Abstract
Indosasa lipoensis, an ornamental garden plant, belongs to the Indosasa genus of the subfamily Bambooaceae within Poaceae. Indosasa lipoensis is endangered and requires protection owing to its relatively narrow distribution area. Chloroplast (cp) genome offers a novel awareness of the evolutionary and genetic variation of higher plants. Herein, we assembled and elucidated the complete cp genome of I. lipoensis, and compared it with four previously published cp genomes from this genus. The I. lipoensis cp genome was 139,655 bp in size, with a typical quadripartite structure, encompassing a large single-copy region (LSC, 83,256 bp), a small single-copy region (SSC, 12,809 bp), and a pair of inverted repeat regions (IR, 21,795 bp). The cp genome consisted of 130 genes with 84 protein-coding genes (CDS), 38 tRNA genes, and 8 rRNA genes. The plastomes were highly conservative, compared to other bamboo species, and exhibited similar patterns of codon usage, number of repeat sequences, and expansion and contraction of the IR boundary. Five hypervariable hotspots were identified as potential DNA barcodes, namely rbcL, petA, petB, trnL-UAG, and ndhE-ndhI, respectively. Phylogenetic analysis based on the complete cp genomes revealed, with high resolution, that I. lipoensis and I. gigantea were most closely related. Overall, these results provided valuable characterization for the future conservation, genetic evaluation, and the breeding of I. lipoensis.
1. Introduction
Bamboos are critical members of the subfamily Bambooaceae of the family Poaceae. Bamboos are the most primitive members of the Poaceae, and it contains the most diverse group of grasses. It has the characteristics of reduced flowering and fruiting, complex and diverse root systems, and a high degree of stem lignification [1]. According to the authoritative statistics of the World Checklist of Bamboo published in 2017, there are 88 genera and 1642 species within the Bambooaceae, with primary distributions in tropical, subtropical, and warm temperate regions [2]. China, a distribution center, and the origin of the worldwide bamboo population, has approximately 39 genera and 500 species of bamboo. It is also the country with the most available bamboo resources, the most widely distributed bamboo forest area, and the largest bamboo production [3]. Bambusoideae plants have a variety of uses, including animal forage, human consumption, architectural, and ornamental purposes [4,5].
The Indosasa genus is among the smaller genera of Bambusoideae. Overall, 15 species of Indosasa have been identified, 13 of which are endemic to China, and are found in the Guizhou, Yunnan, Guangxi, and Hunan Provinces [6,7]. Traditionally, the Indosasa is considered to be closely related, and their morphological characteristics are very similar. The main difference between them lies in the floral structure [7]. Certain species of the Indosasa genus can be used to prepare scaffolding and bamboo utensils, and their shoots are edible.
Indosasa lipoensis C. D. Chu et K. M. Lan is an endemic plant from the Libo County in the Guizhou Province, Southwest China, and it grows to an altitude of about 400–700 m under broad-leaved forest trees or in hilly forests [8]. Indosasa lipoensis is an ornamental garden plant whose shoots are edible, and whose bamboo timber can be used for hedging and utensil preparation. This species has been subjected to severe damage leading to significant reduction in its populations [9]. In 2013, Indosasa lipoensis was recorded as endangered in the Redlist of China’s Biodiversity (http://www.iplant.cn/rep/protlist/4, accessed on 25 December 2022). This listing was announced due to its narrow distribution area, scarce species populations, and severe human disturbance.
In angiosperms, the chloroplast is a critical organelle, that contains its own genetic material, which primarily encodes proteins related to photosynthesis [10]. The chloroplast genomes are usually a circular structure and the gene contents and order are often highly conserved among these higher plants. However, the cp genome can also undergo some specific mutations, such as a loss of IR region, the loss of genes, gene duplication and rearrangement. Chloroplast serves an essential function in multiple biochemical networks, and it has great significance in plant physiology and development due to rich information sites and DNA barcodes [11,12]. The chloroplast (cp) genome is distinct from the nuclear genome and possesses semi-self-help genetic features [13]. Its genome is 75–250 kb in size, and exhibits a typical quadripartite structure containing a large single-copy (LSC) region, a small single-copy (SSC) region and a pair of inverted repeats (IRs) region [14,15], with protein-coding genes (CDS) ranging from 110–130 [16,17]. The cp genomes are especially useful in the study of evolution, plant taxonomy, and biogeographic inference due to its relatively conserved structures [18]. Previously, scholars had hardly used molecular data to classify and phylogenetic studies of Indosasa and its related genera. To date, only four chloroplast genome data have been published in this genus, namely Indosasa gigantea, I. sinica, I. crassiflora, and I. shibataeoides, respectively. The molecular phylogenetic studies of Indosasa and comparative analysis of chloroplast genomes are lacking.
At present, multiple investigations reported on the endemic and endangered bamboo species found in the Guizhou Province, such as, Ampelocalamus scandens, Chimonobambusa lactistriata, and Indocalamus hirsutissimus. However, studies on the cp genome are extremely scarce, and no such study exists on the cp genome of I. lipoensis [19,20,21]. Herein, we assembled, annotated, and compared for the first time the cp genome of I. lipoensis with published cp genomes of other Indosasa species, including chloroplast genome characteristics, comparative genomic, codon usage, repeat sequences, selective pressure, and phylogenetic relationship. Our goals were to: (1) display the characteristic of the Indosasa plastomes; (2) investigate a wealth of information sites in this genus; and (3) elucidate their phylogenetic relationships. Our chloroplast genomic data will provide useful information and resources for the future genetics analyses of I. lipoensis, and contribute to the studies on the phylogenetic and evolutionary relationships of the Indosasa genus.
2. Materials and Methods
2.1. Plant Material, DNA Extraction and Genome Sequencing
Fresh young I. lipoensis leaves were obtained from the Maolan National Nature Reserve, Libo, Guizhou, China (N 25°26′54.78″, E 107°53′12.40″, 550 m), followed by drying in silica gel, then storage at The Natural Museum of Guizhou University (accession number: GB65). The plant material collection followed the guidelines of the Maolan National Nature Reserve. A modified CTAB technique was employed for total genomic DNA extraction from the silica gel-dried leaves. [22]. DNA qualification and quantification were performed via 1% agarose gel electrophoresis and a NanoDrop spectrophotometer, respectively. The DNA sample was dispatched to BMK-Beijing (Beijing, China) for library construction and sequencing on the Illumina HiSeq2500 platform. Following 2 × 150 bp pair-end raw reads with Q30 (Phred quality score ≥ 30) was obtained. Low-quality data and adaptors filtration, clean date was obtained.
2.2. Genome Assembly and Annotation
Using the raw data, we assembled the entire cp genome in the GetOrganelle v1.6.2a software [23]. The chloroplast genome was visualized and urther validating using software Bandage. Moreover, employing the complete I. gigantea (MN917206) sequence as a reference, the plastome sequences were annotated using the script Plastid Genome Annotator (PGA) [24]. The annotation of tRNA was checked in tRNAscan-SE v.2.0, then a manual correction via the software Geneious 10.0.5 [25]. Lastly, the OGDRAW v1.3.1 [26] program was utilized to construct a map of the I. lipoensis cp genome for feature visualization. Accession: OP936084.
2.3. Repeated Sequence and Codon Usage Analysis
The CodonW1.4.4 program [27] was employed for the calculation of the relative synonymous codon usage values (RSCU) of shared CDS. When RSCU value > 1 indicates that the codon is preferred, RSCU value = 1 means that the codon is not preferred, and RSCU value < 1 shows that the codon usage is low. Subsequently, the web-based REPuter [28] was utilized to screen for repeat sequences using parameters as follows: minimal repeat size: 30, maximal computed repeats: 50, and hamming distance: 10. MISA [29], an online software, was employed for Simple Sequence Repeats (SSR) detection. The minimum cut-offs were: 10 repeat units for mononucleotide SSRs, 5 repeat units for dinucleotide SSRs, and 3 repeat units for trinucleotide, tetranucleotide, pentanucleotide, and hexanucleotide SSRs.
2.4. Comparative Analysis of the Entire Cp Genome
We conducted comparative analysis of the genomes of five species of Indosasa, one of which was obtained in this study, and the other four were downloaded by NCBI. Comparison of IR boundaries of the chloroplast genome from I. lipoensis chloroplast regions with four other Indosasa-related species via IRscope [30]. Subsequently, using the I. lipoensis genome as a reference, we visualized the divergent regions in the Shuffle-LAGAN mode in mVISTA [31]. Lastly, the 5-Indosasa species nucleotide diversity was computed using the sliding window analysis in DnaSP [32], employing a window length of 600 bp and a step size of 200 bp.
2.5. Phylogenetic Analysis
Next, to elucidate the phylogenetic status of I. lipoensis within Indosasa, we conducted phylogenetic analysis using 26 complete cp genomes, with 25 cp genomes (including 4 Ampelocalamus, 4 Bambusa, 4 Indosasa, 4 Phyllostachys, 3 Dendrocalamus, 2 Chimonobambusa, 1 Indocalamus, and 1 Bonia) obtained from NCBI, and Oryza rufipogon (NC_005973) and Lolium perenne (AM777385) employed as outgroups. Then, using the Maximum Likelihood (ML) and Bayesian Inference (BI) analyses of the entire cp genome sequences, we generated phylogenies. MAFFT [33] was employed for multiple sequence alignment of cp genomes. Moreover, poorly aligned locations and sites with excess divergence were eliminated from alignment using Gblocks v0.91 [34]. IQtree [35] performed ML analysis, and the bootstrap replicates parameter was adjusted to 1000. The BI analysis employed RAxML-HPC2 on XSEDE [36] as performed on the CIPRES Science Gateway using the ModelFinder for selecting the optimal model in PhyloSuite [37]. The parameter was run over five million generations, with every 1000 generations sampling. All other settings remained as default settings, and the first 25% of each run was eliminated as burn-in.
3. Results
3.1. Composition and Features of the Cp Genome
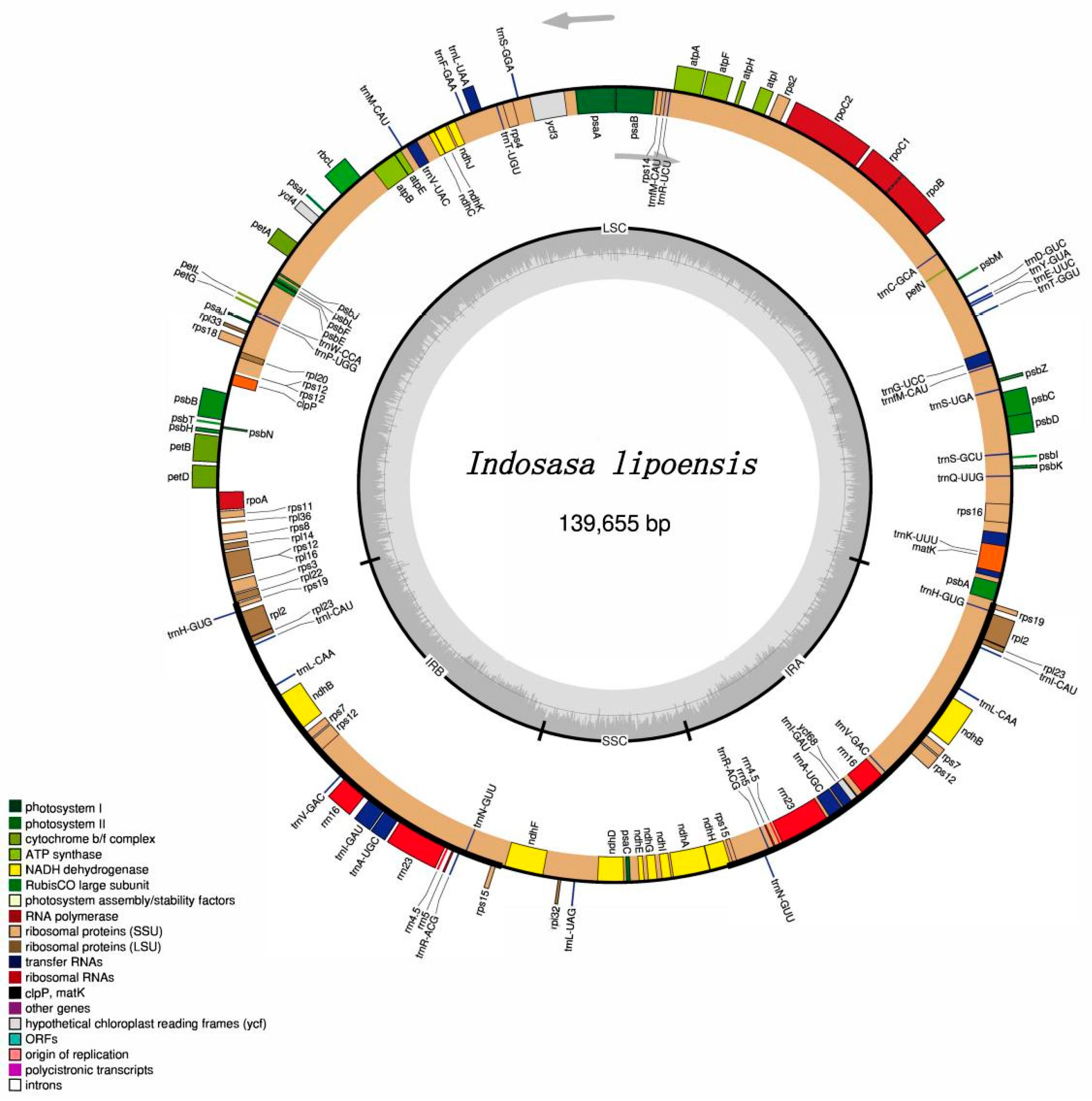
The next-generation sequencing produced raw data of 3G, including 1,026,443,883 clean reads, and 14,168,726 reads used for the assembly of the chloroplast (cp) genome of I. lipoensis. The entire I. lipoensis cp genome was 139,655 bp in length. The plastomes displayed a typical circular quadripartite structure, harboring a large single-copy region (LSC, 83,256 bp), a small single-copy region (SSC, 12,809 bp), and a pair of IRs (21,795 bp) (Figure 1). The total GC content was 38.9%, among which, GCs belonging to the IR, LSC, and SSC regions were 44.2%, 37.0%, and 33.3%, respectively. The cp genome of I. lipoensis encoded 130 genes, with 84 CDS, 38 tRNA genes, and 8 rRNA genes. Among the 130 genes, 77 fragments were associated with self-replication, 45 with photosynthesis, five with other genes (including maturase, envelop membrane protein, c-type cytochrome synthesis gene, translational initiation, protease), and one with unknown function (conserved open reading frames) (Table 1). Overall, we identified 16 genes with intron(s), among them, 14 harbored only one intron (trnA-UGC, rnI-GAU, trnG-UCC, trnL-UAA, trnV-UAC, rpsl16, petB, petD, atpF, ndhA, rps12, rpl2, ndhB, rpl16), while two genes, namely, rps12 and ycf3, harbored two introns. The rps12 gene was trans-spliced, and carried two replicas of the 3′ end in the IR region and two copies of the 5′ end in the LSC region.
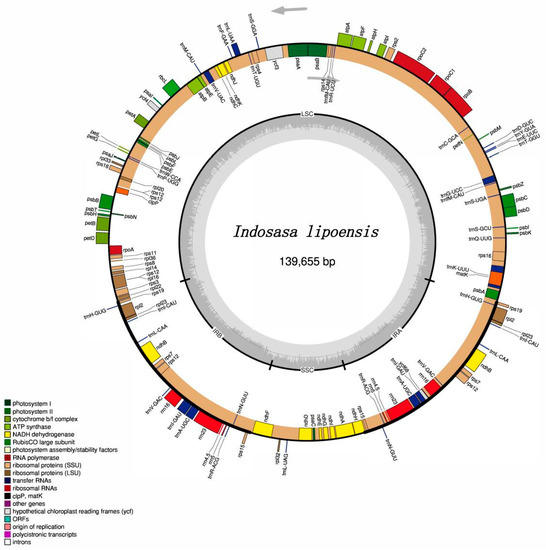
Figure 1.
The gene map of the cp genome of Indosasa lipoensis. Genes outside of the circle are transcribed in the counterclockwise direction, and those within the circle are transcribed in the clockwise direction.

Table 1.
Annotated list of genes identified within the cp genome of Indosasa lipoensis.
3.2. Chloroplast REPEATED SEQUENCES and SSRs
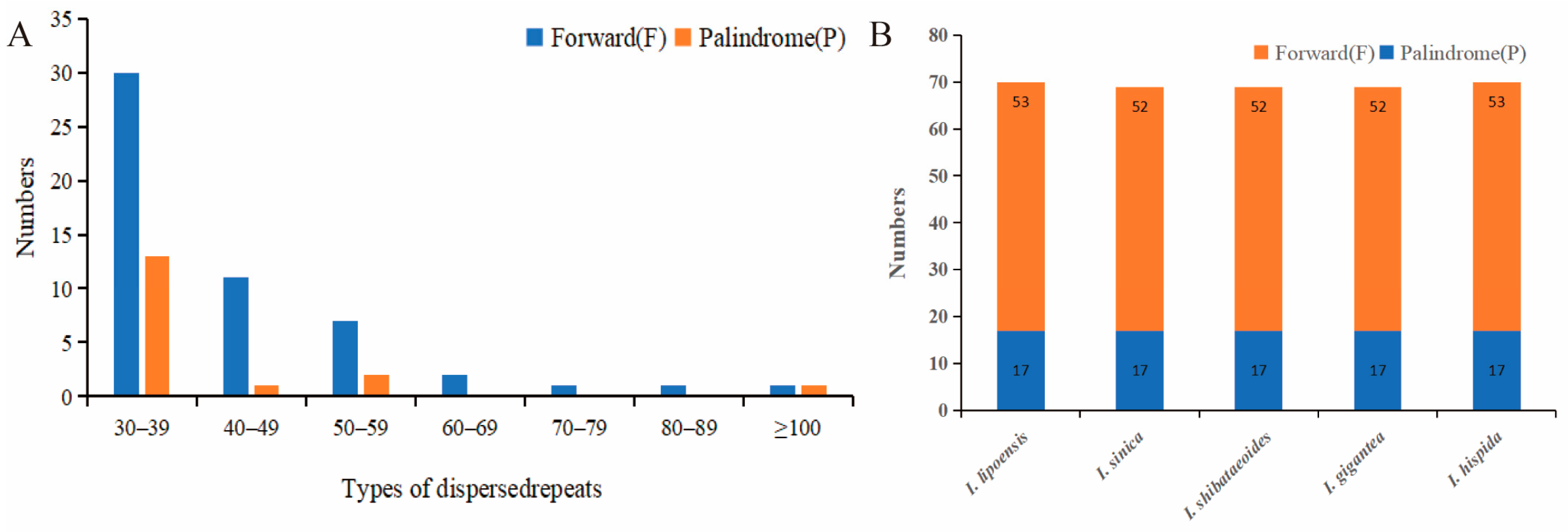
Overall, 70 repeats were identified in I. lipoensis, including 17 palindromic repeats, and 53 forward repeats, with no reverse and complement repeats. In these 70 sequences, 43, 12, 9, 2, 1, and 1 repeats ranged between 30–39 bp, 40–49 bp, 50–59 bp, 60–69 bp, 70–79 bp, and 80–89 bp, respectively. In addition, two were over 100 bp in length (Figure 2). Most repeats were between 30–39 bp (61.4%). Our repeat sequence analysis of five Indosasa genus species revealed 69–70 repeats, with only palindromic and forward repeats. Moreover, both I. lipoensis and I. hispida species contained of 70 repeats, one more than the other species.
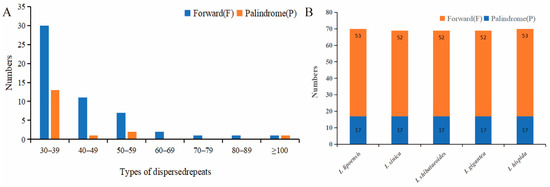
Figure 2.
Repetitive sequences in the cp genome of various species. (A) Quantification of the different types of long repeats in the cp genome of Indosasa lipoensis. (B) Repeat sequence information of the cp genome of the Indosasa genus species.
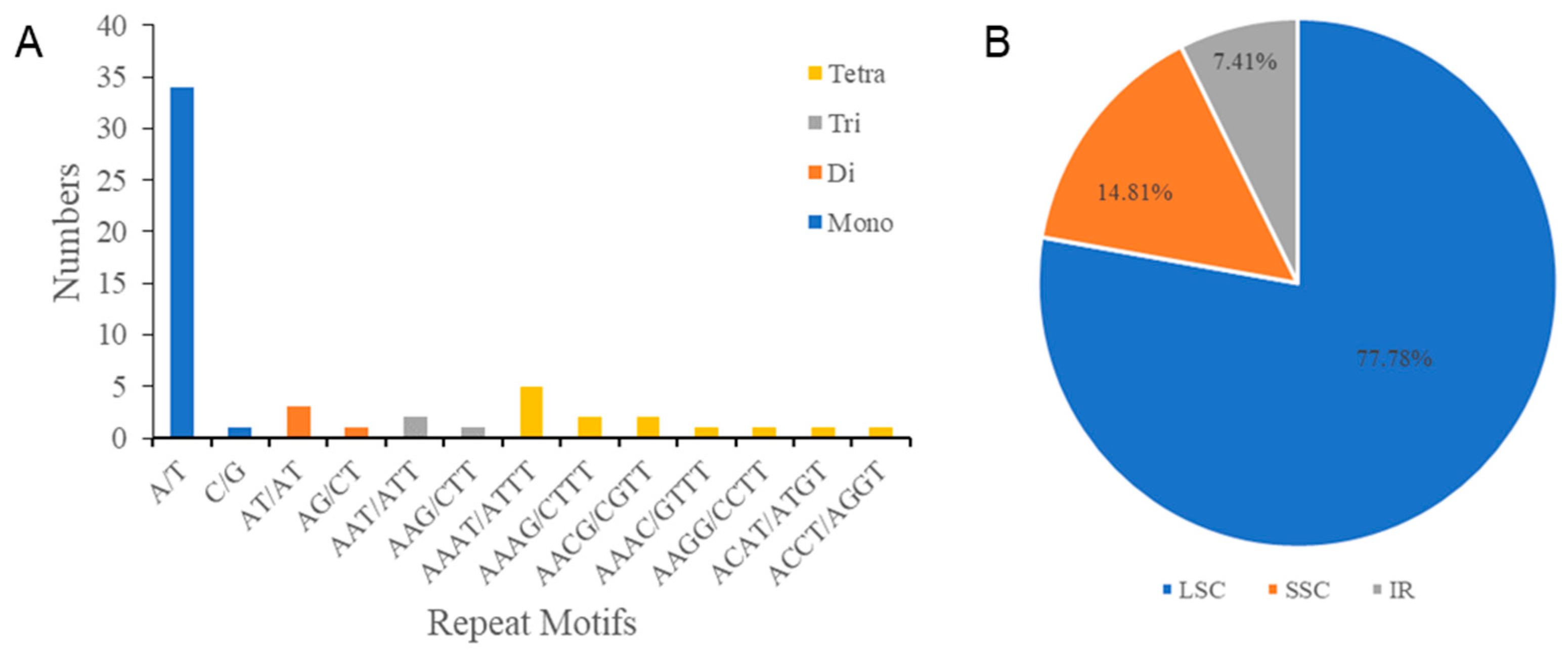
We also identified six categories of SSRs, namely, mononucleotide, dinucleotide, trinucleotide, tetranucleotide, pentanucleotide, and hexanucleotide repeats via MISA analysis of the cp genome of I. lipoensis (Figure 3A). Four categories of 54 SSRs were screened, among which, mononucleotide repeats were the most prevalent, making up 63% of all repeats. No pentanucleotide or hexanucleotide was present in I. lipoensis. Further investigation revealed that most SSRs were identified in the LSC region, with the SSC and IR regions, accounting for 77.78%, 14.81%, and 7.41%, respectively (Figure 3B).
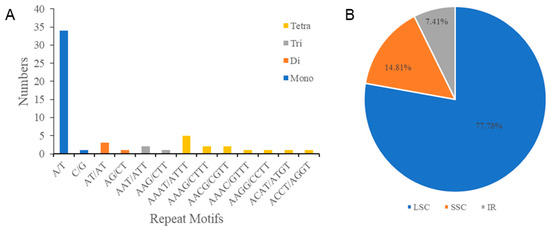
Figure 3.
SSRs in the cp genome of I. lipoensis. (A) Quantification of various categories of SSRs. (B) Percentages of repeats in the LSC, IR, and SSC locations.
3.3. Codon Usage Analyses
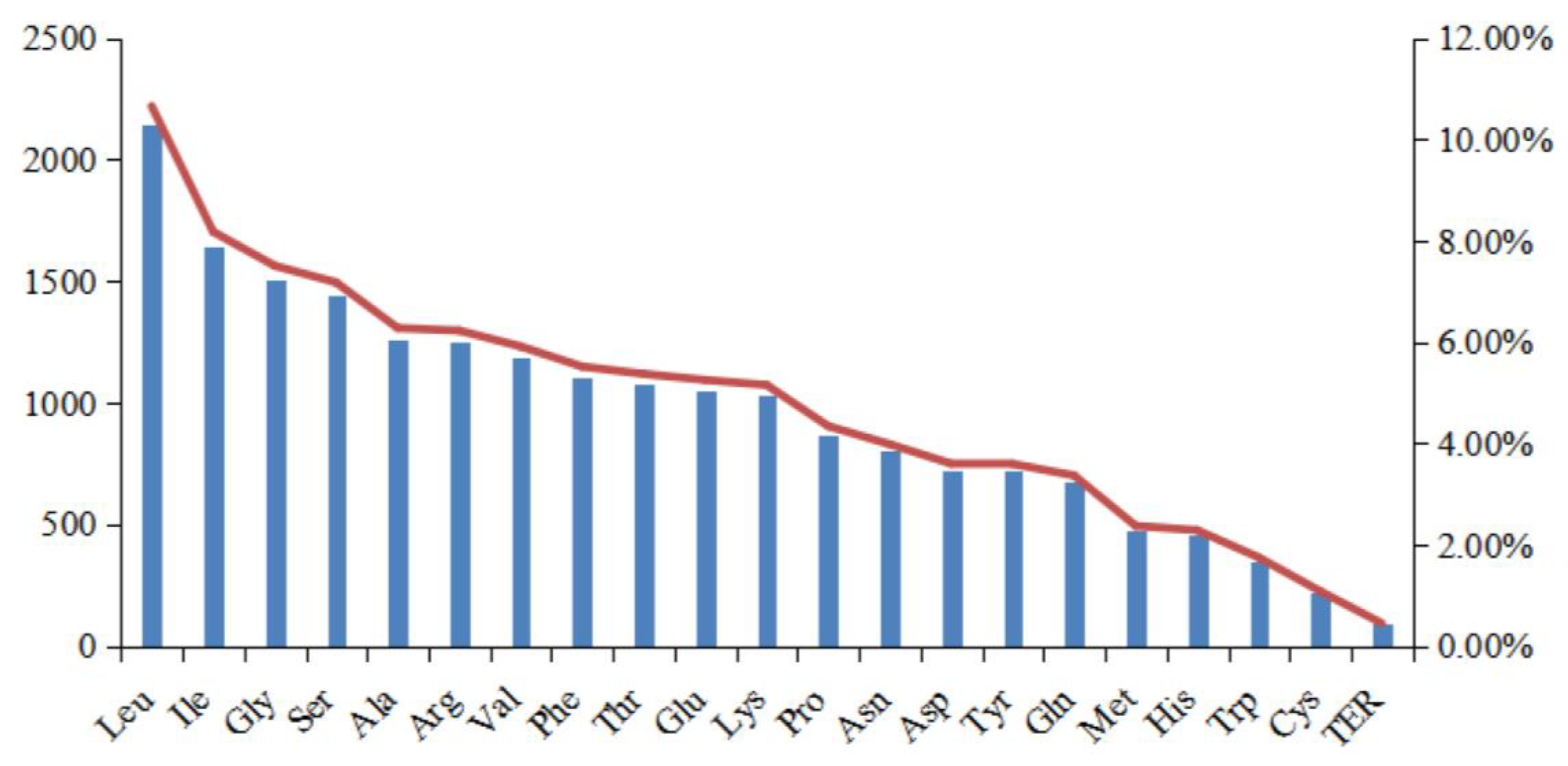
We next identified the rates of codon usage in the cp genome of I. lipoensis, using the CDS (Figure 4 and Table 2). Overall, 20 097 codons were identified, with 64 varying types, including 20 amino acids, and three termination codons. Among them, Leu exhibited the most frequent usage, with a total of 2 141 (10.65%), and Cys exhibited the least, with 217 (1.08%) codons. Both Met (ATG) and Trp (UGG) consisted of one codon type only, with 476 and 351 instances, respectively. The RSCU frequency analysis revealed bias in codon usage, with 31 preferred codons, and RSCU values > 1.0. Lastly, all codons with RSCU values > 1.0 ended in A or U, apart from Leu (UUG).
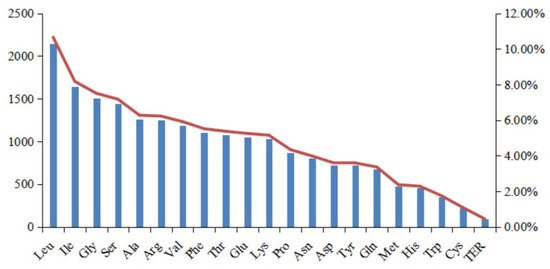
Figure 4.
Amino acid frequencies in the Indosasa lipoensis cp genome protein coding sequences (CDS).

Table 2.
Comparative analysis of chloroplast codon usage bias of Indosasa lipoensis.
3.4. Comparative Genomic Analyses
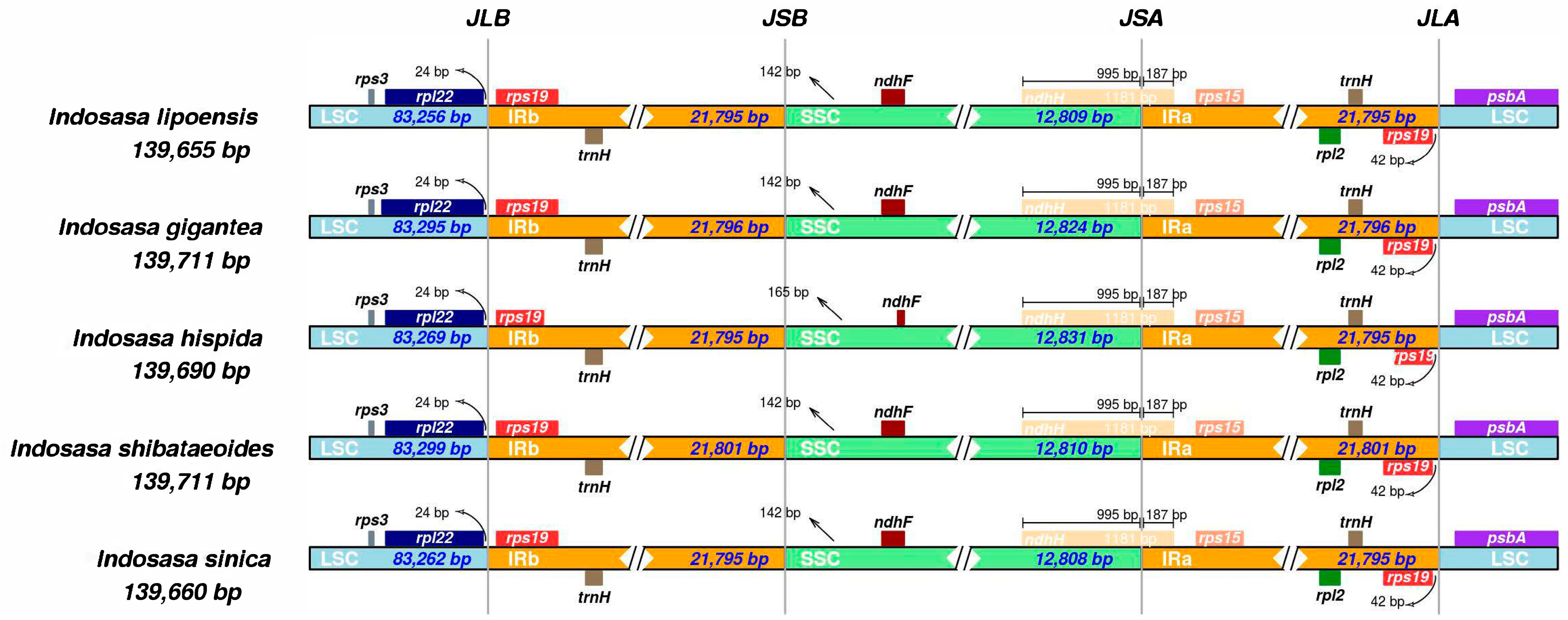
To elucidate the contraction and expansion of the IR boundary, we next assessed the SC and IR boundaries of the five Indosasa species. Based on our analyses, the IR boundaries of the Indosasa genus were highly conserved (Figure 5). The IR length was between 21,795–21,801 bp, the rps19 and trnH genes were present within the IR region, whereas, the spl22 and psbA genes were present within the LSC region. The spl22 gene was 24 bp from the LSC and IRb boundary. Lastly, the SSC and IRa boundary were within the ndhH gene.
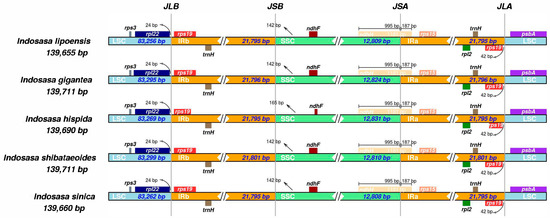
Figure 5.
Comparisons of the LSC, SSC, and IRs junctions among the Indosasa species.
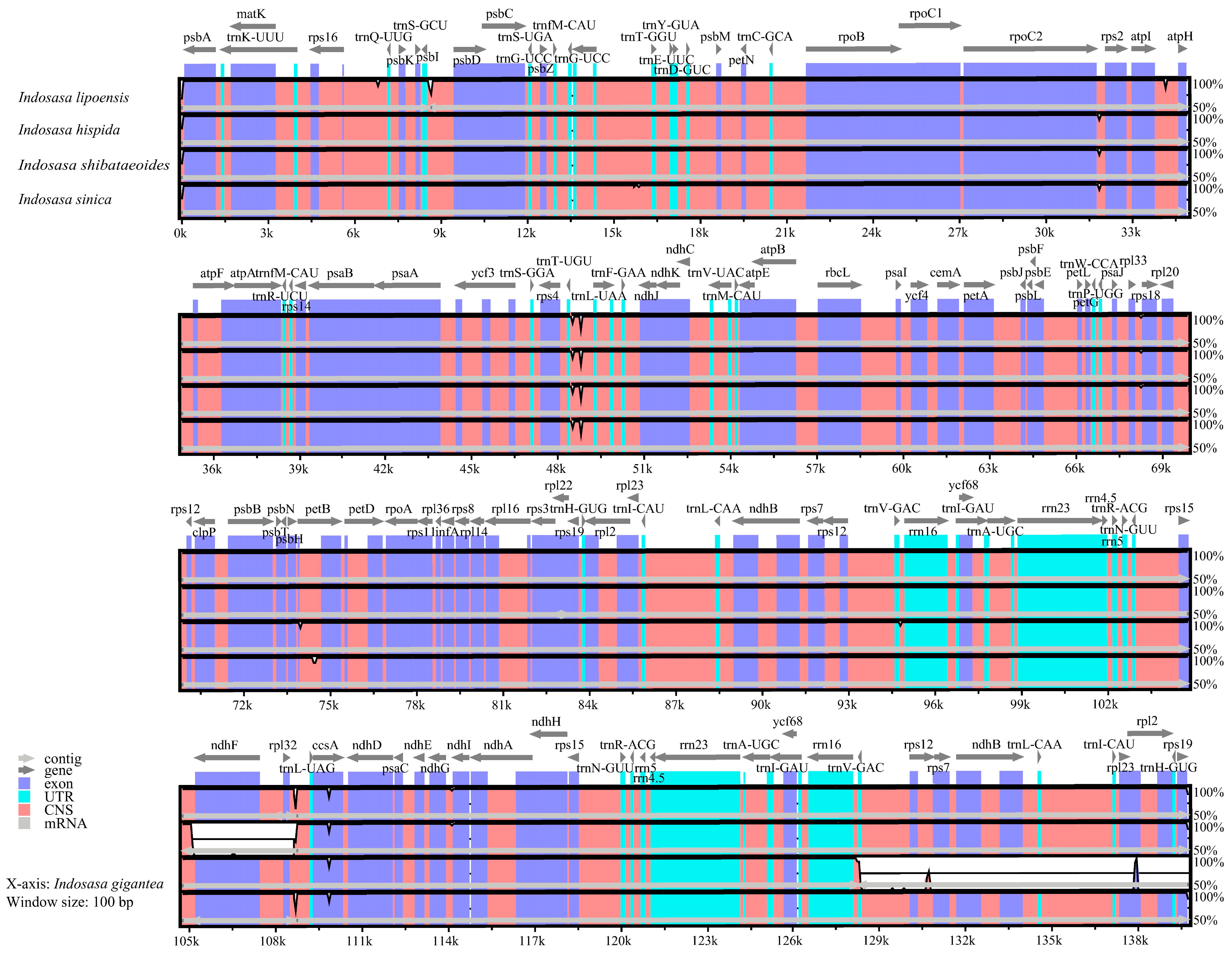
Subsequently, we assessed the cp genome sequences of I. lipoensis with congeneric species using the online alignment tool mVISTA, with I. gigantea as the reference (Figure 6). Based on our analysis, the IRs regions were more conserved, relative to the LSC and SSC regions. Moreover, the non-coding regions were more differentiated, relative to the CDS. The intergenic region variants were present in the rps19-psbA, psbI-psbD, rpoC2-rps2, atpI-atpH, trnT-UAA-trnF-GAA, and rpl32-trnL-UAG regions.
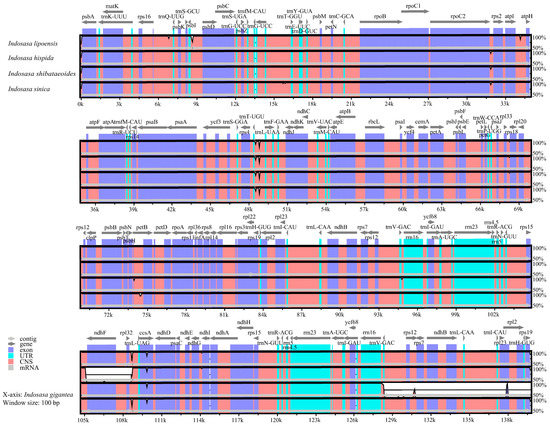
Figure 6.
Comparison of five cp genomes from the Indosasa genus using mVISTA.
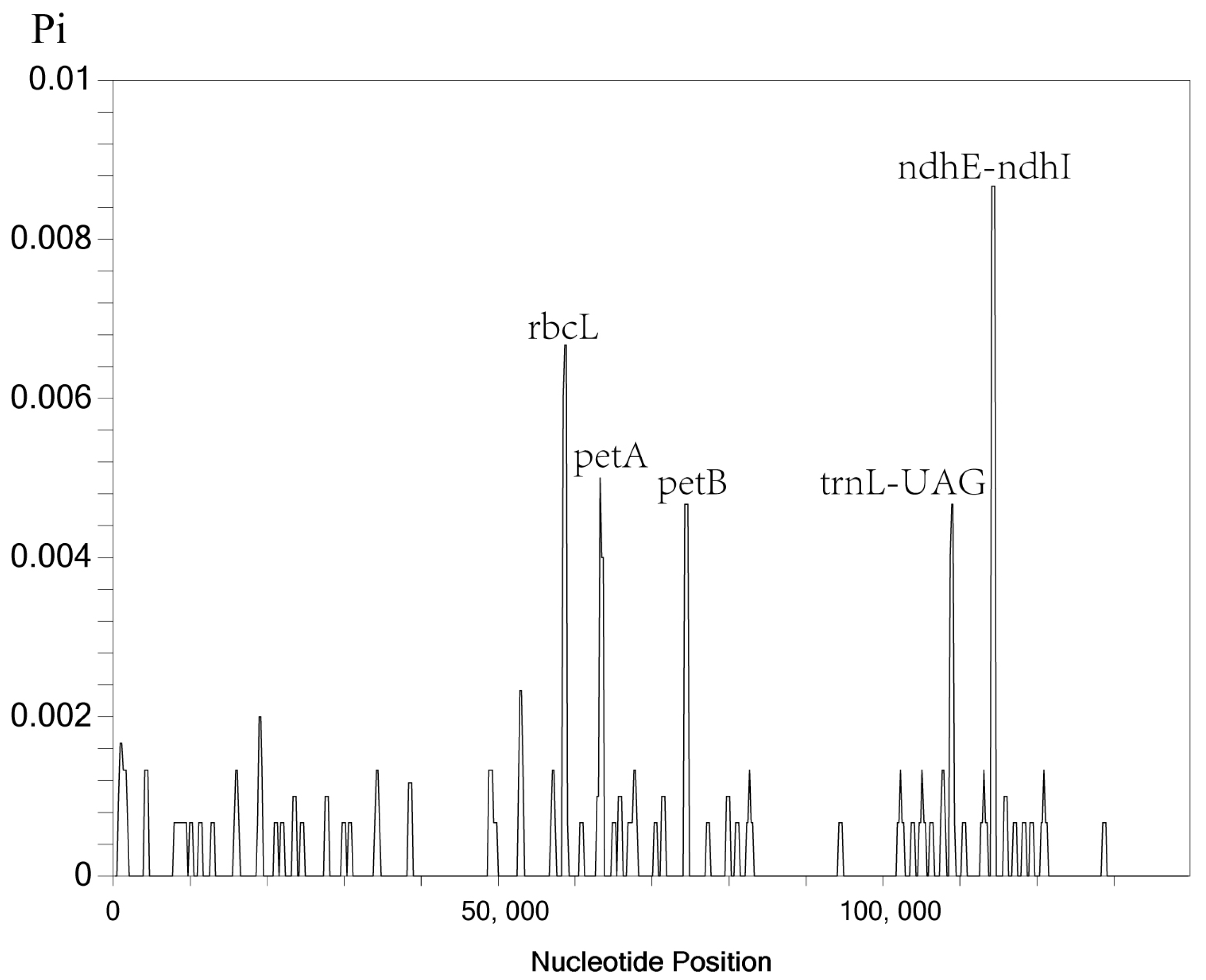
In addition, we also performed nucleotide polymorphism analysis of five Indosasa genus species using the DnaSP software (Figure 7). Our analysis revealed that the Pi values of the Indosasa cp genome varied between 0–0.00867. We detected five highly variable regions (Pi > 0.004), namely, rbcL, petA, petB, trnL-UAG, and ndhE-ndhI. Among these, three regions, namely, rbcL, petA, and petB resided in the LSC region, whereas, two areas, namely, trnL-UAG and ndhE-ndhI, resided in the SSC area. Consistent with the mVISTA results, the more divergent loci were concentrated in the single copy (SC) regions, while the IR regions showed less divergence.
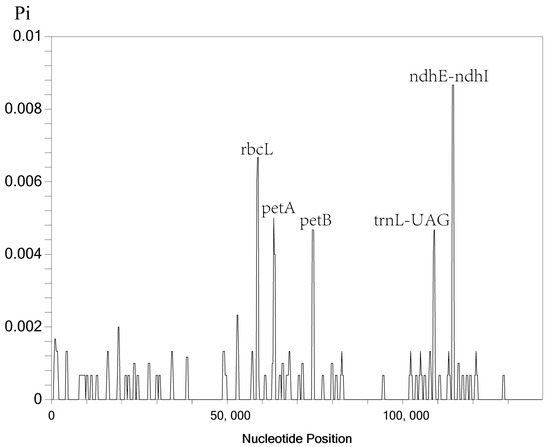
Figure 7.
Sliding window analysis of the Indosasa genus entire cp genome sequences.
3.5. Phylogenetic Analysis
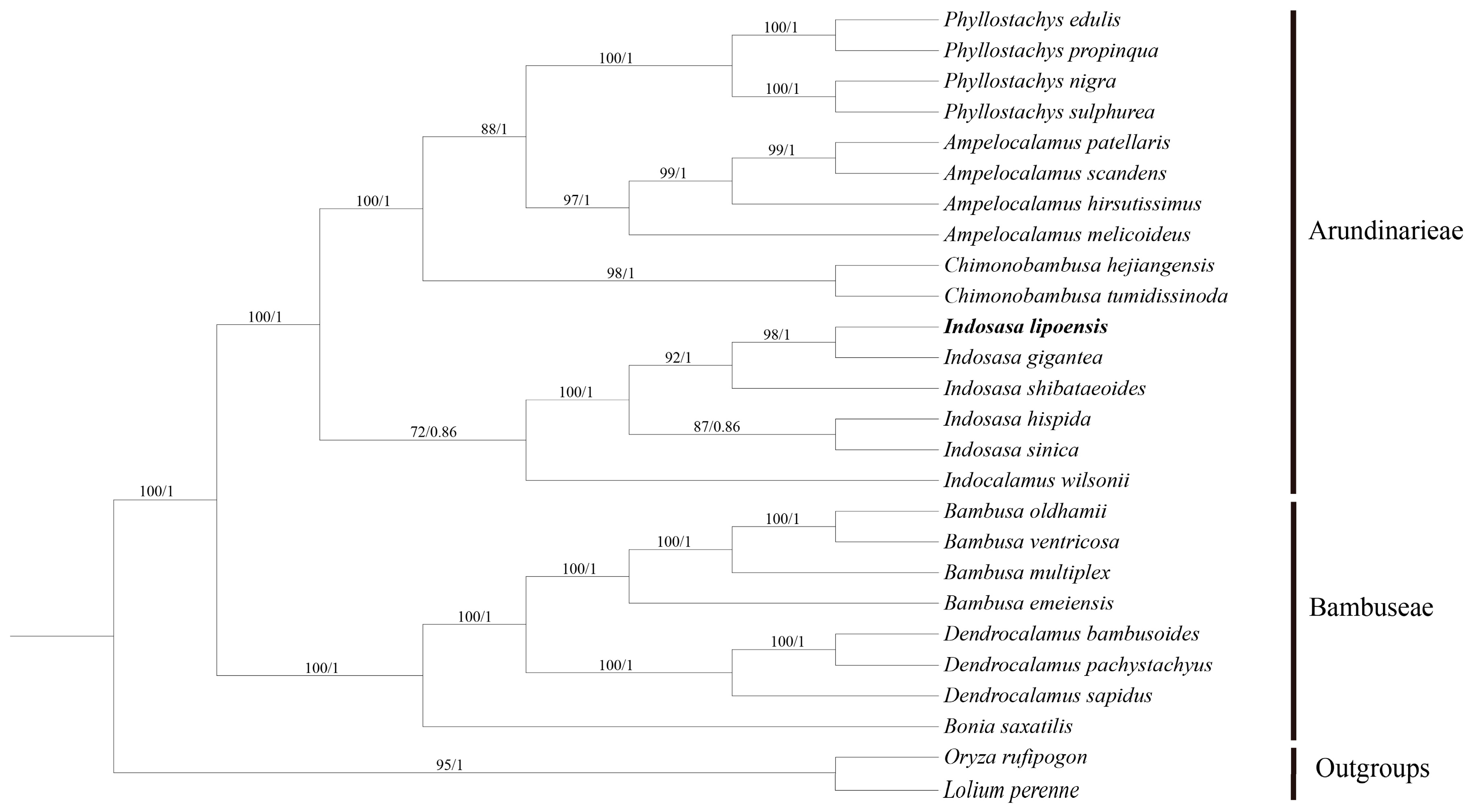
We constructed an ML and BI trees with 26 complete cp genomes to establish the phylogenetic location of I. lipoensis (Figure 8). It was found that topology of the phylogenetic trees built by the ML and BI methods were similar, although with different bootstrap values (BS). The phylogenetic tree divided the subfamily Bambooaceae species into two groups, namely, Arundinarieae and Bambuseae, with 100% BS and 1.0 posterior probability (PP). I. lipoensis and I. gigantea exhibited the closest relationship (BS = 98%, PP = 1.0). Moreover, the five Indosasa species exhibited close associations, and established a monophyletic cohort with 100% BS and 1.0 PP.
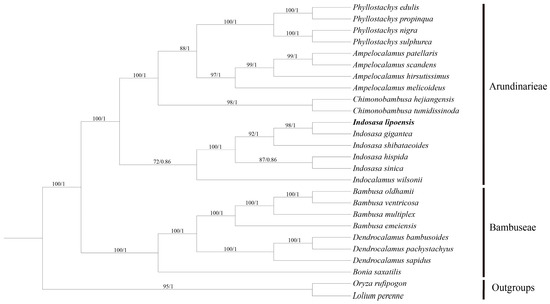
Figure 8.
The maximum likelihood (ML)- and Bayesian Inference (BI)-constructed phylogenetic tree of the cp genomes of 24 Bambooaceae species. Oryza rufipogon and Lolium peremne were employed as outgroups.
4. Discussion
Herein, we newly assembled and assessed the entire plastid genome of I. lipoensis, and compared it with four reported cp sequences of this genus. Similar to the cp genomes of most angiosperms [10], the I. lipoensis genome showed a typical quadripartite structure with an LSC (83,256 bp in length), an SSC (12,809 bp in length), and a pair of IRs (21,795 bp in length). In general, the expansion and shrinkage of the IR region are considered the primary factors that determine variations in the cp genome length [38,39]. The present study found that the IR boundary variations were highly consistent within the Indosasa genus, thus, implying strong conservation during the evolution of the cp genome in this genus.
The GC content is crucial for the stability of the cp genomic structure [40]. Marked enhancement of the GC content (44.2%) was observed in the IR regions in comparison with the LSC (37.0%) and SSC (33.3%) regions, similar findings have been reported for other higher plants [41]. Abundant rRNA and tRNA genes are the main contributors to the elevated GC content of IR regions [42]. Furthermore, the plastid genome of I. lipoensis is also highly conserved in gene content and order without rearrangements or inversions, which is consistent with other species of this genus [43].
The RSCU and amino acid frequencies of the cp genomes of these species are highly consistent with other taxa as well [44,45]. Among these, leucine was the most dominant amino acid, and cysteine was the least. Most codons with RSCU values > 1 ended in an A or U, and is similar to what has been observed with other higher plants.
Long repeat (LR) sequences are essential for the reorganization, rearrangement, and phylogeny of cp genome [46]. A total of 70 LRs were detected among these species; forward repeats being the most prevalent, corroborating with the trend found in other bamboo plants [47,48]. SSRs are useful genetic resources for population genetics and biogeography of closely associated taxa [49,50]. In all, 54 SSR markers were screened among the aforementioned cp sequences, and their distribution was found across the LSC and SSC locations; the IR sites exhibited reduced variability and primarily contained single copy regions [51]. Among the SSRs, the A/T mono-nucleotides were the most dominant, whereas, the tandem G/C repeats occurred less frequently, and this pattern was also present in other higher plants [52,53]. Given these evidences, the aforementioned repeat sequences can be employed for the future examination of genetic structure, differentiation, the five Indosasa species identification, and taxa classification.
Chloroplast genomes are a powerful tool for plant taxonomy, evolutionary relationship, and biogeographic inference, owing to their abundant polymorphic loci [54]. Overall, the IR regions exhibited more conservation, relative to the LSC and SSC sites, and the non-coding sites displayed more divergence, relative to the CDS, which corroborated with prior investigations involving other taxa [55,56]. Based on our analyses in mVISTA, the highly variable regions were present in the intergenic spacer regions, namely, rps19-psbA, psbI-psbD, rpoC2-rps2, atpI-atpH, trnT-UAA-trnF-GAA, and rpl32-trnL-UAG. The nucleotide variability (Pi) values were further employed to estimate the divergence present in the CDS and intergenic regions [57]. Herein, five highly variable locations (Pi > 0.004) were detected and located within the single-copy (SC) sites, namely, rbcL, petA, petB, trnL-UAG, and ndhE-ndhI. These five mutational hotspots can serve as promising DNA barcodes for the future detection and population investigation of various Indosasa species.
Overall, the evolutionary relationships of Indosasa species failed to clearly clarified because of the lack of molecular data. Only four chloroplast genome data have been published in previous studies of the Indosasa gunus, in which they did not have comprehensive genomic comparisons and phylogenetic analyses. In this study, our phylogenetic analysis results indicated that phylogenetic relationships of some species within Indosasa, including all species currently published in this genus. In additional, I. lipoensis has strong nodes of support for its phylogenetic position within the genus. The Indosasa species were all clustered in one branch, which corroborated with the classical stratification data. The I. lipoensis and I. gigantea are sister groups with close genetic relationship, and they provide consistent results in terms of morphological characteristics.
5. Conclusions
Herein, we newly assembled and illustrated the entire cp genome of I. lipoensis, and performed a comprehensive analysis of the cp genome with four previously published cp genomes in this genus. Based on our analysis, the plastid genome was highly conserved with other species of Poaceae in gene content and order. The plastomes of I. lipoensis was consisted of an LSC (83,256 bp), and SSC (12,809 bp), and a pair of IR locations (21,795 bp). Comparative cp genome analysis of the five Indosasa species revealed that all exhibited similar contraction and expansion of the IR boundary. Codon use preferences are highly similar among species in this genus. Moreover, the five sequence divergence regions, namely, rbcL, petA, petB, trnL-UAG, and ndhE-ndhI, can serve as potential biomarkers for future species identification from the Indosasa genus. Phylogenetic relationships, based on the complete cp genome revealed that I. lipoensis and I. gigantea formed the closest relationship with a strongly supported node. Together, these evidences provided significant resources for elucidating the phylogenetic association and genomic evolution of the genus Indosasa.
Author Contributions
Conceptualization, Z.-X.D.; software, M.-L.W. and X.X.; formal analysis, M.-L.W. and R.-R.Y.; writing—original draft preparation, M.-L.W.; writing—review and editing, M.-L.W., Z.-X.D., and G.-Q.G.; funding acquisition, Z.-X.D. and G.-Q.G. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the Breeding of Improved Bamboo Varieties for Shoot (Te-Lin-Yan 2020-17).
Institutional Review Board Statement
Not applicable.
Data Availability Statement
The plastome data supporting this study are openly available in the GenBank nucleotide database. Other data are contained within the article.
Acknowledgments
The authors would like to thank all the reviewers who participated in the review for their assistance during the preparation of this manuscript.
Conflicts of Interest
The authors declare no conflict of interest.
Specimen Collection Statement
The collection of fresh leaves obtained the permission of the nature reserve.
References
- Guo, Z.H.; Li, D.Z. Advances in the Systematics and Biogeography of the Bambusoideae (Gramineae) with Re marks on Some Remaining Problems. Acta Bot. Yunnanica 2002, 24, 431–438. [Google Scholar]
- Fei, B.H. Bamboo Industry Promote Human Life Better. World Bamboo Ratt. 2019, 12, 1–4. [Google Scholar]
- Bi, Y.F.; Cai, H.J.; Wang, A.K.; Wang, Y.K. Progress in Genetic Engineering of Bamboo. Mol. Plant Breed. 2016, 14, 3390–3399. [Google Scholar]
- Reng, C.C.; Jia, Y.L.; Lou, Y.L.; Qiao, S.Y.; Xu, W.J. Analysis of nutritional and functional components of bamboo shoots in Chimonobambusa utilis, Guizhou. Food Ferment. Ind. 2017, 47, 214–221. [Google Scholar]
- Chen, G.; Li, X.; Yu, Y.F.; He, B.; Zhao, L.L. Research on constitutive relationship of flat-pressure laminated moso bamboo lumber for structural application. Build. Struct. 2021, 51, 135–139. [Google Scholar]
- Keng, B.J.; Wang, Z.P. Bambusoideae. In Flora Reipubliaris Sinica; Poaceae, Keng, B.J., Wang, Z.P., Eds.; Science Press: Beijing, China, 1996; Volume 9, pp. 204–205. [Google Scholar]
- Li, D.Z. Poaceae. In The Families and Genera of Chinese Vascular Plants; Science Press: Beijing, China, 2020; Volume I, pp. 616–617. [Google Scholar]
- Qin, L.J.; Ran, J.C.; Yao, Z.M.; Mo, J.W.; Tang, A.X.; Meng, H.L. Survey about Nectar Plant Resources in Maolan Nature Reserve. J. Anhui Agri. Sci. 2012, 40, 14425–14428. [Google Scholar]
- Deng, L.X.; Wang, Z.J. Study on the Resource of primary Bamboos and Their Ornamental Characteristics in Guizhou Province. Guizhou For. Sci. Technol. 2006, 1, 48–54. [Google Scholar]
- Wicke, S.; Schneeweiss, G.M.; DePamphilis, C.W.; Müller, K.F.; Quandt, D. The evolution of the plastid chromosome in land plants: Gene content, gene order, gene function. Plant Mol. Biol. 2011, 76, 273–297. [Google Scholar] [CrossRef]
- Fu, G.; Liu, Y.; Caraballo-Ortiz, M.A.; Zheng, C.; Liu, T.; Xu, Y.; Su, X. Characterization of the Complete Chloroplast Genome of the Dragonhead Herb, Dracocephalum heterophyllum (Lamiaceae), and Comparative Analyses with Related Species. Diversity 2022, 14, 110. [Google Scholar] [CrossRef]
- Zarei, A.; Ebrahimi, A.; Mathur, S.; Lawson, S. The First Complete Chloroplast Genome Sequence and Phylogenetic Analysis of Pistachio (Pistacia vera). Diversity 2022, 14, 577. [Google Scholar] [CrossRef]
- Gu, L.; Hou, Y.; Wang, G.; Liu, Q.; Ding, W.; Weng, Q. Characterization of the chloroplast genome of Lonicera ruprechtiana Regel and comparison with other selected species of Caprifoliaceae. PLoS ONE 2022, 17, e0262813. [Google Scholar] [CrossRef]
- Choi, K.; Hang, Y.; Hong, J.-K. Comparative Chloroplast Genomics and Phylogenetic Analysis of Persicaria amphibia (Polygonaceae). Diversity 2022, 14, 641. [Google Scholar] [CrossRef]
- Wu, M.L.; Liu, Y.J.; Xu, X.; Zhu, X.; Gou, G.Q.; Dai, Z.X. The complete chloroplast genome sequence of Chimonobambusa luzhiensis, an endangered species endemic to Guizhou Province, China. Mitochondrial DNA Part B 2022, 7, 1360–1361. [Google Scholar] [CrossRef] [PubMed]
- Li, D.M.; Zhao, C.Y.; Liu, X.F. Complete chloroplast genome sequences of Kaempferia galanga and Kaempferia elegans: Molecular structures and comparative analysis. Molecules 2019, 24, 474. [Google Scholar] [CrossRef] [PubMed]
- Daniell, H.; Lin, C.S.; Yu, M.; Chang, W.J. Chloroplast genomes: Diversity, evolution, and applications in genetic engineering. Genome Biol. 2016, 17, 134. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Lanfear, R. Long-Reads Reveal That the Chloroplast Genome Exists in Two Distinct Versions in Most Plants. Genome Biol. Evol. 2019, 11, 3372–3381. [Google Scholar] [CrossRef] [PubMed]
- Zhong, H.M.; Gou, G.S.; Liu, Y.J.; Zhu, X.; Dai, Z.X. Population Status and Protection Evaluation of Endemic Bamboo Species Ampelocalamus Scandens in Chishui River Wate. J. Mt. Agric. Biol. 2021, 40, 71–74. [Google Scholar]
- Hu, X.P.; Wei, T.L.; Zhu, X.; Dai, Z.X. Population Status and Protection Evaluation of Endemic Bamboo Species Indocalamus hirsutissimus in Wangmo County. J. Mt. Agric. Biol. 2022, 41, 67–70. [Google Scholar]
- Wu, M.L.; Wei, T.L.; Liu, Y.J.; Zhu, X.; Dai, Z.X. The Current Situation of the Wild Resources of the Endemic Species of Chimonobambusa lactistriata in Guizhou. J. Mt. Agric. Biol. 2022, 41, 64–68. [Google Scholar]
- Doyle, J.J.; Doyle, J.L. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem. Bull. 1987, 19, 11–15. [Google Scholar]
- Jin, J.J.; Yu, W.B.; Yang, J.B.; Song, Y.; De Pamphilis, C.W.; Yi, T.S.; Li, D.Z. GetOrganelle: A fast and versatile toolkit for accurate de novo assembly of organelle genomes. Genome Biol. 2020, 21, 241. [Google Scholar] [CrossRef] [PubMed]
- Qu, X.J.; Moore, M.J.; Li, D.Z.; Yi, T.S. PGA: A software package for rapid, accurate, and flexible batch annotation of plastomes. Plant Methods 2019, 15, 50. [Google Scholar] [CrossRef] [PubMed]
- Kearse, M.; Moir, R.; Wilson, A.; Stones-Havas, S.; Cheung, M.; Sturrock, S.; Buxton, S.; Cooper, A.; Markowitz, S.; Duran, C.; et al. Geneious Basic: An integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 2012, 28, 1647–1649. [Google Scholar] [CrossRef] [PubMed]
- Lohse, M.; Drechsel, O.; Kahlau, S.; Bock, R. OrganellarGenomeDRAW—A suite of tools for generating physical maps of plastid and mitochondrial genomes and visualizing expression data sets. Nucleic Acids Res. 2013, 41, W575–W581. [Google Scholar] [CrossRef]
- Peden, J.F. Analysis of Codon Usage. Ph.D. Thesis, University of Nottingham, Nottingham, UK, 1999. [Google Scholar]
- Kurtz, S.; Choudhuri, J.V.; Ohlebusch, E.; Schleiermacher, C.; Stoye, J.; Giegerich, R. REPuter: The manifold applications of repeat analysis on a genomic scale. Nucleic Acids Res. 2001, 29, 4633–4642. [Google Scholar] [CrossRef]
- Beier, S.; Thiel, T.; Münch, T.; Scholz, U.; Mascher, M. MISA-web: A web server for microsatellite prediction. Bioinformatics 2017, 33, 2583–2585. [Google Scholar] [CrossRef]
- Amiryousefi, A.; Hyvönen, J.; Poczai, P. IRscope: An online program to visualize the junction sites of chloroplast genomes. Bioinformatics 2018, 34, 3030–3031. [Google Scholar] [CrossRef]
- Kurtz, S.; Phillippy, A.; Delcher, A.L.; Smoot, M.; Shumway, M.; Antonescu, C.; Salzberg, S.L. Versatile and open software for comparing large genomes. Genome Biol. 2004, 5, R12. [Google Scholar] [CrossRef]
- Librado, P.; Rozas, J. DnaSP v5: A software for comprehensive analysis of DNA polymorphism data. Bioinformatics 2009, 25, 1451–1452. [Google Scholar] [CrossRef]
- Katoh, K.; Standley, D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef]
- Talavera, G.; Castresana, J. Improvement of phylogenies after removing divergent and ambiguously aligned blocks from protein sequence alignments. Syst. Biol. 2007, 56, 564–577. [Google Scholar] [CrossRef]
- Nguyen, L.T.; Schmidt, H.A.; von Haeseler, A.; Minh, B.Q. IQ-TREE: A fast and efective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 2015, 32, 268–274. [Google Scholar] [CrossRef] [PubMed]
- Stamatakis, A. RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 2014, 30, 1312–1313. [Google Scholar] [CrossRef]
- Zhang, D.; Gao, F.; Jakovlić, I.; Zou, H.; Zhang, J.; Li, W.X.; Wang, G.T. PhyloSuite: An integrated and scalable desktop platform for streamlined molecular sequence data management and evolutionary phylogenetics studies. Mol. Ecol. Resour. 2019, 20, 348–355. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.X.; Hu, G.X.; Hu, G.W. Comparative genomics and phylogenetic relationships of two endemic and endangered species (Handeliodendron bodinieri and Eurycorymbus cavaleriei) of two monotypic genera within Sapindales. BMC Genom. 2022, 23, 27. [Google Scholar] [CrossRef] [PubMed]
- Du, Y.P.; Bi, Y.; Yang, F.P.; Zhang, M.F.; Chen, X.Q.; Xue, J.; Zhang, X.H. Complete chloroplast genome sequences of Lilium: Insights into evolutionary dynamics and phylogenetic analyses. Sci. Rep. 2017, 7, 5751. [Google Scholar] [CrossRef]
- Jin, S.; Daniell, H. The Engineered Chloroplast Genome Just Got Smarter. Trends Plant Sci. 2015, 20, 622–640. [Google Scholar] [CrossRef] [PubMed]
- Guo, S.; Guo, L.; Zhao, W.; Xu, J.; Li, Y.; Zhang, X.; Shen, X.; Wu, M.; Hou, X. Complete chloroplast genome sequence and phylogenetic analysis of Paeonia ostii. Molecules 2018, 23, 246. [Google Scholar] [CrossRef]
- Huang, Y.; Wang, J.; Yang, Y.; Fan, C.; Chen, J. Phylogenomic Analysis and Dynamic Evolution of Chloroplast Genomes in Salicaceae. Front Plant Sci. 2017, 8, 1050. [Google Scholar] [CrossRef]
- Tu, D. The complete chloroplast genome of Indosasa hispida ‘Rainbow’ (Poaceae, Bambuseae): An ornamental bamboo species in horticulture. Mitochondrial DNA Part B 2022, 7, 619–621. [Google Scholar] [CrossRef]
- Huo, Y.; Gao, L.; Liu, B.; Yang, Y.; Kong, S.; Sun, Y.; Yang, Y.; Wu, X. Complete chloroplast genome sequences of four Allium species: Comparative and phylogenetic analyses. Sci. Rep. 2019, 9, 12250. [Google Scholar] [CrossRef] [PubMed]
- Gui, L.; Jiang, S.; Xie, D.; Yu, L.; Huang, Y.; Zhang, Z.; Liu, Y. Analysis of complete chloroplast genomes of Curcuma and the contribution to phylogeny and adaptive evolution. Gene 2020, 30, 144355. [Google Scholar] [CrossRef] [PubMed]
- Chumley, T.W.; Palmer, J.D.; Mower, J.P.; Fourcade, H.M.; Calie, P.J.; Boore, J.L.; Jansen, R.K. The complete chloroplast genome sequence of Pelargonium hortorum: Organization and evolution of the largest and most highly rearranged chloroplast genome of land plants. Mol. Biol. Evol. 2006, 23, 2175–2190. [Google Scholar] [CrossRef]
- Xia, H.; Liu, X.; Wang, Y.; Li, X.; Wang, J.; Jin, C. The complete chloroplast genome sequence of Bambusa stenoaurita (Bambusoideae). Mitochondrial DNA B 2021, 6, 2184–2185. [Google Scholar] [CrossRef]
- Zheng, X.; Yang, M.; Ding, Y.L.; Lin, S.Y. The complete chloroplast genome sequence of Acidosasa gigantea (Bambusoideae: Arundinarieae): An ornamental bamboo species endemic to China. Mitochondrial DNA B 2020, 5, 1119–1121. [Google Scholar] [CrossRef]
- Zhou, J.; Hu, Y.P.; Yu, Z.Y.; Li, J.J.; Xu, M.Y.; Guo, Q.R. The complete chloroplast genome of a solid type of Phyllostachys nidularia (Bambusoideae: Poaceae), a species endemic to China. Mitochondrial DNA B 2021, 6, 978–979. [Google Scholar]
- Gao, J.; Gao, L.Z. The complete chloroplast genome sequence of the Phyllostachys sulphurea (Poaceae: Bambusoideae). Mitochondrial DNA A 2016, 27, 983–985. [Google Scholar] [CrossRef]
- Luo, Y.; He, J.; Lyu, R.; Xiao, J.; Li, W.; Yao, M.; Pei, L.; Cheng, J.; Li, J.; Xie, L. Comparative Analysis of Complete Chloroplast Genomes of 13 Species in Epilobium, Circaea, and Chamaenerion and Insights Into Phylogenetic Relationships of Onagraceae. Front. Genet. 2021, 12, 730495. [Google Scholar] [CrossRef]
- Tang, C.; Chen, X.; Deng, Y.; Geng, L.; Ma, J.; Wei, X. Complete chloroplast genomes of Sorbus sensu stricto (Rosaceae): Comparative analyses and phylogenetic relationships. BMC Plant Biol. 2022, 22, 495. [Google Scholar] [CrossRef] [PubMed]
- Krawczyk, K.; Myszczyński, K.; Nobis, M.; Sawicki, J. Insights into adaptive evolution of plastomes in Stipa L. (Poaceae). BMC Plant Biol. 2022, 22, 525. [Google Scholar] [CrossRef]
- Li, Y.; Zhang, L.; Wang, T.; Zhang, C.; Wang, R.; Zhang, D.; Xie, Y.; Zhou, N.; Wang, W.; Zhang, H.; et al. The complete chloroplast genome sequences of three lilies: Genome structure, comparative genomic and phylogenetic analyses. J. Plant Res. 2022, 135, 723–737. [Google Scholar] [CrossRef] [PubMed]
- Xi, J.; Lv, S.; Zhang, W.; Zhang, J.; Wang, K.; Guo, H.; Hu, J.; Yang, Y.; Wang, J.; Xia, G.; et al. Comparative plastomes of Carya species provide new insights into the plastomes evolution and maternal phylogeny of the genus. Front. Plant Sci. 2022, 13, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Wen, F.; Hong, X.; Li, Z.; Mi, Y.; Zhao, B. Comparative chloroplast genome analyses of Paraboea (Gesneriaceae): Insights into adaptive evolution and phylogenetic analysis. Front. Plant Sci. 2022, 13, 1019831. [Google Scholar] [CrossRef]
- Tyrrell, C.D.; Santos-Gonçalves, A.P.; Londoño, X.; Clark, L.G. Molecular phylogeny of the arthrostylidioid bamboos (Poaceae: Bambusoideae: Bambuseae: Arthrostylidiinae) and new genus Didymogonyx. Mol. Phylogenet. Evol. 2012, 65, 136–148. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).