (Applied Biosystems, Waltham, MA, USA).
GenAlEx 6.501 [36] was used to calculate genetic diversity indices, and AMOVA and pairwise population differentiation (FST) (using 999 permutations). A neighbor-joining tree was constructed based on Nei’s standard genetic distance with 1000 bootstrap replications using POPTREEW [37], which was visualized using MEGA11 [38]. STRUCTURE 2.3.4 [39] was used to determine the clustering of the populations using a 100,000 burn-in period, followed by 200,000 MCMC steps, and K was determined for 1 to 11 with 10 runs for each value of K. The admixture model was used without sampling locations (LOCPRIOR). The optimal value of K was determined using the ΔK method [40] using STRUCTURE HARVESTER [41], and the probability was determined by K using median values of Ln (Pr Data) implemented via the CLUMPAK server [42]. Coefficients of membership to the most likely K cluster number were visualized using Distruct via the CLUMPAK server.
3. Results
Of the 16 SSR markers tested on a subset of samples, twelve (TPSSR16, TPSSR17, TPSSR40, TPSSR50, RCS0883, RCS1928, RCS1225, RCS1897, RCS2667, RCS3666, TPSSR09, and TPSSR46) amplified fragments that were able to be unambiguously genotyped, and all DNA samples were subsequently genotyped with these markers. Two markers (TPSSR09 and TPSSR46) were not amplified in 63% and 47% of DNA samples, and were excluded from further analysis. In addition, two markers (RCS1928 and RCS1897) were monomorphic in all analyzed samples, and so were also excluded from further analysis. Therefore, all samples were genotyped with eight SSR markers (Table S2).
The genetic diversity revealed by the analyzed SSR markers was low, with the number of alleles identified by each marker ranging from 2 to 16. However, rare alleles were prevalent, with 40 of the 56 alleles found having a frequency of less than 0.05. With the exception of the marker TPSSR50, the observed heterozygosity was lower or the same as the expected heterozygosity (Table S3). This could possibly indicate the presence of null alleles; however, it is more likely that this result is a consequence of the biological and reproductive characteristics of the analyzed populations. The other genetic diversity parameters of the analyzed populations were also low, with the exception of the cv. ‘Palestine’ (Figure 2, Table S4). Three markers were monomorphic in all populations except for the cv. ‘Palestine’ (RCS0883, RCS1225, and RCS3666).
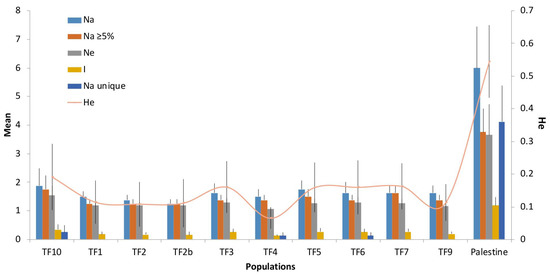
Figure 2.
Genetic diversity parameters (mean over all loci) of the analyzed T. fragiferum populations. Na—number of alleles, Na ≥ 5%—number of alleles with a frequency equal to or more than 5%, Ne—effective number of alleles, I—information index, Na unique—number of unique alleles in each population, and He—expected heterozygosity.
Genetic differentiation of the analyzed populations was high; overall, FST was 0.380 (p < 0.001). Removing individuals from the highly polymorphic and differentiated variety ‘Palestine’ from analysis did not have a large impact on the overall FST between the remaining populations, which was 0.375 (p < 0.001). This differentiation was mainly due to the locus TPSSR40. Pairwise population FST values were above 0.05, with the exception of TF5–TF6 (0.038), TF5–TF7 (0.017), and TF6–TF7 (0.007) (Table 1). Unique (private) alleles were identified for the cv. ‘Palestine’ at all loci, at frequencies ranging from 0.021 to 0.583. Private alleles were also found in the population TF10 (TPSSR40, 2 alleles, f = 0.130, and 0.413), as well as the population TF4 (TPSSR40, f = 0.021), and TF6 (TPSSR16, f = 0.042).

Table 1.
Pairwise FST values between the analyzed T. fragiferum populations.
The differentiation of the cultivar ‘Palestine’ and the clustering of the populations TF5, TF6, and TF7 can be observed in the neighbor-joining dendrogram based on pairwise Nei genetic distances (Figure 3). Using the ΔK method, the analysis of the Structure software results indicated that the most likely number of clusters was K = 5, with K = 4 also having a high ΔK value (Figure 4). Structure analysis also indicated that the cv. ‘Palestine’ was highly differentiated from the other populations. Populations TF5, TF6, and TF7 were clustered together, which also corresponded to their close geographical proximity (Figure 1). Interestingly, despite their close geographical proximity, the populations TF2, TF2B, TF3 and TF4 were differentiated, which may be a reflection of the dispersal mechanisms of T. fragiferum, and the differing provenances of the founders of these populations. The populations TF9 and TF10 (from Denmark and Estonia, respectively), were not highly differentiated from the Latvian populations. Again, this could be a consequence of the dispersal mechanisms of this species, or alternatively, a result of the low genetic diversity revealed using the SSR markers.
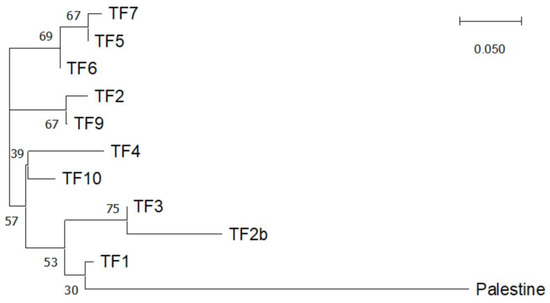
Figure 3.
Neighbor-joining tree based on Nei genetic distances between populations. Numbers indicate bootstrap values (%).
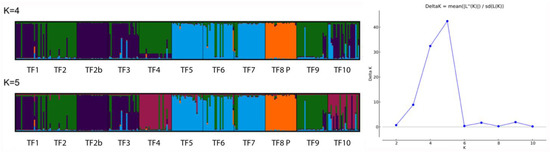
Figure 4.
Plots of coefficients of membership for K = 4 and K = 5 (the most likely number of clusters as determined by the ΔK method).
4. Discussion
Within the northeast area of the distribution range of T. fragiferum, including Latvia, this species is found in small, geographically isolated populations that are associated with water reservoirs [8]. Populations from sites with similar plant-available mineral nutrient concentrations in soil as well as with similar mineral nutrient concentrations in leaves had similar phenotypic characteristics (e.g., TF2, TF2b, TF3 (located on river shores), or TF5, TF6, TF7 (located in coastal areas) [8]. Genetic analysis performed in the present study confirmed the relatively close genetic association between TF5, TF6, and TF7 (Table 1). However, in spite of the close geographical proximity of TF2, TF2b, and TF3, as well as their similar physiological properties, these populations showed pronounced genetic differentiation (Table 1). In contrast, plants from the genetically well differentiated populations TF2 and TF7 (Table 1) showed similar stress tolerance-related characteristics, e.g., responses to cutting, soil waterlogging and trampling [21], and substrate salinity [23], as well as rhizobial inoculation and nitrogen treatment [9].
In the populations analyzed in this study, the local adaptation to salinity in coastal populations in comparison to river populations might have been expected, which could also be influenced by the possible propagule dispersion by water routes. However, the T. fragiferum accessions from the most saline habitats (TF1, TF5, TF6, and TF9) were not genetically clustered in one group (Figure 3 and Figure 4). In contrast, the TF1 and TF9 populations clustered together based on the multivariate analysis of physiological responses to the increasing substrate salinity [23]. This suggests that the genetic structure identified by the utilized markers does not reflect an adaptation of the populations to particular ecological conditions. This is not surprising, given the small number and presumed adaptive neutrality of the markers used. Genetic variation and adaptation potential are not always directly correlated, and rapid adaptation can occur without significant genetic variation [43]. Knowledge about the role of epigenetic processes in shaping the adaptation of local plant populations is increasing [44], but a thorough understanding of the importance of these mechanisms is still in its infancy. In addition, genetic diversity of neutral sequences might not fully reflect adaptive variation, e.g., protein-coding genes and other regulatory elements [45]. Consequently, local genetic adaptation as a mechanism for the formation of ecotypes might not be reflected in the genetic diversity revealed by selectively neutral SSR markers.
In general, there was no obvious correspondence of the genetic clusters identified with the geographic location, with the exception of populations TF5, TF6, and TF7, located on the northern coast of the Gulf of Riga. The four populations near Riga (TF2, TF2b, TF3, and TF4) were differentiated from each other (the lowest pairwise FST value was 0.112 between TF2b and TF3). While the three northern Latvian populations are not contiguous, they are all located on the seashore, and there are no barriers to prevent gene flow between them. In contrast, the four population near Riga are located in more urbanized areas, and are in the vicinity of the Baltic Sea as well as rivers, which could affect dispersal patterns. Anthropogenic factors may have influenced these populations, including establishment and subsequent dispersal, which could be reflected in the relatively high differentiation between these populations. These four populations near Riga were more similar to geographically distant populations, e.g., TF2 and TF9 (Denmark), and TF4 and TF10 (Estonia). Further investigation of additional Baltic Sea region T. fragiferum populations will assist in clarifying the population structure of T. fragiferum populations within the larger region, as well as identifying possible dispersal routes.
T. fragiferum is a species capable of clonal reproduction due to the ability of creeping stolons to form adventitious roots at nodes [5]. The proportion of vegetative reproduction vs. sexual reproduction has not been estimated for any clover species, but, based on the low genetic diversity within the analyzed wild populations (Figure 2), it is possible that in the small, geographically isolated populations of T. fragiferum, clonal propagation is prevalent. The use of markers with low levels of variability in studies of clonal plants might result in erroneous estimates of low diversity within populations [46]. However, while the genetic diversity within the analyzed T. fragiferum populations was low, many of the populations were genetically well differentiated. This suggests that the low levels of genetic diversity within the analyzed populations is likely a reflection of clonal propagation and/or reproductive isolation within these marginal and isolated micropopulations, rather than the low variability of the utilized SSR markers. In addition, a high genetic diversity was detected within the cv. ‘Palestine’.
The genetic diversity detected by the SSR markers in this study was in general lower than reported in previous analyses of T. fragiferum using the same markers [31]. This could be a reflection of the populations analyzed—in the previous study, eight Iranian T. fragiferum populations were analyzed. However, the intrapopulation genetic diversity was not reported, nor was the mode of reproduction (generative or vegetative). In this study, the polymorphism of the previously utilized RCS markers was lower than the TPSSR markers (Table S3). The further analysis of additional SSR markers developed for various Trifolium species can identify additional informative markers for use in genetic analyses of T. fragiferum. A previous study utilized isozymes to analyze Polish T. fragiferum populations [30], and populations from Northern Poland had the lowest genetic diversity, while the eastern populations had the highest, which was explained by more favorable climatic conditions in the eastern region. In addition, genetic diversity was higher in populations growing in meadows and pastures as opposed to those growing in more marginal and suboptimal areas. This is in agreement with the results of this study, as all the analyzed wild T. fragiferum populations with low genetic diversity were growing in isolated micropopulations, subjected to high abiotic stress conditions. In contrast, the genetic diversity of the cv. ‘Palestine’ was relatively high.
The ex situ conservation of seeds in gene banks or of plants in field collections at botanic gardens or other sites is complementary to in situ conservation, and both approaches are required to effectively conserve and utilize plants’ genetic resources [47]. In situ conservation is dynamic, and allows populations to adapt to changing environmental conditions, but can be difficult to implement, and requires ongoing monitoring to ensure secure conservation. Ex situ conservation can ensure the relatively simple and cost-effective conservation of plants’ genetic diversity, and can facilitate the distribution of material to breeders, researchers, and other end users. However, this is a static form of conservation, and plant collection and regeneration protocols need to be to be optimized to maximize the genetic diversity maintained in ex situ collections. Given the small population sizes and their location in marginal areas, both in situ and ex situ conservation approaches should be applied to preserve the genetic diversity of Latvian and Baltic T. fragiferum populations. Further research on the extent of clonal reproduction within Latvian and Baltic T. fragiferum populations is needed to optimize sampling strategies. The collection and maintenance of samples for ex situ collections (either as seeds or field collections), ensuring a representative coverage of the genetic and adaptive diversity, will be difficult. Results from this and previous studies indicate that the genetic differentiation of Latvian T. fragiferum populations is not always associated with geographic location or the physiological and other adaptive traits of populations. Therefore, in situ conservation is also crucial to ensure the long-term survival of Latvian and Baltic T. fragiferum populations, maintaining the current levels of genetic and adaptive diversity.
The assessment of genetic diversity and population structure is required for the development of efficient in situ and ex situ conservation strategies. Genetic analyses are needed to establish a baseline for the development of sampling strategies and monitoring of changes in conserved populations over time. Genetic diversity within species ensures that they can evolve and adapt to environmental changes, as well as providing a resource for breeding and cultivation. For long-term in situ conservation and use in breeding, adaptive genetic diversity is more important than neutral genetic diversity. However, the evaluation of adaptive diversity can be difficult and resource intensive; therefore, using neutral genetic diversity as a proxy for adaptive genetic diversity can provide useful information for the development of in situ conservation strategies [25]. AFLP markers have been used to provide baseline genetic data for Brassica species in the United Kingdom [48], and also identify factors influencing the genetic diversity of natural populations. Genetic analyses of both wild and domesticated accessions of leafy kale in Italy allowed for the determination of the proportion of genetic diversity conserved in domesticated accessions, and to identify areas with the highest variability [49]. Genetic data can complement assessments of population size and threat status to prioritize populations for in situ conservation, as reported for the Portuguese endemic wild relative of ornamental carnations Dianthus cintranus Boiss. and Reut. subsp barbatus R. Fern. and Franco [25].
Therefore, this initial genetic assessment of wild T. fragiferum populations in Latvia establishes a baseline for future monitoring activities, provides information for the development of an in situ conservation strategy, and identifies further research directions for T. fragiferum in Latvia and the Baltic region.
5. Conclusions
The results of this study of wild Latvian T. fragiferum populations, and previous studies of the physiological diversity of these populations in response to abiotic and biotic factors [13,14,15,16], indicate that these isolated micropopulations are both genetically differentiated and have differing physiological reactions. Further genetic analyses utilizing the non-destructive sampling of individuals within populations can determine the extent of the clonal propagation. The analysis of additional populations from the Baltic Sea region will provide additional information about the regional dispersal patterns of this species. The results of this study can be used for the development of in situ conservation strategies for T. fragiferum in Latvia and the Baltic Sea region. The relatively high genetic differentiation of the analyzed populations and the variability of their physiological responses indicates that the conservation of a large proportion of these populations is needed to maximize the preservation of T. fragiferum diversity in the Baltic Sea region. The analysis of genetic and physiological parameters is crucial to determine the diversity, differentiation and adaptation of T. fragiferum populations in the Baltic Sea region, and to develop efficient in situ and ex situ conservation strategies.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/d15040473/s1, Table S1. Details of the analysed Trifolium fragiferum populations; Table S2. Genotypes of the analysed T. fragiferum populations; Table S3. Genetic diversity parameters of the analysed SSR markers; Table S4: Mean genetic diversity parameters (over all loci) of the analysed T. fragiferum populations.
Author Contributions
U.A.-O. and G.I. designed the study. U.A.-O. and A.J. obtained plant material. D.E.R. obtained and analyzed molecular data. D.E.R. and G.I. wrote the first draft. U.A.-O. and A.J. critically read and edited the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
The study was supported by the Latvian Science Council project lzp-2020/2-0349 “Molecular, physiological and ecological evaluation of Latvian genetic resources of valuable wild legume species, Trifolium fragiferum, in a context of sustainable agriculture”.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The data is contained within the manuscript and Supplementary Materials.
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
References
- Zhang, H.; Mittal, N.; Leamy, L.J.; Barazani, O.; Song, B.-H. Back into the wild—Apply untapped genetic diversity of wild relatives for crop improvement. Evol. Appl. 2017, 10, 5–24. [Google Scholar] [CrossRef] [PubMed]
- Dempewolf, H.; Baute, G.; Anderson, J.; Kilian, B.; Smith, C.; Guarino, L. Past and future use of wild relatives in crop breeding. Crop Sci. 2017, 57, 1070–1082. [Google Scholar] [CrossRef]
- Maxted, N.; Kell, S.; Ford-Lloyd, B.; Dulloo, E.; Toledo, Á. Toward the systematic conservation of global crop wild relative diversity. Crop Sci. 2012, 52, 774–785. [Google Scholar] [CrossRef]
- Zhang, H.; Yasmin, F.; Song, B.-H. Neglected treasures in the wild—Legume wild relatives in food security and human health. Curr. Opin. Plant Biol. 2019, 49, 17–26. [Google Scholar] [CrossRef] [PubMed]
- Huber, H.; Wiggerman, L. Shade avoidance in the clonal herb Trifolium fragiferum: A field study with experimentally manipulated vegetation height. Plant Ecol. 1997, 130, 53–62. [Google Scholar] [CrossRef]
- Legume Phylogeny Working Group (LPWG); Andrella, G.C.; Atahuachi Burgos, M.; Bagnatori Sartori, Â.L.; Balan, A.; Bandyopadhyay, S.; Barbosa Pinto, R.; Barrett, R.; Boatwright, J.S.; Broich, S.L.; et al. The World Checklist of Vascular Plants (WCVP): Fabaceae. In Catalogue of Life Checklist; Bánki, O., Roskov, Y., Döring, M., Ower, G., Vandepitte, L., Hobern, D., Remsen, D., Schalk, P., DeWalt, R.E., Keping, M., et al., Eds.; The Royal Botanic Gardens, Kew: Richmond, UK, 2022. [Google Scholar] [CrossRef]
- Townsend, C.E. Miscellaneous perennial clovers. In Clover Science and Technology; Taylor, J.L., Ed.; ASA/CSSA/SSSA: Madison, WI, USA, 1985; pp. 563–578. [Google Scholar]
- Andersone-Ozola, U.; Jēkabsone, A.; Karlsons, A.; Romanovs, M.; Ievinsh, G. Soil chemical properties and mineral nutrition of Latvian accessions of Trifolium fragiferum, a crop wild relative plant species. Environ. Exp. Biol. 2021, 19, 245–254. [Google Scholar]
- Jēkabsone, A.; Andersone-Ozola, U.; Karlsons, A.; Neiceniece, L.; Romanovs, M.; Ievinsh, G. Dependence on nitrogen availability and rhizobial symbiosis of different accessions of Trifolium fragiferum, a crop wild relative legume species, as related to physiological traits. Plants 2022, 11, 1141. [Google Scholar] [CrossRef]
- Dabkevičiene, G.; Dabkevičius, Z. Evaluation of wild red clover (Trifolium pratense L.) ecotypes and hybrid populations (Trifolium pratense L. × Trifolium diffusum Ehrh.) for clover rot resistance (Sclerotinia trifoliorum Erikss.). Biologija 2005, 3, 54–58. [Google Scholar]
- Rancane, S.; Jansone, B.; Sparnina, M. The evaluation of genetic resources of forage legumes collected from natural grassland. In Sustainable Grassland Productivity, Proceedings of the 21st General Meeting of the European Grassland Federation, Badajoz, Spain, 3–6 April 2006; Lloveras, J., Gonzalez-Rodriguez, A., Vazquez-Yanez, O., Pineiro, J., Santamaria, O., Olea, L., Poblaciones, M.J., Eds.; European Grassland Federation: Madrid, Spain, 2006; pp. 327–329. [Google Scholar]
- Bērziņa, I.; Zhuk, A.; Veinberga, I.; Rashal, I.; Ruņģis, D. Genetic fingerprinting of Latvian red clover (Trifolium pratense L.) varieties using simple sequence repeat (SSR) markers: Comparison over time and space. Latv. J. Agron. 2008, 11, 28–33. [Google Scholar]
- Paplauskienė, V.; Dabkevičienė, G. A study of genetic diversity in Trifolium hybridum varieties using morphological characters and ISSR markers. Zemdirb.–Agric. 2012, 99, 313–318. [Google Scholar]
- Lemežienė, N.; Stukonis, V.; Kemešytė, V.; Norkevičienė, E. Wild and semi natural ecotypes of perennial grasses and legumes–for breeding purposes. In Breeding Grasses and Protein Crops in the Era of Genomics; Brazauskas, G., Statkevičiūtė, G., Jonavičienė, K., Eds.; Springer International Publishing AG: Cham, Switzerland, 2018; pp. 88–95. [Google Scholar]
- Rūsiņa, S. 1630* Boreal Baltic coastal meadows. In European Union Protected Habitats in Latvia. Interpretation Manual; Auniņš, A., Ed.; Latvian Fund for Nature, Ministry of Environmental Protection and Regional Development: Riga, Latvia, 2013; pp. 55–57. [Google Scholar]
- Heywood, V.H.; Zohary, D. A catalogue of the wild relatives of cultivated plants native to Europe. Flora Mediterr. 1995, 5, 375–415. [Google Scholar]
- Fitzgerald, H.; Palmé, A.; Asdal, Å.; Endresen, D.; Kiviharju, E.; Lund, B.; Rasmussen, M.; Thorjömsson, H.; Weibull, J. A regional approach to Nordic crop wild relative in situ conservation planning. Plant Genet. Resour. Charact. Util. 2019, 17, 196–207. [Google Scholar] [CrossRef]
- Nichols, P.G.H.; Revell, C.K.; Humphries, A.W.; Howie, J.H.; Hall, E.J.; Sandral, G.A.; Ghamkhar, K.; Harris, C.A. Temperate pasture legumes in Australia—Their history, current use, and future prospects. Crop Pasture Sci. 2012, 63, 691–725. [Google Scholar] [CrossRef]
- Gerard, P.J.; Aalders, L.T.; Hardwick, S.; Wilson, D.J. Investigation into the contrasting production of eight perennial clover cultivars in the first two years at field sites in in Waikato and Canterbury. N. Z. J. Agric. Res. 2022, 65, 271–289. [Google Scholar] [CrossRef]
- Rumbaugh, M.D.; Pendery, B.M.; James, D.W. Variation in the salinity tolerance of strawberry clover (Trifolium fragiferum L.). Plant Soil 1993, 153, 265–271. [Google Scholar] [CrossRef]
- Andersone-Ozola, U.; Jēkabsone, A.; Purmale, L.; Romanovs, M.; Ievinsh, G. Abiotic stress tolerance of coastal accessions of a promising forage legume species, Trifolium fragiferum. Plants 2021, 10, 1552. [Google Scholar] [CrossRef] [PubMed]
- Ievinsh, G.; Karlsons, A.; Jēkabsone, A.; Andersone-Ozola, U. Heavy metal tolerance and accumulation potential of coastal accessions of Trifolium fragiferum, a promising forage species. In Proceedings of the 10th International Scientific Conference Rural Development 2021, Vytautas Magnus University Agriculture Academy, Kaunas, Lithuania, 26–28 September 2021; pp. 214–219. [Google Scholar] [CrossRef]
- Jēkabsone, A.; Andersone-Ozola, U.; Karlsons, A.; Romanovs, M.; Ievinsh, G. Effect of salinity on growth, ion accumulation and mineral nutrition of different accessions of a crop wild relative legume species, Trifolium fragiferum. Plants 2022, 11, 797. [Google Scholar] [CrossRef] [PubMed]
- Heywood, V.; Casas, A.; Ford-Lloyd, B.; Kell, S.; Maxted, N. Conservation and sustainable use of crop wild relatives. Agric. Ecosyst. Environ. 2007, 121, 245–255. [Google Scholar] [CrossRef]
- Brehm, J.M.; Ford-Lloyd, B.V.; Maxted, N.; Martins-Loução, M.A. Using neutral genetic diversity to prioritize crop wild relative populations: A Portuguese endemic case study for Dianthus cintranus Boiss. & Reut. subsp. barbatus R. Fern. & Franco. In Agrobiodiversity Conservation: Securing the Diversity of Crop Wild Relatives and Landraces; CABI: Wallingford, UK, 2012; pp. 193–210. [Google Scholar]
- Mondini, L.; Nooorani, A.; Pagnotta, M.A. Assessing plant genetic diversity by molecular tools. Diversity 2009, 1, 19–35. [Google Scholar] [CrossRef]
- Zhu, C.; Gore, M.; Buckler, E.S.; Yu, J. Status and prospects of association mapping in plants. Plant Genome 2008, 1, 5–20. [Google Scholar] [CrossRef]
- Zalapa, J.E.; Cuevas, H.; Zhu, H.; Steffan, S.; Senalik, D.; Zeldin, E.; McCown, B.; Harbut, R.; Simon, P. Using next-generation sequencing approaches to isolate simple sequence repeat (SSR) loci in the plant sciences. Am. J. Bot. 2012, 99, 193–208. [Google Scholar] [CrossRef] [PubMed]
- Kalia, R.K.; Raj, M.K.; Kalia, S.; Singh, R.; Dhawan, A.K. Microsatellite markers: An overview of the recent progress in plants. Euphytica 2011, 177, 309–334. [Google Scholar] [CrossRef]
- Bulińska-Radomska, Z. Enzyme polymorphism and adaptation in strawberry clover (Trifolium fragiferum L.). Genet. Resour. Crop Evol. 2000, 47, 197–205. [Google Scholar] [CrossRef]
- Haerinasab, M.; Rahiminejad, M.R.; Ellison, N.W. Transferability of simple sequence repeat (SSR) markers developed in red clover (Trifolium pratense L.) to some Trifolium species. Iranian J. Sci. Technol. Transact. A Sci. 2016, 40, 59–62. [Google Scholar] [CrossRef]
- Haerinasab, M.; Ali-Farsangi, F.; Bordbar, F.; Farouji, A. Genetic diversity and infraspecific relationships of Trifolium fragiferum L. in Iran. Iranian J. Sci. Technol. Transact. A Sci. 2020, 44, 345–354. [Google Scholar] [CrossRef]
- Porebski, S.; Bailey, L.G.; Baum, B.R. Modification of a CTAB DNA extraction protocol for plants containing high polysaccharide and polyphenol components. Plant Mol. Biol. Report. 1997, 15, 8–15. [Google Scholar] [CrossRef]
- Kölliker, R.; Enkerli, J.; Widmer, F. Characterization of novel microsatellite loci for red clover (Trifolium pratense L.) from enriched genomic libraries. Mol. Ecol. Notes 2006, 6, 50–53. [Google Scholar] [CrossRef]
- Sato, S.; Isobe, S.; Asamizu, E.; Ohmido, N.; Kataoka, R.; Nakamura, Y.; Kaneko, T.; Sakurai, N.; Okumura, K.; Klimenko, I.; et al. Comprehensive structural analysis of the genome of red clover (Trifolium pratense L.). DNA Res. 2005, 12, 301–364. [Google Scholar] [CrossRef]
- Peakall, R.; Smouse, P.E. GenAlEx 6.5: Genetic analysis in Excel. Population genetic software for teaching and research—An update. Bioinformatics 2012, 28, 2537–2539. [Google Scholar] [CrossRef]
- Takezaki, N.; Nei, M.; Tamura, K. POPTREEW: Web version of POPTREE for constructing population trees from allele frequency data and computing some other quantities. Mol. Biol. Evol. 2014, 31, 1622–1624. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular Evolutionary Genetics Analysis Version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef] [PubMed]
- Pritchard, J.K.; Stephens, M.; Donnelly, P. Inference of population structure using multilocus genotype data. Genetics 2000, 155, 945–959. [Google Scholar] [CrossRef] [PubMed]
- Evanno, G.; Regnaut, S.; Goudet, J. Detecting the number of clusters of individuals using the software STRUCTURE: A simulation study. Mol. Ecol. 2005, 14, 2611–2620. [Google Scholar] [CrossRef] [PubMed]
- Earl, D.A.; von Holdt, B.M. STRUCTURE HARVESTER: A website and program for visualizing STRUCTURE output and implementing the Evanno method. Conserv. Genet. Resour. 2012, 4, 359–361. [Google Scholar] [CrossRef]
- Kopelman, N.M.; Mayzel, J.; Jakobsson, M.; Rosenberg, N.A.; Mayrose, I. Clumpak: A program for identifying clustering modes and packaging population structure inferences across K. Mol. Ecol. Resour. 2015, 15, 1179–1191. [Google Scholar] [CrossRef] [PubMed]
- Mable, B.K. Conservation of adaptive potential and functional diversity: Integrating old and new approaches. Conserv. Genet. 2018, 20, 89–100. [Google Scholar] [CrossRef]
- Abratowska, A.; Wasowicz, P.; Bednarek, P.T.; Telka, J.; Wierzbicka, M. Morphological and genetic distinctiveness of metallicolous and non-metallicolous populations of Armeria maritima s.l. (Plumbaginaceae) in Poland. Plant Biol. 2012, 14, 586–595. [Google Scholar] [CrossRef]
- Kirk, H.; Freeland, J.R. Applications and implications of neutral versus non-neutral markers in molecular ecology. Int. J. Mol. Sci. 2011, 12, 3966–3988. [Google Scholar] [CrossRef]
- Arnaud-Haond, S.; Alberto, F.; Teixeira, S.; Procaccini, G.; Serrao, E.A.; Duarte, C.M. Assessing genetic diversity in clonal organisms: Low diversity or low resolution? Combining power and cost efficiency in selecting markers. J. Hered. 2005, 96, 434–440. [Google Scholar] [CrossRef]
- Magos-Brehm, J.; Kell, S.; Thormann, I.; Gaisberger, H.; Dulloo, M.E.; Maxted, N. Interactive Toolkit for Crop Wild Relative Conservation Planning Version 1.0; University of Birmingham: Birmingham, UK; Bioversity International: Rome, Italy, 2017; Available online: www.cropwildrelatives.org/conservation-toolkit/ (accessed on 1 June 2022).
- Watson-Jones, S.J.; Maxted, N.; Ford-Lloyd, B.V. Population baseline data for monitoring genetic diversity loss for 2010: A case study for Brassica species in the UK. Biol. Conserv. 2006, 132, 490–499. [Google Scholar] [CrossRef]
- Maggioni, L.; von Bothmer, R.; Poulsen, G.; Branca, F.; Bagger Jørgensen, R. Genetic diversity and population structure of leafy kale and Brassica rupestris Raf. in south Italy. Hereditas 2014, 151, 145–158. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).