Assessment of the Genetic Diversity and Population Structure of the Peruvian Andean Legume, Tarwi (Lupinus mutabilis), with High Quality SNPs
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Material and DNA Extraction
2.2. Genotyping by Sequencing Data
2.3. Phylogenetic Analysis
2.4. Population Structure and Genetic Diversity
3. Results
3.1. Sequencing and Distribution of SNPs
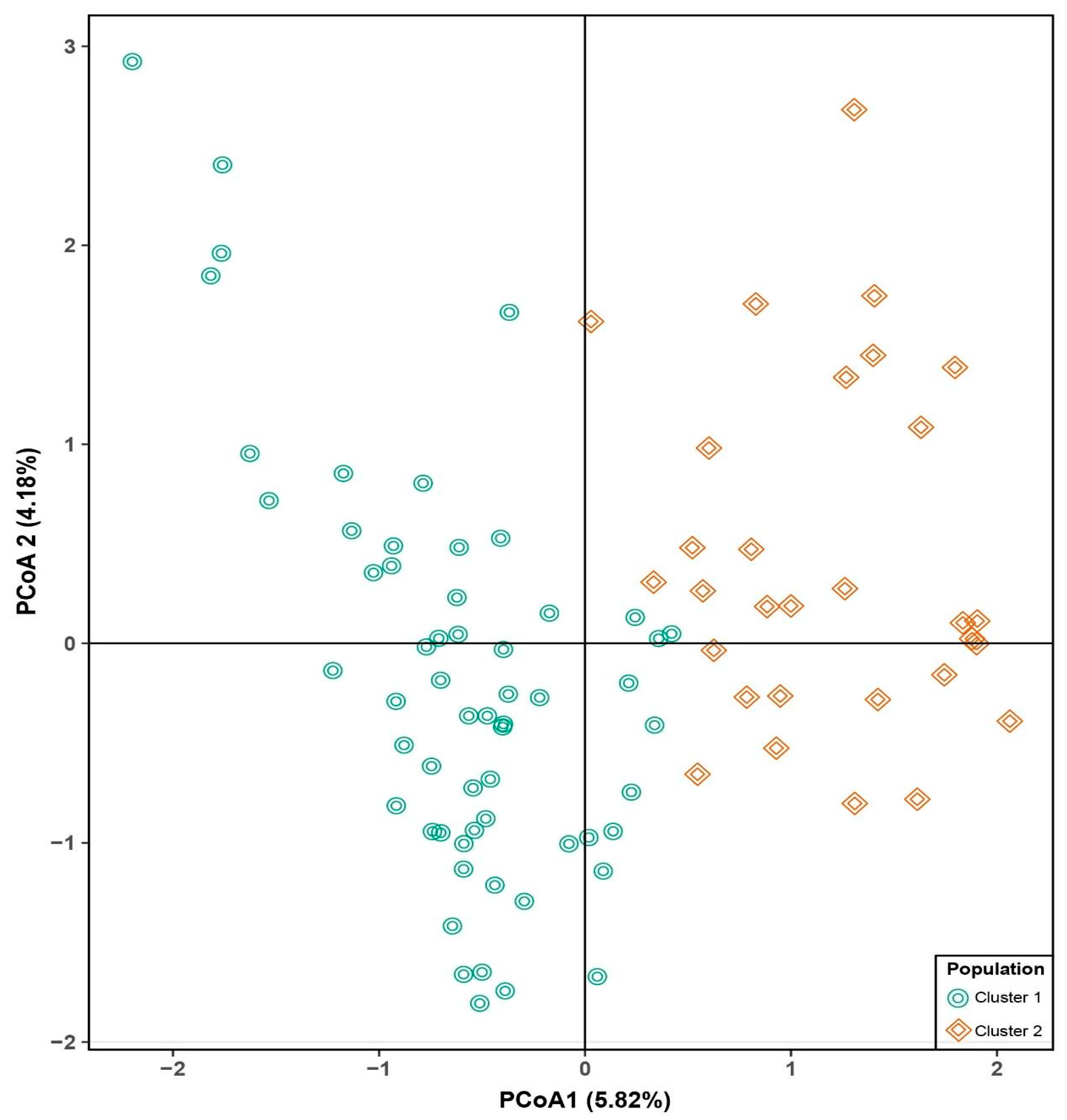
3.2. Population Structure and Genetic Relationships
3.3. Genetic Diversity of the Tarwi Collection
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Caligari, P.D.S.; Römer, P.; Rahim, M.A.; Huyghe, C.; Neves-Martins, J.; Sawicka-Sienkiewicz, E.J. The Potential of Lupinus mutabilis as a crop. In Linking Research and Marketing Opportunities for Pulses in the 21st Century, Proceedings of the Third International Food Legumes Research Conference, Adelaide, Australia, 22–26 September 1997; Springer: Dordrecht, The Netherlands, 2000; pp. 569–573. [Google Scholar] [CrossRef]
- Atchison, G.W.; Nevado, B.; Eastwood, R.J.; Contreras-Ortiz, N.; Reynel, C.; Madriñán, S.; Filatov, D.A.; Hughes, C.E. Lost crops of the incas: Origins of domestication of the Andean pulse crop Tarwi, Lupinus mutabilis. Am. J. Bot. 2016, 103, 1592–1606. [Google Scholar] [CrossRef] [PubMed]
- Neves Martins, J.M.; Talhinhas, P.; Bruno de Sousa, R. Yield and seed chemical composition of Lupinus mutabilis in Portugal. Rev. Ciências Agrárias 2016, 39, 518–525. [Google Scholar] [CrossRef]
- Carvajal-Larenas, F.E.; Linnemann, A.R.; Nout, M.J.R.; Koziol, M.; van Boekel, M.A.J.S. Lupinus mutabilis: Composition, Uses, Toxicology, and Debittering. Crit. Rev. Food Sci. Nutr. 2016, 56, 1454–1487. [Google Scholar] [CrossRef] [PubMed]
- Borek, S.; Ratajczak, W.; Ratajczak, L. Regulation of storage lipid metabolism in developing and germinating lupin (Lupinus spp.) seeds. Acta Physiol. Plant. 2015, 37, 119. [Google Scholar] [CrossRef]
- Guilengue, N.; Alves, S.; Talhinhas, P.; Neves-Martins, J. Genetic and genomic diversity in a tarwi (Lupinus mutabilis sweet) germplasm collection and adaptability to mediterranean climate conditions. Agronomy 2020, 10, 21. [Google Scholar] [CrossRef]
- Adomas, B.; Galek, R.; Gas-Smereka, M.; Helios, W.; Hurej, M.; Kotecki, A.; Kozak, M.; Malarz, W.; Okorski, A.; Agnieszka, P.; et al. Adaptation of the Andean lupin (Lupinus mutabilis Sweet) to natural conditions of south-western Poland; Wydawnictwo Uniwersytetu Przyrodniczego we Wrocławiu: Wroclaw, Poland, 2015; ISBN 9788377172353. [Google Scholar]
- Jacobsen, E.; Mujica, A. El tarwi (Lupinus mutabilis Sweet) y sus parientes silvestres. In Botánica Económica de los Andes Centrales; Universidad Mayor de San Andrés: La Paz, Bolivia, 2006; pp. 458–482. [Google Scholar] [CrossRef]
- Martínez Flores, L.A.; Ruivenkamp, G.; Jongerden, J. Fitomejoramiento y racionalidad social: Los efectos no intencionales de la liberación de una semilla de lupino (Lupinus mutabilis Sweet) en Ecuador. Antípoda. Rev. Antropol. Arqueol. 2016, 26, 71–91. [Google Scholar] [CrossRef]
- Camillo, M.F.; Pozzobon, M.T.; Schifino-Wittm, M.T. Chromosome numbers in South American andean spcecies of Lupinus (Leguminosae). Bonplandia 2006, 15, 113–119. [Google Scholar]
- Chirinos-Arias, M.C.; Jiménez, J.E.; Vilca-Machaca, L.S. Analysis of Genetic Variability among thirty accessions of Andean Lupin (Lupinus mutabilis Sweet) using ISSR molecular markers. Sci. Agropecu. 2015, 6, 17–30. [Google Scholar] [CrossRef]
- Ruiz-Gil, P.J.; Paramo-Ortíz, E.E.; De La Luna-Bonilla, Ó.Á. ¿Entrecruces entre Lupinus domesticados y silvestres? El tarwi del Perú P. Desde el Herb. CICY 2019, 11, 195–200. [Google Scholar]
- Kroc, M.; Koczyk, G.; Święcicki, W.; Kilian, A.; Nelson, M.N. New evidence of ancestral polyploidy in the Genistoid legume Lupinus angustifolius L. (narrow-leafed lupin). Theor. Appl. Genet. 2014, 127, 1237–1249. [Google Scholar] [CrossRef]
- Soltis, P.S.; Soltis, D.E. Plant Molecular Systematics: Macromolecular Approaches: Inferences of Phylogeny and Evolutionary Processes. Evol. Biol. 1995, 28, 139–194. [Google Scholar] [CrossRef]
- Melchinger, A.E.; Gumber, R.K. Overview of heterosis and heterotic groups in agronomic crops. Concepts Breed. Heterosis Crop. Plants 2015, 25, 29–44. [Google Scholar] [CrossRef]
- Elshire, R.J.; Glaubitz, J.C.; Sun, Q.; Poland, J.A.; Kawamoto, K.; Buckler, E.S.; Mitchell, S.E. A robust, simple genotyping-by-sequencing (GBS) approach for high diversity species. PLoS ONE 2011, 6, e19379. [Google Scholar] [CrossRef]
- Beebe, S.; Rengifo, E.; Gaitan, E.; Duque, M.C.; Tohme, J. Diversity and origin of Andean landraces of common bean. Crop. Sci. 2001, 41, 854–862. [Google Scholar] [CrossRef]
- Uitdewilligen, J.G.A.M.L.; Wolters, A.M.A.; D’hoop, B.B.; Borm, T.J.A.; Visser, R.G.F.; van Eck, H.J. A Next-Generation Sequencing Method for Genotyping-by-Sequencing of Highly Heterozygous Autotetraploid Potato. PLoS ONE 2013, 8, e62355. [Google Scholar] [CrossRef]
- Ramstein, G.P.; Lipka, A.E.; Lu, F.; Costich, D.E.; Cherney, J.H.; Buckler, E.S.; Casler, M.D. Genome-wide association study based on multiple imputation with low-depth sequencing data: Application to biofuel traits in reed canarygrass. G3 Genes Genomes Genet. 2015, 5, 891–909. [Google Scholar] [CrossRef]
- Wong, M.M.L.; Gujaria-Verma, N.; Ramsay, L.; Yuan, H.Y.; Caron, C.; Diapari, M.; Vandenberg, A.; Bett, K.E. Classification and characterization of species within the genus lens using genotyping-by-sequencing (GBS). PLoS ONE 2015, 10, e0122025. [Google Scholar] [CrossRef]
- Lu, F.; Romay, M.C.; Glaubitz, J.C.; Bradbury, P.J.; Elshire, R.J.; Wang, T.; Li, Y.; Li, Y.; Semagn, K.; Zhang, X.; et al. High-resolution genetic mapping of maize pan-genome sequence anchors. Nat. Commun. 2015, 6, 6914. [Google Scholar] [CrossRef]
- Liu, H.; Bayer, M.; Druka, A.; Russell, J.R.; Hackett, C.A.; Poland, J.; Ramsay, L.; Hedley, P.E.; Waugh, R. An evaluation of genotyping by sequencing (GBS) to map the Breviaristatum-e (ari-e) locus in cultivated barley. BMC Genom. 2014, 15, 104. [Google Scholar] [CrossRef]
- Spindel, J.; Wright, M.; Chen, C.; Cobb, J.; Gage, J.; Harrington, S.; Lorieux, M.; Ahmadi, N.; McCouch, S. Bridging the genotyping gap: Using genotyping by sequencing (GBS) to add high-density SNP markers and new value to traditional bi-parental mapping and breeding populations. Theor. Appl. Genet. 2013, 126, 2699–2716. [Google Scholar] [CrossRef]
- Jarquín, D.; Kocak, K.; Posadas, L.; Hyma, K.; Jedlicka, J.; Graef, G.; Lorenz, A. Genotyping by sequencing for genomic prediction in a soybean breeding population. BMC Genom. 2014, 15, 740. [Google Scholar] [CrossRef] [PubMed]
- Lipka, A.E.; Lu, F.; Cherney, J.H.; Buckler, E.S.; Casler, M.D.; Costich, D.E. Accelerating the switchgrass (Panicum virgatum L.) breeding cycle using genomic selection approaches. PLoS ONE 2014, 9, e112227. [Google Scholar] [CrossRef]
- Poland, J.; Endelman, J.; Dawson, J.; Rutkoski, J.; Wu, S.; Manes, Y.; Dreisigacker, S.; Crossa, J.; Sánchez-Villeda, H.; Sorrells, M.; et al. Genomic Selection in Wheat Breeding using Genotyping-by-Sequencing. Plant Genome 2012, 5, 103–113. [Google Scholar] [CrossRef]
- Doyle, J.J.; Doyle, J.L. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem. Bull. 1987, 19, 11–15. [Google Scholar]
- Bradbury, P.J.; Zhang, Z.; Kroon, D.E.; Casstevens, T.M.; Ramdoss, Y.; Buckler, E.S. TASSEL: Software for association mapping of complex traits in diverse samples. Bioinformatics 2007, 23, 2633–2635. [Google Scholar] [CrossRef] [PubMed]
- Glaubitz, J.C.; Casstevens, T.M.; Lu, F.; Harriman, J.; Elshire, R.J.; Sun, Q.; Buckler, E.S. TASSEL-GBS: A high capacity genotyping by sequencing analysis pipeline. PLoS ONE 2014, 9, e90346. [Google Scholar] [CrossRef]
- Hane, J.K.; Ming, Y.; Kamphuis, L.G.; Nelson, M.N.; Garg, G.; Atkins, C.A.; Bayer, P.E.; Bravo, A.; Bringans, S.; Cannon, S.; et al. A comprehensive draft genome sequence for lupin (Lupinus angustifolius), an emerging health food: Insights into plant–microbe interactions and legume evolution. Plant Biotechnol. J. 2017, 15, 318–330. [Google Scholar] [CrossRef]
- Li, H.; Durbin, R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 2009, 25, 1754–1760. [Google Scholar] [CrossRef]
- Danecek, P.; Auton, A.; Abecasis, G.; Albers, C.A.; Banks, E.; DePristo, M.A.; Handsaker, R.E.; Lunter, G.; Marth, G.T.; Sherry, S.T.; et al. The variant call format and VCFtools. Bioinformatics 2011, 27, 2156–2158. [Google Scholar] [CrossRef]
- Zheng, X.; Levine, D.; Shen, J.; Gogarten, S.M.; Laurie, C.; Weir, B.S. A high-performance computing toolset for relatedness and principal component analysis of SNP data. Bioinformatics 2012, 28, 3326–3328. [Google Scholar] [CrossRef]
- Team, R.C. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2020; p. 326864. [Google Scholar]
- Prevosti, A.; Ocaña, J.; Alonso, G. Distances between populations of Drosophila subobscura, based on chromosome arrangement frequencies. Theor. Appl. Genet. 1975, 45, 231–241. [Google Scholar] [CrossRef]
- Kamvar, Z.N.; Tabima, J.F.; Grünwald, N.J. Poppr: An R package for genetic analysis of populations with clonal, partially clonal, and/or sexual reproduction. PeerJ 2014, 2014, e281. [Google Scholar] [CrossRef]
- Dray, S.; Dufour, A.B. The ade4 package: Implementing the duality diagram for ecologists. J. Stat. Softw. 2007, 22, 1–20. [Google Scholar] [CrossRef]
- Purcell, S.; Neale, B.; Todd-Brown, K.; Thomas, L.; Ferreira, M.A.R.; Bender, D.; Maller, J.; Sklar, P.; De Bakker, P.I.W.; Daly, M.J.; et al. PLINK: A tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 2007, 81, 559–575. [Google Scholar] [CrossRef] [PubMed]
- Lischer, H.E.L.; Excoffier, L. PGDSpider: An automated data conversion tool for connecting population genetics and genomics programs. Bioinformatics 2012, 28, 298–299. [Google Scholar] [CrossRef] [PubMed]
- Pritchard, J.K.; Stephens, M.; Donnelly, P. Inference of population structure using multilocus genotype data. Genetics 2000, 155, 945–959. [Google Scholar] [CrossRef]
- Earl, D.A.; vonHoldt, B.M. STRUCTURE HARVESTER: A website and program for visualizing STRUCTURE output and implementing the Evanno method. Conserv. Genet. Resour. 2012, 4, 359–361. [Google Scholar] [CrossRef]
- Evanno, G.; Regnaut, S.; Goudet, J. Detecting the number of clusters of individuals using the software STRUCTURE: A simulation study. Mol. Ecol. 2005, 14, 2611–2620. [Google Scholar] [CrossRef]
- Francis, R.M. pophelper: An R package and web app to analyse and visualize population structure. Mol. Ecol. Resour. 2017, 17, 27–32. [Google Scholar] [CrossRef] [PubMed]
- Arbizu, C.I.; Ellison, S.L.; Senalik, D.; Simon, P.W.; Spooner, D.M. Genotyping-by-sequencing provides the discriminating power to investigate the subspecies of Daucus carota (Apiaceae). BMC Evol. Biol. 2016, 16, 234. [Google Scholar] [CrossRef] [PubMed]
- Martínez-Flores, F.; Crespo, M.B.; Simon, P.W.; Ruess, H.; Reitsma, K.; Geoffriau, E.; Allender, C.; Mezghani, N.; Spooner, D.M. Subspecies Variation of Daucus carota Coastal (“Gummifer”) Morphotypes (Apiaceae) Using Genotyping-by-Sequencing. Syst. Bot. 2020, 45, 688–702. [Google Scholar] [CrossRef]
- Govindaraj, M.; Vetriventhan, M.; Srinivasan, M. Importance of genetic diversity assessment in crop plants and its recent advances: An overview of its analytical perspectives. Genet. Res. Int. 2015, 2015, 688–702. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Zhang, X.; Li, H.; Zheng, H.; Zhang, J.; Olsen, M.S.; Varshney, R.K.; Prasanna, B.M.; Qian, Q. Smart breeding driven by big data, artificial intelligence, and integrated genomic-enviromic prediction. Mol. Plant 2022, 15, 1664–1695. [Google Scholar] [CrossRef]
- Thomson, M.J.; Septiningsih, E.M.; Suwardjo, F.; Santoso, T.J.; Silitonga, T.S.; McCouch, S.R. Genetic diversity analysis of traditional and improved Indonesian rice (Oryza sativa L.) germplasm using microsatellite markers. Theor. Appl. Genet. 2007, 114, 559–568. [Google Scholar] [CrossRef]
- Mammadov, J.; Aggarwal, R.; Buyyarapu, R.; Kumpatla, S. SNP markers and their impact on plant breeding. Int. J. Plant Genomics 2012, 2012, 728398. [Google Scholar] [CrossRef]
- Persson, K.; Von Bothmer, R. Genetic diversity amongst landraces of rye (Secale cereale L.) from northern Europe. Hereditas 2002, 136, 29–38. [Google Scholar] [CrossRef]
- Martin-Sanz, A.; Caminero, C.; Jing, R.; Flavell, A.J.; Perez de la Vega, M. Genetic diversity among Spanish pea (Pisum sativum L.) landraces, pea cultivars and the World Pisum sp. core collection assessed by retrotransposon-based insertion polymorphisms (RBIPs). Spanish J. Agric. Res. 2011, 9, 166. [Google Scholar] [CrossRef]
- Bracco, M.; Lia, V.V.; Hernández, J.C.; Poggio, L.; Gottlieb, A.M. Genetic diversity of maize landraces from lowland and highland agro-ecosystems of Southern South America: Implications for the conservation of native resources. Ann. Appl. Biol. 2012, 160, 308–321. [Google Scholar] [CrossRef]
- Hour, A.L.; Hsieh, W.H.; Chang, S.H.; Wu, Y.P.; Chin, H.S.; Lin, Y.R. Genetic Diversity of Landraces and Improved Varieties of Rice (Oryza sativa L.) in Taiwan. Rice 2020, 13, 82. [Google Scholar] [CrossRef]
- Aesomnuk, W.; Ruengphayak, S.; Ruanjaichon, V.; Sreewongchai, T.; Malumpong, C.; Vanavichit, A.; Toojinda, T.; Wanchana, S.; Arikit, S. Estimation of the genetic diversity and population structure of thailand’s rice landraces using snp markers. Agronomy 2021, 11, 995. [Google Scholar] [CrossRef]
- Lorello, I.M.; Lampasona, S.C.G.; Peralta, I.E. Genetic diversity of squash landraces (Cucurbita maxima) collected in Andean valleys of Argentina. Rev. Fac. Ciencias Agrar. 2020, 52, 293–313. [Google Scholar]
- Özkan, G.; Haliloglu, K.; Türkoglu, A.; Özturk, H.I.; Elkoca, E.; Poczai, P. Determining Genetic Diversity and Population Structure of Common Bean (Phaseolus vulgaris L.) Landraces from Türkiye Using SSR Markers. Genes 2022, 13, 1410. [Google Scholar] [CrossRef] [PubMed]
- Tehseen, M.M.; Tonk, F.A.; Tosun, M.; Istipliler, D.; Amri, A.; Sansaloni, C.P.; Kurtulus, E.; Mubarik, M.S.; Nazari, K. Exploring the Genetic Diversity and Population Structure of Wheat Landrace Population Conserved at ICARDA Genebank. Front. Genet. 2022, 13, 900572. [Google Scholar] [CrossRef] [PubMed]
- Raman, R.; Cowley, R.B.; Raman, H.; Luckett, D.J. Analyses Using SSR and DArT Molecular Markers Reveal that Ethiopian Accessions of White Lupin (Lupinus albus L.) Represent a Unique Genepool. Open J. Genet. 2014, 04, 87–98. [Google Scholar] [CrossRef]
- El-Harty, E.; Ashrie, A.; Ammar, M.; Alghamdi, S. Genetic variation among egyptian white lupin (Lupinus albus L.) genotypes. Turk. J. Field Crops 2016, 21, 148–155. [Google Scholar] [CrossRef]
- Atnaf, M.; Yao, N.; Martina, K.; Dagne, K.; Wegary, D.; Tesfaye, K. Molecular genetic diversity and population structure of Ethiopian white lupin landraces: Implications for breeding and conservation. PLoS ONE 2017, 12, e0188696. [Google Scholar] [CrossRef]
- Ji, Y.; Liu, R.; Hu, J.; Huang, Y.; Wang, D.; Li, G.; Rahman, M.M.; Zhang, H.; Wang, C.; Li, M.; et al. Genetic diversity analysis for narrow-leafed lupin (Lupinus angustifolius L.) by SSR markers. Mol. Biol. Rep. 2020, 47, 5215–5224. [Google Scholar] [CrossRef]
- Skorupski, J.; Szenejko, M.; Gruba-Tabaka, M.; Śmietana, P.; Panicz, R. Inferring population structure and genetic diversity of the invasive alien nootka lupin in Iceland. Polar Res. 2021, 40, 1–13. [Google Scholar] [CrossRef]
- Eltaher, S.; Sallam, A.; Belamkar, V.; Emara, H.A.; Nower, A.A.; Salem, K.F.M.; Poland, J.; Baenziger, P.S. Genetic diversity and population structure of F3:6 Nebraska Winter wheat genotypes using genotyping-by-sequencing. Front. Genet. 2018, 9, 76. [Google Scholar] [CrossRef]
- Huaringa, A.; De la Cruz, N.; Camarena, F.; Caycho, P.; Mostacero, E.; Miguel, P. Phenotypic evaluation and preliminary yield of lupin ecotype (Lupinus mutabilis sweet) grown in Carhuaz, Ancash. Ancash. In Abstract Book XV ILC 2019: Developing Lupin Crop into a Modern and Sustainable Food and Feed Source; Fundación PROINPA: Quillacollo, Bolivia, 2019. [Google Scholar]
- Mujica, A.; Moscoso, G. La planta del tarwi. In Lupinus mutabilis (Tarwi): Leguminosa Andina con Gran Potencial Industrial; Zavaleta, A.I., Ed.; Universidad Nacional Mayor de San Marcos, Universidad del Perú, Decana de América Fondo Editorial: Lima, Peru, 2018; ISBN 9789972466205. [Google Scholar]
- MINAGRI. Manejo Agronómico. Prácticas de Conservación de Suelo, Producción, Comercialización y Perspectivas de Granos Andinos; MINAGRI: Lima, Peru, 2018; p. 86. [Google Scholar]
- Li, B.; Suzuki, J.I.; Hara, T. Latitudinal variation in plant size and relative growth rate in Arabidopsis thaliana. Oecologia 1998, 115, 293–301. [Google Scholar] [CrossRef]
- Zhou, L.; Yu, H.; Yang, K.; Chen, L.; Yin, W.; Ding, J. Latitudinal and Longitudinal Trends of Seed Traits Indicate Adaptive Strategies of an Invasive Plant. Front. Plant Sci. 2021, 12, 657813. [Google Scholar] [CrossRef] [PubMed]
- Fijen, T.P.M.; Morra, E.; Kleijn, D. Pollination increases white and narrow-leaved lupin protein yields but not all crop visitors contribute to pollination. Agric. Ecosyst. Environ. 2021, 313, 107386. [Google Scholar] [CrossRef]
- Kelemu, S.; Shiferaw, E.; Hailu, F. Genetic diversity and population structure of Lupinus albus (l.) from the Amhara region of ethiopia using seed storage protein markers. J. Agric. Sci. 2022, 67, 1–11. [Google Scholar] [CrossRef]
- Nadathur, S.R.; Wanasundara, J.P.D.; Scanlin, L. Proteins in the Diet: Challenges in Feeding the Global Population; Elsevier Inc.: Amsterdam, The Netherlands, 2017; ISBN 9780128027769. [Google Scholar]
- Shrestha, S.; van’t Hag, L.; Haritos, V.S.; Dhital, S. Lupin proteins: Structure, isolation and application. Trends Food Sci. Technol. 2021, 116, 928–939. [Google Scholar] [CrossRef]
- Vishnyakova, M.A.; Kushnareva, A.V.; Shelenga, T.V.; Egorova, G.P. Alkaloids of narrow-leaved lupine as a factor determining alternative ways of the crop’s utilization and breeding. Vavilovskii Zhurnal Genet. Sel. 2020, 24, 625–635. [Google Scholar] [CrossRef] [PubMed]
- Gulisano, A.; Alves, S.; Rodriguez, D.; Murillo, A.; van Dinter, B.J.; Torres, A.F.; Gordillo-Romero, M.; de Lourdes Torres, M.; Neves-Martins, J.; Paulo, M.J.; et al. Diversity and Agronomic Performance of Lupinus mutabilis Germplasm in European and Andean Environments. Front. Plant Sci. 2022, 13, 903661. [Google Scholar] [CrossRef] [PubMed]
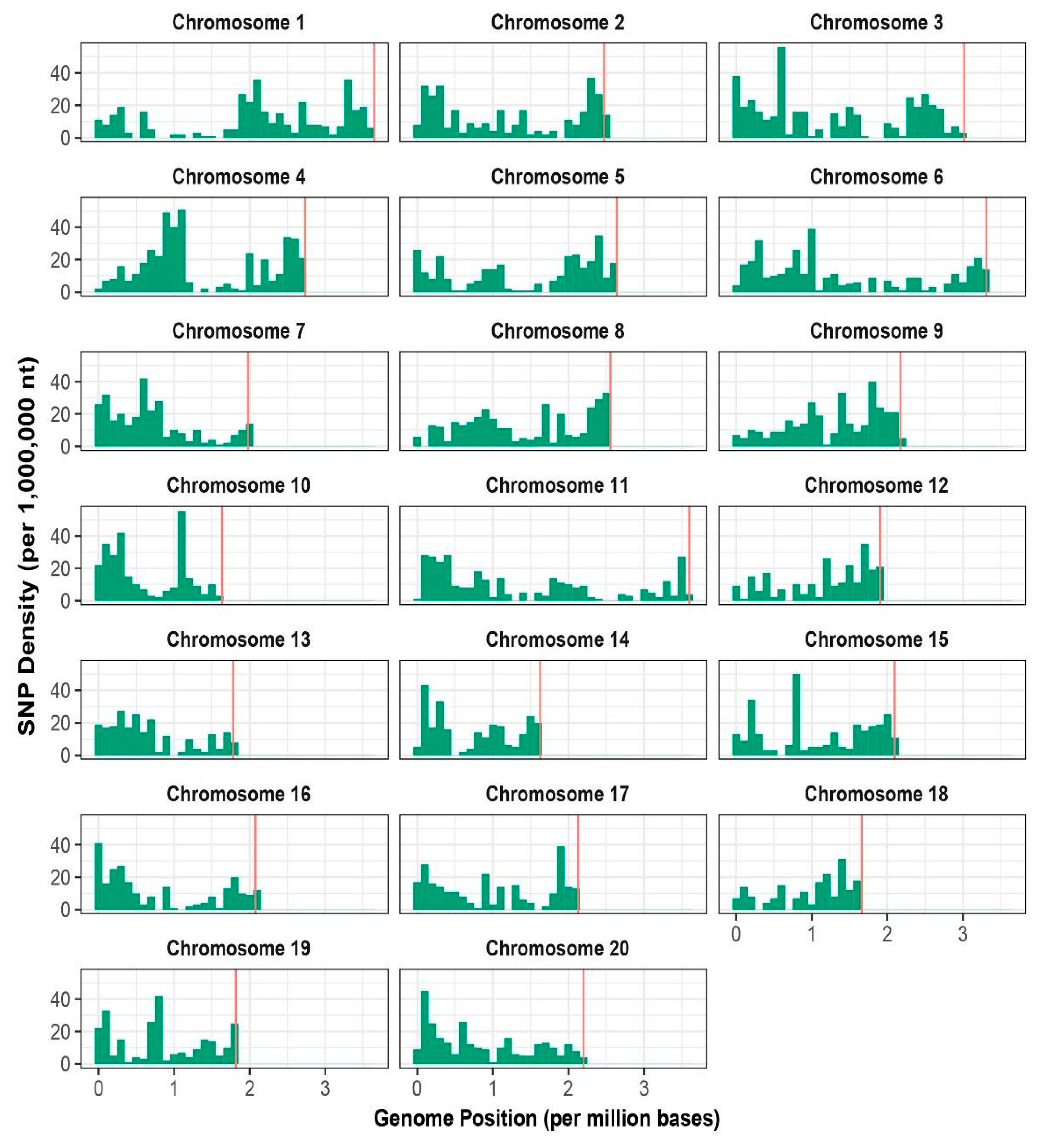


| Chromosomes | No. of SNPs | % SNPs | Total Length (Mb) | Density (Kb) |
|---|---|---|---|---|
| 1 | 363 | 6.13 | 36.46 | 100.43 |
| 2 | 322 | 5.44 | 24.70 | 76.70 |
| 3 | 410 | 6.92 | 30.15 | 73.54 |
| 4 | 430 | 7.26 | 23.77 | 63.57 |
| 5 | 303 | 5.12 | 26.38 | 87.05 |
| 6 | 344 | 5.81 | 33.11 | 96.25 |
| 7 | 294 | 4.96 | 19.78 | 67.29 |
| 8 | 330 | 5.57 | 25.52 | 77.34 |
| 9 | 331 | 5.59 | 21.75 | 65.72 |
| 10 | 273 | 4.61 | 16.34 | 59.86 |
| 11 | 319 | 5.39 | 35.96 | 112.74 |
| 12 | 237 | 4.00 | 19.07 | 80.45 |
| 13 | 230 | 3.88 | 17.82 | 77.48 |
| 14 | 250 | 4.22 | 16.25 | 65.01 |
| 15 | 280 | 4.73 | 20.96 | 74.87 |
| 16 | 244 | 4.12 | 20.79 | 85.19 |
| 17 | 252 | 4.26 | 21.30 | 84.52 |
| 18 | 184 | 3.11 | 16.59 | 90.15 |
| 19 | 248 | 4.19 | 18.16 | 73.23 |
| 20 | 278 | 4.69 | 21.99 | 79.10 |
| Department (Region) | Cluster 1 | Cluster 2 |
|---|---|---|
| Ancash (north) | 1 | 13 |
| Apurimac (south) | 3 | 1 |
| Cajamarca (north) | 1 | 2 |
| Cusco (south) | 32 | 1 |
| Huancavelica (center) | 3 | 1 |
| Huanuco (center) | 1 | 2 |
| Junin (center) | 5 | 2 |
| La Libertad (north) | 0 | 5 |
| Puno (south) | 13 | 3 |
| Total | 59 | 30 |
| Cluster | Number of Accessions | Na | AR | Ho | He | H | Fis | Fst |
|---|---|---|---|---|---|---|---|---|
| 1 | 59 | 2 | 1.997 | 0.641 | 0.421 | 4.08 | −0.524 | |
| 2 | 30 | 1.999 | 1.998 | 0.657 | 0.433 | 3.4 | −0.518 | |
| Mean | 1.999 | 1.999 | 0.649 | 0.427 | 3.74 | −0.521 | 0.019 | |
| Source of Variation | df | SS | MS | Est. Var. | % |
|---|---|---|---|---|---|
| Between clusters | 1 | 4960.23 | 4960.23 | 95.48 | 7.59 |
| Within clusters | 87 | 101,150.54 | 1162.65 | 1162.65 | 92.41 |
| Total | 88 | 106,110.77 | 1205.80 | 1258.13 | 100 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Huaringa-Joaquin, A.; Saldaña, C.L.; Saravia, D.; García-Bendezú, S.; Rodriguez-Grados, P.; Salazar, W.; Camarena, F.; Injante, P.; Arbizu, C.I. Assessment of the Genetic Diversity and Population Structure of the Peruvian Andean Legume, Tarwi (Lupinus mutabilis), with High Quality SNPs. Diversity 2023, 15, 437. https://doi.org/10.3390/d15030437
Huaringa-Joaquin A, Saldaña CL, Saravia D, García-Bendezú S, Rodriguez-Grados P, Salazar W, Camarena F, Injante P, Arbizu CI. Assessment of the Genetic Diversity and Population Structure of the Peruvian Andean Legume, Tarwi (Lupinus mutabilis), with High Quality SNPs. Diversity. 2023; 15(3):437. https://doi.org/10.3390/d15030437
Chicago/Turabian StyleHuaringa-Joaquin, Amelia, Carla L. Saldaña, David Saravia, Sady García-Bendezú, Pedro Rodriguez-Grados, Wilian Salazar, Felix Camarena, Pedro Injante, and Carlos I. Arbizu. 2023. "Assessment of the Genetic Diversity and Population Structure of the Peruvian Andean Legume, Tarwi (Lupinus mutabilis), with High Quality SNPs" Diversity 15, no. 3: 437. https://doi.org/10.3390/d15030437
APA StyleHuaringa-Joaquin, A., Saldaña, C. L., Saravia, D., García-Bendezú, S., Rodriguez-Grados, P., Salazar, W., Camarena, F., Injante, P., & Arbizu, C. I. (2023). Assessment of the Genetic Diversity and Population Structure of the Peruvian Andean Legume, Tarwi (Lupinus mutabilis), with High Quality SNPs. Diversity, 15(3), 437. https://doi.org/10.3390/d15030437









