The Contact Zone of Phylogenetic Lineages of Freshwater Fish in Arctic Eurasia: Genetic Polymorphism of Coregonid Populations
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Collection
2.2. mtDNA Extraction and Sequencing
2.3. Phylogenetic and Phylogeographic Analyses
3. Results
3.1. Phylogenetic Lineages and Their Contact Zones in Considered Cisco Species
3.2. Phylogenetic Lineages and Their Contact Zones in Whitefish
4. Discussion
4.1. The Level of Population Polymorphism in the Contact Zones of Coregonids Phylogenetic Lineages
4.2. Possible Reasons for the Formation of the Observed Patterns of Contact Zones in Considered Cisco Species and Whitefish
4.3. Comparison of Our Own Results with other Studies of the Phylogeography of the Whitefish and Considered Ciscoes Species of Eurasia
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Taberlet, P.; Fumagalli, L.; Wust-Saucy, A.-G.; Cosson, J.-F. Comparative phylogeography and postglacial colonization routes in Europe. Mol. Ecol. 1998, 7, 453–464. [Google Scholar] [CrossRef] [PubMed]
- Martinson, D.G.; Pisias, N.G.; Hays, J.D.; Imbrie, J.D.; Moore, T.C.; Shackleton, N.J. Age dating and orbital theory of the ice ages: Development of a high resolution 0–300,000-year chronostratigraphy. Quat. Res. 1987, 27, 1–29. [Google Scholar] [CrossRef]
- Dawson, A.G. Ice Age Earth. Late Quaternary Geology and Climate, 1st ed.; Routledge Press: London, UK, 1992. [Google Scholar]
- Bernatchez, L.; Wilson, C.C. Comparative phylogeography of Nearctic and Palearctic fishes. Mol. Ecol. 1998, 7, 431–452. [Google Scholar] [CrossRef]
- Petit, J.R.; Jouzel, J.; Raynaud, D.; Barkov, N.I.; Barnola, J.-M.; Basile, I.; Bender, M.; Chappellaz, J.; Davis, M.; Delaygue, G.; et al. Climate and atmospheric history of the past 420,000 years from the Vostok ice core, Antarctica. Nature 1999, 399, 429–436. [Google Scholar] [CrossRef]
- Pielou, E.C. After the Ice Age: The Return of Life to Glaciated North America; The University of Chicago Press: Chicago, IL, USA, 1991. [Google Scholar]
- Hewitt, H.G.M. Some genetic consequences of ice ages, and their role in divergence and speciation. Biol. J. Linn. Soc. 1996, 58, 247–276. [Google Scholar] [CrossRef]
- Hewitt, H.G.M. The genetic legacy of the quaternary ice ages. Nature 2000, 405, 907–913. [Google Scholar] [CrossRef]
- Behnke, R.J. The systematic of salmonid fishes of recently deglaciated lakes. J. Fish. Res. Broad Can. 1972, 29, 639–671. [Google Scholar] [CrossRef]
- Hewitt, H.G.M. Speciation, hybrid zones and phylogeography—Or seeing genes in space and time. Mol. Ecol. 2001, 10, 537–549. [Google Scholar] [CrossRef]
- Avise, J.C. Molecular Markers, Natural History, and Evolution, 2nd ed.; Sinauer Associates, Inc. Publishers: Sunderland, MA, USA, 2004. [Google Scholar]
- Durand, J.D.; Templeton, A.R.; Guinand, B.; Imsiridou, A.; Bouvet, Y. Nested clade and phylogeographic analyses of the chub, Leuciscus cephalus (Teleostei, Cyprinidae), in Greece: Implications for Balkan Peninsula biogeography. Mol. Phylogenet. Evol. 1999, 13, 566–580. [Google Scholar] [CrossRef]
- Hewitt, G.M. Genetic consequences of climatic oscillations in the Quaternary. Phil. Trans. R. Soc. Lond. B 2004, 359, 183–195. [Google Scholar] [CrossRef]
- Yamamoto, S.; Morita, K.; Kitano, S.; Watanabe, K.; Koizumi, I.; Maekawa, K.; Takamura, K. Phylogeography of white-spotted charr (Salvelinus leucomaenis) inferred from mitochondrial DNA sequences. Zool. Sci. 2004, 21, 229–240. [Google Scholar] [CrossRef] [PubMed]
- Kikko, T.; Kuwahara, M.; Iguchi, K.; Kurumi, S.; Yamamoto, S.; Kai, Y.; Nakayama, K. Mitochondrial DNA population structure of the white-spotted charr (Salvelinus leucomaenis) in the Lake Biwa water system. Zool. Sci. 2008, 25, 146–153. [Google Scholar] [CrossRef] [PubMed]
- Weider, L.J.; Hobæk, A. Phylogeography and arctic biodiversity: A review. Ann. Zool. Fenn. 2000, 37, 217–231. [Google Scholar]
- Bernatchez, L.; Renaut, S.; Whiteley, A.R.; Derome, N.; Jeukens, J.; Landry, L.; Lu, G.; Nolt, A.W.; Østbye, K.; Rogers, S.M.; et al. On the origin of species: Insights from the ecological genomics of lake whitefish. Phil. Trans. R. Soc. B 2010, 365, 1783–1800. [Google Scholar] [CrossRef] [PubMed]
- Renaut, S.; Maillet, N.; Normandeau, E.; Sauvage, C.; Derome, N.; Rogers, S.M.; Bernatchez, L. Genome-wide patterns of divergence during speciation: The lake whitefish case study. Phil. Trans. R. Soc. B 2012, 367, 354–363. [Google Scholar] [CrossRef]
- April, J.; Hanner, R.H.; Mayden, R.L.; Bernatchez, L. Metabolic rate and climatic fluctuations shape continental wide pattern of genetic divergence and biodiversity in fishes. PLoS ONE 2013, 8, e70296. [Google Scholar] [CrossRef]
- Gagnaire, P.-A.; Pavey, S.A.; Normandeau, E.; Bernatchez, L. The genetic architecture of reproducrive isolation during speciation-with-gene-flow in lake whitefih species pairs assessed by RAD sequencing. Evolution 2013, 67, 2483–2497. [Google Scholar] [CrossRef]
- Skurikhina, L.A.; Oleinik, A.G.; Kukhlevsky, A.D.; Kovpak, N.E.; Frolov, S.V.; Sendek, D.S. Phylogeography and demographic history of the Pacific smelt Osmerus dentex inferred from mitochondrial DNA variation. Polar Biol. 2018, 41, 877–896. [Google Scholar] [CrossRef]
- Levin, B.A.; Casal-López, M.; Simonov, E.; Dgebuadze, Y.Y.; Mugue, N.S.; Tiunov, A.V.; Doadrio, I.; Golubtsov, A.S. Adaptive radiation of barbs of the genus Labeobarbus (Cyprinidae) in an East African river. Freshw. Biol. 2019, 64, 1721–1736. [Google Scholar] [CrossRef]
- Artamonova, V.S.; Bardukov, N.V.; Aksenova, O.V.; Ivanova, T.S.; Ivanov, M.V.; Kirillova, E.A.; Koulish, A.V.; Lajus, D.L.; Malyutina, A.M.; Pashkov, A.N.; et al. Round-the world voyage of the threespine stickleback (Gasterosteus aculeatus): Phylogeographic data covering the entire species range. Water 2022, 14, 2484. [Google Scholar] [CrossRef]
- Denys, G.P.J.; Persat, H.; Dettai, A.; Geiger, M.F.; Freyhof, J.; Fesquet, J.; Keith, P. Genetic and morphological discrimination of three species of ninespined stickleback Pungitius spp. (Teleostei, Gasterosteidae) in France with the revalidation of Pungitius vulgaris (Mauduyt, 1848). J. Zool. Syst. Evol. Res. 2018, 56, 77–101. [Google Scholar] [CrossRef]
- Artaev, O.N.; Ermakov, O.A.; Vekhov, D.A.; Konovalov, A.F.; Levina, M.A.; Pozdeev, I.V.; Ruchin, A.B.; Alyushin, I.V.; Iljin, V.Y.; Levin, B.A. Genetic screening of distribution pattern of roaches Rutilus rutilus and R. lacustris (Cyprinidae) in broad range of secondary contact (Volga basin). Inland Water Biol. 2021, 14, 205–214. [Google Scholar] [CrossRef]
- Borovikova, E.A.; Artamonova, V.S. Vendace (Coregonus albula) and least cisco (Coregonus sardinella) are a single species: Evidence from revised data on mitochondrial and nuclear DNA polymorphism. Hydrobiologia 2021, 848, 4241–4262. [Google Scholar] [CrossRef]
- Petrosino, G.; Tancioni, L.; Turani, M.; Rakaj, A.; Ciuffardi, L.; Rossi, A.R. Phylogeography of Sarmarutilus rubilio (Cypriniformes: Leuciscidae): Complex genetic structure, clue to a new cryptic species and further insights into roaches phylogeny. Genes 2022, 13, 1071. [Google Scholar] [CrossRef] [PubMed]
- Takács, P.; Bánó, B.; Czeglédi, I.; Erős, T.; Ferincz, Á.; Gál, B.; Bánó-Kern, B.; Kovács, B.; Nagy, A.A.; Nyeste, K.; et al. The mixed phylogenetics origin of northern pike (Esox lucius Linnaeus 1758) populations in the Middle Danubian drainage. BMC Zool. 2022, 7, 28–40. [Google Scholar] [CrossRef]
- Swenson, N.G.; Howard, D.J. Clustering of contact zones, hybrid zones, and phylogeographic breaks in North America. Am. Nat. 2005, 166, 581–591. [Google Scholar] [CrossRef] [PubMed]
- Costedoat, C.; Gilles, A. Quaternary patterns of freshwater fishes in Europe: Comparative phylogeography and conservation perspective. Open Conserv. Biol. J. 2009, 3, 36–48. [Google Scholar] [CrossRef]
- Bernatchez, L. The evolutionary history of brown trout (Salmo trutta L.) inferred from phylogeographic, nested clade, and mismatch analyses of mitochondrial DNA variation. Evolution 2001, 55, 351–379. [Google Scholar]
- Cortey, M.; Vera, M.; Pla, C.; Garsía-Marín, J.-L. Northern and Southern expansions of Atlantic brown trout (Salmo trutta) populations during the Pleistocene. Biol. J. Linn. Soc. 2009, 97, 904–917. [Google Scholar] [CrossRef]
- D’Agaro, E.; Gibertoni, P.P.; Esposito, S. Broun trout (Salmo trutta) phylogenetics. In Prime Archives in Applied Sciences; Henninger, H., Suzdaltsev, A., Eds.; Vide Leaf: Hyderbad, India, 2022. [Google Scholar]
- Brunner, P.C.; Douglas, M.R.; Osinov, A.; Wilson, C.C.; Bernatchez, L. Holarctic phylogeography of Arctic charr (Salvelinus alpinus L.) inferred from mitochondrial DNA sequences. Evolution 2001, 55, 573–586. [Google Scholar] [CrossRef]
- Moore, J.-S.; Bajno, R.; Reist, J.D.; Taylor, E.B. Post-glacial recolonization of the North American Arctic by Arctic char (Salvelinus alpinus): Genetic evidence of multiple northern refugia and hybridization between glacial lineages. J. Biogeogr. 2015, 42, 2089–2100. [Google Scholar] [CrossRef]
- Makhrov, A.A.; Bolotov, I.N.; Spitsyn, V.M.; Gofarov, M.Y.; Artamonova, V.S. Resident and anadromous forms of Arctic charr (Salvelinus alpinus) from North-East Europe: An example of high ecological variability without speciation. Dokl. Ross. Akad. Nauk. 2019, 485, 242–246. (In Russian) [Google Scholar]
- Jacobsen, M.W.; Jensen, N.W.; Nygaard, R.; Præbel, K.; Jónsson, B.; Nielsen, N.H.; Pujolar, J.M.; Fraser, D.J.; Bernatchez, L.; Hansen, M.M. A melting pot in the Arctic: Analysis of mitogenome variation in Arctic char (Salvelinus alpinus) reveals a 1000-km contact zone between highly divergent lineages. Ecol. Freshw. Fish 2022, 31, 330–346. [Google Scholar] [CrossRef]
- Osinov, A.G.; Volkov, A.A.; Pavlov, D.A. Secondary contact, hybridization, and diversification in Arctic charr (Salvelinus alpinus (L.) species complex) from lakes of the Norilo-Pyasinskaya water system, Taimyr: How many forms exist there? Hydrobiologia 2022, 849, 2521–2547. [Google Scholar] [CrossRef]
- Shubin, P.N.; Zakharov, A.B. Hybridization of the European Thymallus thumallus (L.) and Arctic Thymallus arcticus (Pallas) graylings (Thymallidae) in the zone of secondary contact. J. Ichthyol. 1984, 24, 502–504. (In Russian) [Google Scholar]
- Koskinen, M.T.; Nilsson, J.; Veselov, A.J.; Potutkin, A.G.; Ranta, E.; Primmer, C.R. Microsatellite data resolve phylogeographic patterns in European grayling, Thymallus thymallus, Salmonidae. Heredity 2002, 88, 391–401. [Google Scholar] [CrossRef]
- Gum, B.; Gross, R.; Kuehn, R. Mitochondrial and nuclear DNA phylogeography of European grayling (Thymallus thymallus): Evidence for secondary contact zones in central Europe. Mol. Ecol. 2005, 14, 1707–1725. [Google Scholar] [CrossRef]
- Weiss, S.; Knizhin, I.; Kirillov, A.; Froufe, E. Phenotypic and genetic differentiation of two major phylogeographical lineages of arctic grayling Thymallus arcticus in the Lena River, and surrounding Arctic drainages. Biol. J. Linn. Soc. 2006, 88, 511–525. [Google Scholar] [CrossRef]
- Weiss, S.; Knizhin, I.; Romanov, V.; Kopun, T. Secondary contact between two divergent lineages of grayling Thymallus in the lower Enisey basin and its taxonomic implications. J. Fish Biol. 2007, 71, 371–386. [Google Scholar] [CrossRef]
- Ponomareva, E.; Volkov, A.; Ponomareva, M.; Makeenko, G.; Shubina, E. European grayling (Thymallus thymallus) mtDNA control region haplotypes diversity and postglacial colonization of Russian European North. In Proceedings of the XVI European Congress of Ichthyology, Lausanne, Switzerland, 2–6 September 2019. [Google Scholar] [CrossRef]
- Artamonova, V.S.; Makhrov, A.A.; Popov, I.Y.; Spitsyn, V.M. European smelt (Osmerus eperlanus (Linnaeus, 1758)) on Kolguyev Island, Barents Sea, and factors limiting its spread through the Arctic. Contemp. Probl. Ecol. 2020, 13, 127–131. [Google Scholar] [CrossRef]
- Semenova, A.V.; Stroganov, A.N.; Ponomareva, E.V.; Afanas’ev, K.I.; Vilkina, O.V. Large-scale genetic structure and diversity of Arctic rainbow smelt Osmerus dentex Steindachner et Kner, 1870 throughout its distributional range based on microsatellites. Polar Biol. 2021, 44, 927–940. [Google Scholar] [CrossRef]
- Skog, A.; Vøllestad, L.A.; Stenseth, N.C.; Kasumyan, A.; Jakobsen, K.S. Circumpolar phylogeography of the northern pike (Esox lucius) and its relationship to the Amur pike (E. reichertii). Front. Zool. 2014, 11, 67–74. [Google Scholar] [CrossRef]
- Levin, B.A.; Simonov, E.P.; Ermakov, O.A.; Levina, M.A.; Interesova, E.A.; Kovalchuk, O.M.; Malinina, Y.A.; Mamilov, N.S.; Mustafayev, N.J.; Pilin, D.V.; et al. Phylogeny and phylogeography of the roaches, genus Rutilus (Cyprinidae), at the Eastern part of its range as inferred from mtDNA analysis. Hydrobiologia 2017, 788, 33–46. [Google Scholar] [CrossRef]
- Van Houdt, J.K.J.; de Cleyn, L.; Perretti, A.; Volckaert, F.A.M. A mitogenic view on the evolutionary history of the Holarctic freshwater gadoid, burbot (Lota lota). Mol. Ecol. 2005, 14, 2445–2457. [Google Scholar] [CrossRef]
- Mäkinen, H.S.; Merilä, J. Mitochondrial DNA phylogeography of the three-spined stickleback (Gasterosteus aculeatus) in Europe—Evidence for multiple glacial refugia. Mol. Phylogenet. Evol. 2008, 46, 167–182. [Google Scholar] [CrossRef] [PubMed]
- Guo, B.; Fang, B.; Shikano, T.; Momigliano, P.; Wang, C.; Kravchenko, A.; Merilä, J. A phylogenomic perspective on diversity, hybridization and evolutionary affinities in the stickleback genus Pungitius. Mol. Ecol. 2019, 28, 4046–4064. [Google Scholar] [CrossRef]
- Feng, X.; Merilä, J.; Löytynoja, A. Complex population history affects admixture analyses in nine-spined sticklebacks. Mol. Ecol. 2022, 31, 5386–5401. [Google Scholar] [CrossRef] [PubMed]
- Nesbø, C.L.; Fossheim, T.; Vøllestad, L.A.; Jacobsen, K.S. Genetic divergence and phylogeographic relationships among European perch (Perca fluviatilis) populations reflect glacial refugia and postglacial colonization. Mol. Ecol. 1999, 8, 1387–1404. [Google Scholar] [CrossRef]
- Behrmann-Godel, J.; Gerlach, G.; Eckmann, R. Postglacial colonization shows evidence for sympatric population splitting of Eurasian perch (Perca fluviatilis L.) in Lake Constance. Mol. Ecol. 2004, 13, 491–497. [Google Scholar] [CrossRef]
- Ragauskas, A.; Butkauskas, D.; Prakas, P.; Gadliauskiene, K.; Gajduchenko, H.; Grauda, D. Complex phylogeographic relationships among the Eurasian perch (Perca fluviatilis) populations in the eastern part of the Baltic Sea region. Hydrobiologia 2020, 847, 925–938. [Google Scholar] [CrossRef]
- Kontula, T.; Väinölä, R. Postglacial colonization of Northern Europe by distinct phylogeographic lineages of the bullhead, Cottus gobio. Mol. Ecol. 2001, 10, 1983–2002. [Google Scholar] [CrossRef] [PubMed]
- Volckaert, F.A.M.; Hänfling, B.; Hellemans, B.; Carvalho, G.R. Timing of the population dynamics of bullhead Cottus gobio (Teleostei: Cottidae) during the Pleistocene. J. Evol. Biol. 2002, 15, 930–944. [Google Scholar] [CrossRef]
- Kontula, T. Phylogeography and Evolution of Freshwater Cottid Fishes. Ph.D. Thesis, The University of Helsinki, Helsinki, Finland, 3 October 2003. [Google Scholar]
- Remington, C.L. Suture-zones of hybrid interaction between recently joined biotas. In Evolutionary Biology; Dobzhansky, T., Hecht, M.K., Steere, W.C., Eds.; Plenum: New York, NY, USA, 1968; pp. 321–428. [Google Scholar]
- Hewitt, G.M. Post-glacial recolonization of European biota. Biol. J. Linn. Soc. 1999, 68, 87–112. [Google Scholar] [CrossRef]
- Makhrov, A.A.; Bolotov, I.N.; Artamonova, V.S.; Borovikova, E.A. Mountain regions are the putative place of origin of many arctic animal and plant forms. Zool. Z. 2019, 98, 1291–1303. [Google Scholar] [CrossRef]
- Makhrov, A.A.; Bolotov, I.N.; Vinarski, M.V.; Artamonova, V.S. Origin of north and central European cold-water species glacial relicts: Four dispersal waves from Asia. Biol. Vnutr. Vod. 2022, 6, 615–639. (In Russian) [Google Scholar]
- Sapota, M.R. The round goby (Neogobius melanostomus) in the Gulf of Gdańsk—A species introduction into the Baltic Sea. Hydrobiologia 2004, 514, 219–224. [Google Scholar] [CrossRef]
- Bernatchez, L.; Dodson, J.J. Phylogenetic relationships among Palearctic and Nearctic whitefish (Coregonus sp.) populations as revealed by mitochondrial DNA variation. Can. J. Fish. Aquat. Sci. 1994, 51 (Suppl. S1), 240–251. [Google Scholar] [CrossRef]
- Wilson, C.C.; Hebert, P.D.N. Phylogeography and postglacial dispersal of lake trout (Salvelinus namaycush) in North America. Can. J. Fish. Aquat. Sci. 1998, 55, 1010–1024. [Google Scholar] [CrossRef]
- Østbye, K.; Bernatchez, L.; Næsje, T.F.; Himberg, M.K.-J.; Hindar, K. Evolutionary history of the European whitefish Coregonus lavaretus (L.) species complex as inferred from mtDNA phylogeography and gill-raker numbers. Mol. Ecol. 2005, 14, 4371–4387. [Google Scholar] [CrossRef]
- Hindar, K.; Ryman, N.; Ståhl, G. Genetic differentiation among local populations and morphotypes of Arctic charr, Salvelinus alpinus. Biol. J. Linn. Soc. 1986, 27, 269–285. [Google Scholar] [CrossRef]
- Østbye, K.; Amundsen, P.-A.; Bernatchez, L.; Klementsen, A.; Knudsen, R.; Kristoffersen, R.; Næsje, T.F.; Hindar, K. Parallel evolution of ecomorphological traits in the European whitefish Coregonus lavaretus (L.) species complex during postglacial time. Mol. Ecol. 2006, 15, 3983–4001. [Google Scholar] [CrossRef] [PubMed]
- Levin, B.A.; Simonov, E.; Dgebuadze, Y.Y.; Levina, M.; Golubtsov, A.S. In the rivers: Multiple adaptive radiations of cyprinid fishes (Labeobarbus) in Ethiopian Highlands. Sci. Rep. 2020, 10, 7192–7205. [Google Scholar] [CrossRef] [PubMed]
- Borovikova, E.A.; Makhrov, A.A. Systematic position and origin of European whitefishes (Coregonus, Coregonidae, Osteichthyes). Genetic approach. Usp. Sovrem. Biol. 2009, 129, 58–66. (In Russian) [Google Scholar]
- Sendek, D.S.; Novoselov, A.P.; Boznak, E.I. Genetic differentiation of coregonid fishes in Pechora River. Contemp. Probl. Ecol. 2016, 9, 166–171. [Google Scholar] [CrossRef]
- Ilmast, N.; Sendek, D.; Zuykova, E.; Milyanchuk, N.; Savosin, D.; Borisovskaya, A.; Alekseev, M.; Bochkarev, N. Morphological and genetic variability of the mass whitefish forms in Lake Onega. KnE Life Sci. 2020, 5, 141–151. [Google Scholar] [CrossRef]
- Sendek, D.S.; Bochkarev, N.A.; Savosin, D.S.; Borisovskaya, A.A.; Ilmast, N.V. Current state of sympatric whitefish from Lake Pyaozero, Kovda river basin, Karelia. IOP Conf. Ser. Earth Environ. Sci. 2020, 539, 012195. [Google Scholar] [CrossRef]
- Kohlmann, K.; Kempter, J.; Kersten, P.; Sadowski, J. Haplotype variability at the mitochondrial ND-1 gene region of Coregonus lavaretus from Polish lakes. Adv. Limnol. 2007, 60, 47–57. [Google Scholar]
- Cronin, M.A.; Spearman, W.J.; Wilmot, R.L.; Patton, J.C.; Bickham, J.W. Mitochondrial DNA variation in chinook (Oncorhynchus tshawytscha) and chum salmon (O. keta) detected by restriction enzyme analysis of polymerase chain reaction (PCR) products. Can. J. Fish. Aquat. Sci. 1993, 50, 708–715. [Google Scholar] [CrossRef]
- Politov, D.V.; Gordon, N.Y.; Afanasiev, K.I.; Altukhov, Y.P.; Bickham, J.W. Identification of palearctic coregonid fish species using mtDNA and allozyme genetic markers. J. Fish Biol. 2000, 57, 51–71. [Google Scholar] [CrossRef]
- Bochkarev, N.A.; Zuykova, E.I.; Katokhin, A.V. Morphology and mitochondrial DNA variation of the Siberian whitefish Coregonus lavaretus pidschian (Gmelin) in the upstream water bodies of the Ob and Yenisei rivers. Evol. Ecol. 2011, 25, 557–572. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef]
- Librado, P.; Rozas, J. DNASP v.5: A Software for comprehensive analysis of DNA polymorphism data. Bioinfromatics 2009, 25, 1451–1452. [Google Scholar]
- Tamura, K.; Nei, M. Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol. Biol. Evol. 1993, 10, 512–526. [Google Scholar]
- Bandelt, H.J.; Forster, P.; Röhl, A. Median-joining networks for inferring intraspecific phylogenies. Mol. Biol. Evol. 1999, 16, 37–48. [Google Scholar] [CrossRef] [PubMed]
- Glez-Peña, D.; Gómes-Blanco, D.; Reboiro-Jato, M.; Fdez-Riverola, F.; Posada, D. ALTER: Program-oriented format conversion of DNA and protein alignments. Nucleic Acids Res. 2010, 38, W14–W18. [Google Scholar] [CrossRef] [PubMed]
- Lanfear, R.; Calcott, B.; Ho, S.Y.W.; Guindon, S. PartitionFinder: Combined selection of partitioning schemes and substitution models for phylogenetic analyses. Mol. Biol. Evol. 2012, 29, 1695–1701. [Google Scholar] [CrossRef] [PubMed]
- Ronquist, F.; Teslenko, M.; van der Mark, P.; Ayres, D.L.; Darling, A.; Höhna, S.; Larget, B.; Liu, L.; Suchard, M.A.; Huelsenbeck, J.P. MrBayes 3.2: Efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 2012, 61, 539–542. [Google Scholar] [CrossRef]
- Rambaut, A. FigTree v1.4: Tree Figure Drawing Tool. 2008. Available online: http://tree.bio.ed.ac.uk/software/figtree/ (accessed on 16 December 2022).
- Rogers, A.R.; Harpending, H. Population growth makes waves in the distribution of pairwise genetic differences. Mol. Biol. Evol. 1992, 9, 552–569. [Google Scholar] [PubMed]
- Rogers, A.R. Genetic evidence for a Pleistocene population expansion. Evolution 1995, 49, 608–615. [Google Scholar] [CrossRef]
- Tajima, F. Statistical methods for testing the neutral mutation hypothesis by DNA polymorphism. Genetics 1989, 123, 585–595. [Google Scholar] [CrossRef]
- Tajima, F. The effect of change in population size on DNA polymorphism. Genetics 1989, 123, 597–601. [Google Scholar] [CrossRef] [PubMed]
- Ramos-Onsins, S.E.; Rozas, J. Statistical properties of new neutrality tests against population growth. Mol. Biol. Evol. 2002, 19, 2092–2100. [Google Scholar] [CrossRef] [PubMed]
- Fu, Y.-X. Statistical tests of neutrality against population growth, hitchhiking and background selection. Genetics 1997, 143, 557–570. [Google Scholar] [CrossRef] [PubMed]
- Excoffier, L.; Laval, G.; Schneider, S. Arlequin ver 3.01. An Integrated Software Package for Population Genetics Data Analysis; University of Bern: Bern, Switzerland, 2006. [Google Scholar]
- Borovikova, E.A. Special traits of the genetic structure and origin of the population of vendace Coregonus albula of Pleshcheyevo Lake. Biol. Bull. 2017, 44, 245–250. [Google Scholar] [CrossRef]
- Borovikova, E.A.; Artamonova, V.S. Morphological specificities of vendace (Salmoniformes: Salmonidae: Coregoninae: Coregonus albula) population in Lake Pleshcheyevo (the Volga River basin): Relationships of two phylogenetic lineages in a new zone of secondary contact. Org. Divers. Evol. 2018, 18, 355–366. [Google Scholar] [CrossRef]
- Avise, J.C. Molecular population structure and the biogeographic history of a regional fauna: A case history with lessons for conservation biology. Oikos 1992, 63, 62–76. [Google Scholar] [CrossRef]
- Borovikova, E.A. Phylogeography of the Ciscoes Coregonus albula (L.) and C. sardinella Valenciennes in European North of Russia. Ph.D. Thesis, RUDN University, Moscow, Russia, 2009; 175p. (In Russia). [Google Scholar]
- Mangerud, J.; Astakhov, V.; Jakobsson, M.; Svendsen, J.I. Huge ice-lakes in Russia. J. Quat. Sci. 2001, 16, 773–777. [Google Scholar] [CrossRef]
- Mangerud, J.; Astakhov, V.; Murray, A.; Svendsen, J.I. The chronology of a large ice-dammed lake and the Barents-Kara ice sheet advances, northern Russia. Glob. Planet. Change 2001, 31, 319–334. [Google Scholar] [CrossRef]
- Mangerud, J.; Jakobsson, M.; Alexanderson, H.; Astakhov, V.; Clarke, G.K.C.; Henriksen, M.; Hjort, C.; Krinner, G.; Lunkka, J.-P.; Möller, P.; et al. Ice-dammed lakes and rerouting of the drainage of northern Eurasia during the Last Glaciation. Quat. Sci. Rev. 2004, 23, 1313–1332. [Google Scholar] [CrossRef]
- Nikulina, Y.S. Morphological and Molecular-Genetic Features of Least Cisco Coregonus sardinella Valenciennes from Different Types Water Bodies of the Putorana Plateau and Adjacent Regions. Ph.D. Thesis, Papanin Institute for Biology of Inland Waters RAS, Borok, Russia, 2020; 163p. (In Russian). [Google Scholar]
- Zadelenov, V.A.; Glushchenko, L.A.; Andrushchenko, P.Y.; Matasov, V.V.; Shadrin, E.N. Morpho-ecological characteristics of humpback whitefish (Coregonus lavaretus (L.)) from Sobachye Lake (Putorana Plateau). Vestn. Rybohozjajstvennoj Nauk. 2016, 3, 45–50. (In Russian) [Google Scholar]
- Borovikova, E.A.; Kodukhova, Y.V. Specific features of morphology, ecology, and mitochondrial DNA polymorphism of the whitefish (Coregonus lavaretus L.) from the Keret’ River as a new object for artificial propagation. Contemp. Probl. Ecol. 2018, 11, 259–270. [Google Scholar] [CrossRef]
- Blanchet, S.; Reyjol, Y.; April, J.; Mandrak, N.E.; Rodrígues, M.A.; Bernatchez, L.; Magnan, P. Phenotypic and phylogenetic correlates of geographic range size in Canadian freshwater fishes. Global Ecol. Biogeogr. 2013, 22, 1083–1094. [Google Scholar] [CrossRef]
- Ferguson, A.; Reed, T.E.; Cross, T.F.; McGinnity, P.; Prodöhl, P.A. Anadromy, potamodromy and residency in brown trout Salmo trutta: The role of genes and the environment. J. Fish Biol. 2019, 95, 692–718. [Google Scholar] [CrossRef] [PubMed]
- Hershey, A.E.; Beaty, S.; Fortino, K.; Keyse, M.; Mou, P.P.; O’Brien, W.J.; Ulseth, A.J.; Gettel, G.A.; Lienesch, P.W.; Luecke, C.; et al. Effect of landscape factors on fish distribution in arctic Alaskan lakes. Freshw. Biol. 2006, 51, 39–55. [Google Scholar] [CrossRef]
- Borovikova, E.A.; Makhrov, A.A. Adaptivnye vozmozhnosti populyacij i istoriya ih formirovaniya: Uspekh v rasprostranenii lososevyh zavisit ot razmera lednikovyh refugiumov (Adaptive capabilities of populations and the history of their formation: Success in the distribution of salmonids depends on the size of glacial refugia). In Sovremennye Problemy Evolyucii i Ekologii, Proceedings of the International Conference, Ulyanovsk, Russia, 7–9 April 2014; Ulyanovsk State Pedagogical University: Ulyanovsk, Russia, 2014. (In Russian) [Google Scholar]
- Carvalho, G.R. Evolutionary aspects of fish distribution: Genetic variability and adaptation. J. Fish Biol. 1993, 43, 53–73. [Google Scholar] [CrossRef]
- Campbell, M.A.; Joslin, S.E.K.; Goodbla, A.M.; Willmes, M.; Hobbs, J.A.; Lewis, L.S.; Finger, A.J. Polygenic discrimination of migratory phenotypes in an estuarine forage fish. G3 2022, 12, jkac133. [Google Scholar] [CrossRef]
- Grosswald, M.G.; Hughes, T.J. The Russian component of an Arctic ice sheet during the Last Glaciation Maximum. Quat. Sci. Rev. 2002, 21, 121–146. [Google Scholar] [CrossRef]
- Krinner, G.; Mangerud, J.; Jacobsson, M.; Crucifix, M.; Ritz, C.; Svendsen, J.I. Enhanced ice sheet growth in Eurasia owing to adjacent ice-dammed lakes. Nature 2004, 427, 429–432. [Google Scholar] [CrossRef]
- Svendsen, J.I.; Alexanderson, H.; Astakhov, V.I.; Demidov, I.; Dowdeswell, J.A.; Funder, S.; Gataullin, V.; Henriksen, M.; Hjort, C.; Houmark-Nielsen, M.; et al. Late Quaternary ice sheet history of northern Eurasia. Quat. Sci. Rev. 2004, 23, 1229–1271. [Google Scholar] [CrossRef]
- Panin, A.V.; Astakhov, V.I.; Lotsari, E.; Komatsu, G.; Lang, J.; Winsemann, J. Middle and Late Quaternary glacial lake-outburst floods, drainage diversions and reorganization of fluvial systems in northwestern Eurasia. Earth Sci. Rev. 2020, 201, 103069. [Google Scholar] [CrossRef]
- Borovikova, E.A.; Malina, J.I. Phylogeography of common whitefish (Coregonus lavaretus L.) of Northwestern Russia. Contemp. Probl. Ecol. 2018, 11, 286–296. [Google Scholar] [CrossRef]
- Sendek, D.S. Intra-species alliance of the European whitefish Coregonus lavaretus L. and vendace Coregonus albula L. from the Russian part of the Gulf of Finland and the largest lakes of the Eastern Baltic basin. Oceanology 2012, 52, 790–796. [Google Scholar]
- Sendek, D.S.; Ivanov, E.V.; Khodulov, V.V.; Novoselov, A.P.; Matkovsky, A.K.; Ljutikov, A.A. Genetic differentiation of coregonid populations in Subarctic areas. Adv. Limnol. 2013, 64, 223–246. [Google Scholar] [CrossRef]
- Sendek, D.S. Phylogenetic relationships in vendace and least cisco, and their distribution areas in western Eurasia. Ann. Zool. Fennici 2021, 58, 289–306. [Google Scholar] [CrossRef]
- Sendek, D.S. Electrophoretic studies of Coregonid fishes from across Russia. Arch. Hydroboil. Spec. Issues Adv. Limnol. 2002, 57, 35–55. [Google Scholar]
- Hansen, M.M.; Mensberg, K.-L.D.; Berg, S. Postglacial recolonization patterns and genetic relationships among whitefish (Coregonus sp.) populations in Denmark, inferred from mitochondrial DNA and microsatellite markers. Mol. Ecol. 1999, 8, 239–252. [Google Scholar] [CrossRef]
- Hansen, M.M.; Nielsen, E.E.; Mensberg, K.-L.D. Underwater but not out of sight: Genetic monitoring of effective population size in the endangered North Sea houting (Coregonus oxyrhynchus). Can. J. Fish. Aquat. Sci. 2006, 63, 780–787. [Google Scholar] [CrossRef]
- Bochkarev, N.A. Sigi kompleksa Coregonus lavaretus (Pisces: Coregonidae) iz vodoemov Sibiri: Filogeografiya i filogeniya. (Coregonid fishes of Coregonus lavaretus complex (Pisces: Coregonidae) of the Siberian reservoirs: Phylogeography and phylogeny). Ph.D. Thesis, Institute of Systematics and Ecology of Animals of Siberian Branch of RAS, Novosibirsk, Russia, 25 October 2022. (In Russian). [Google Scholar]
- Politov, D.V.; Baldina, S.N.; Gordon, N.Y. Molecular marking of genetic resources of coregonid fish from Siberia. Rybovod. Rybn. Hozjajstvo 2008, 8, 32–35. [Google Scholar]
- Baldina, S.N.; Gordon, N.Y.; Politov, D.V. Genetic differentiation of some coregonid fish species of Siberia. In Biologia, Biotekhnika Rezvedenia i Sostoyanie Zapasov Sigovykh Ryb (Biology, Breeding Biotechnology and the State of Coregonid Stocks), Proceedings of the VII International Scientific and Production Meeting, Tyumen’, Russia, 16–18 February 2010; Litvinenko, A.I., Reshetnikov, Y.S., Eds.; Gosrybtsentr: Tyumen’, Russia, 2010; pp. 5–9. (In Russian) [Google Scholar]
- Borovikova, E.A.; Makhrov, A.A. Taxonomy and origin of whitefish and ciscoes (Coregonus) in Europe: A morphoecological approach. Trans. Karelian Res. Cent. Russ. Acad. Sci. 2013, 6, 105–115. (In Russian) [Google Scholar]
- Chertoprud, E.S.; Novichkova, A.A.; Novikov, A.A.; Fefilova, E.B.; Vorobjeva, L.D.; Pechenkin, D.S.; Glubokov, A.I. Assemblages of meiobenthic and planktonic microcrustaceans (Cladocera and Copepoda) from small water bodies of mountain Subarctic (Putorana Plateau, Middle Siberia). Diversity 2022, 14, 492. [Google Scholar] [CrossRef]
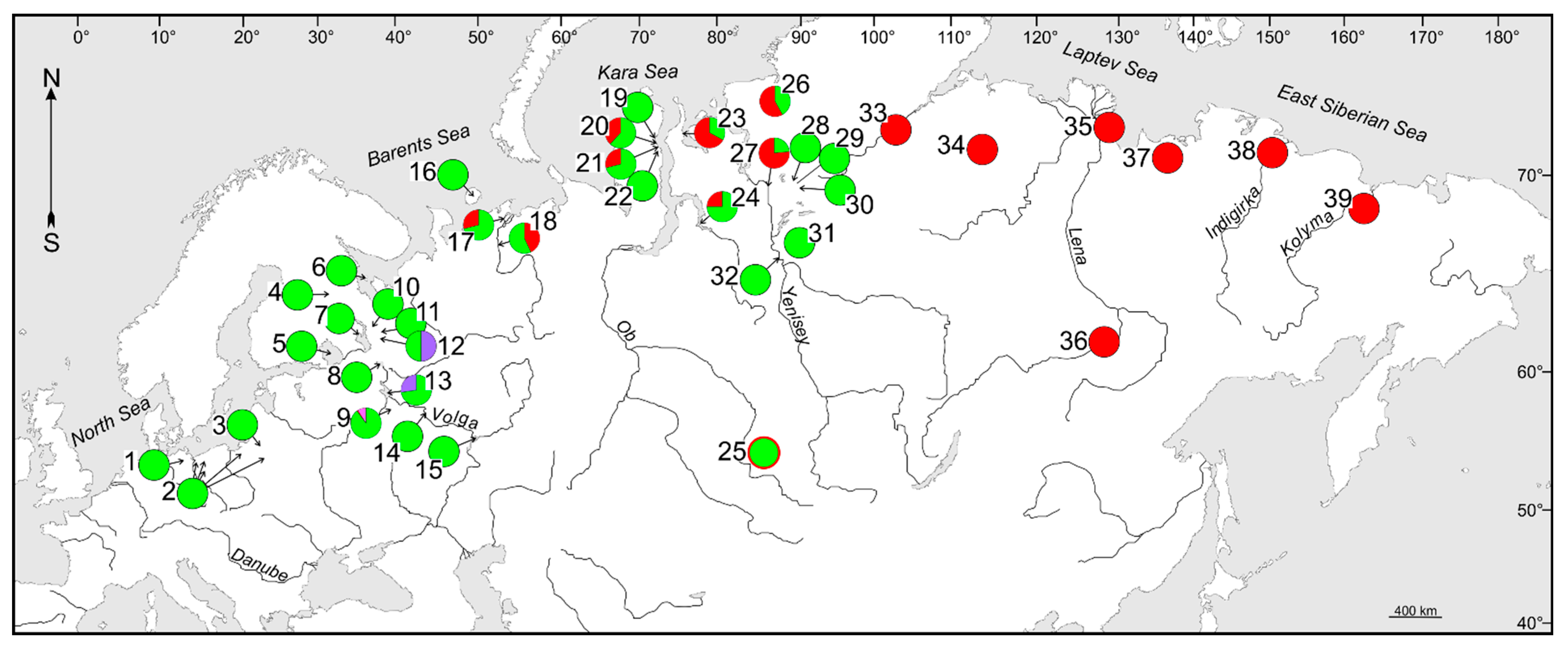
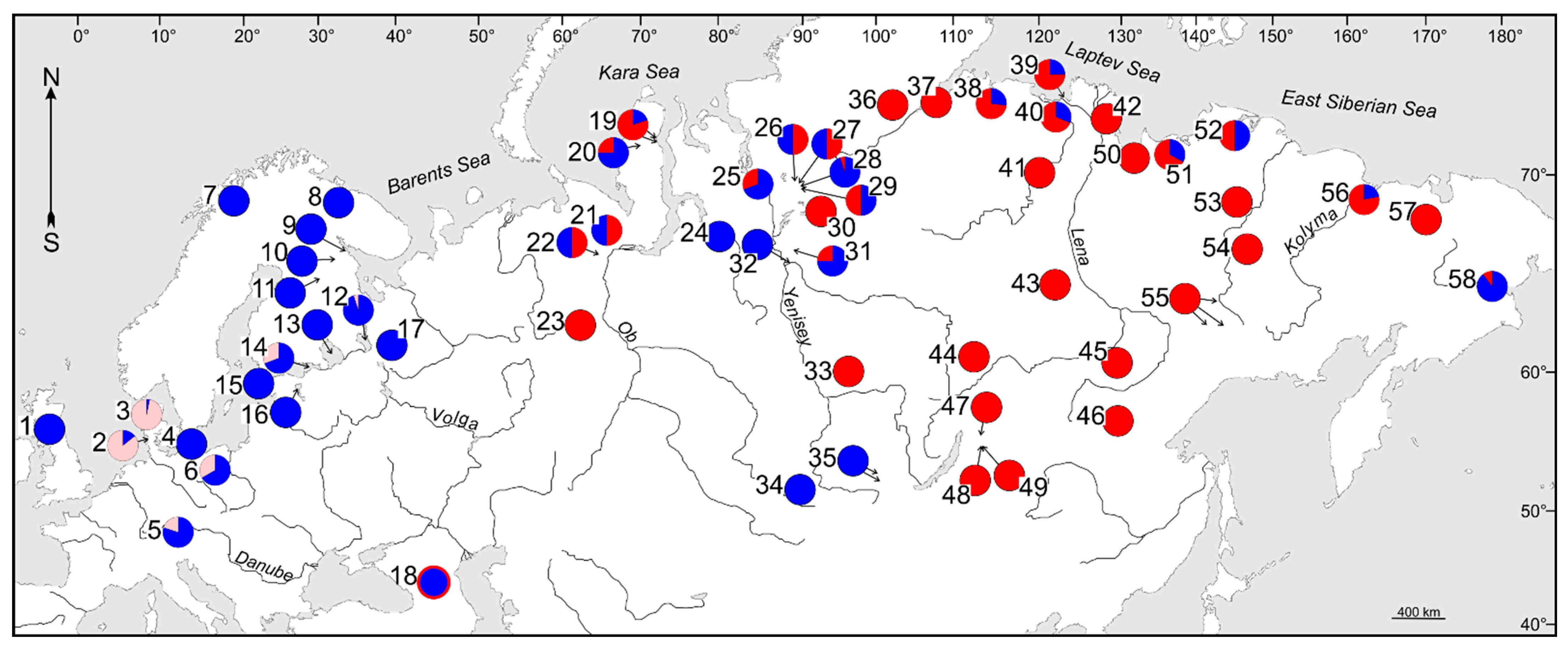
| Species | NL | Contact Zone in Arctic Eurasia | Source of Data |
|---|---|---|---|
| Salmo trutta | 5–7 | Great Britain, the basins of the North and Baltic Seas, southern Europe | [31,32,33] |
| Salvelinus alpinus | 5 | Western Greenland, lakes of the Norilo-Pyasinskaya water system (Taymyr Peninsula), the Beringian region | [34,35,36,37,38] |
| Thymallus arcticus | 2 | for T. thymallus: basins of the Danube and Weser rivers, Lake Constance area; for T. arcticus and T. thymallus: the northern European regions (Norway, Sweden, Finland), the Kola Peninsula, the northern Dvina River, basins of the Pechora and Ob rivers; for T. thymallus and other Siberian Thymallus sp.: Lake Khantayskoye | [39,40,41,42,43,44] |
| Thymallus thymallus | 3 | ||
| Osmerus mordax | 7 | the Kandalaksha Bay, the Kolguyev Island | [21,45,46] |
| Osmerus eperlanus | |||
| Esox lucius | 3 | basins of the Baltic and North Seas, the Middle Danubian drainage, southern Europe | [28,47] |
| Rutilus sp. | 2 | Aegean and Baltic Seas’ basins, the White Sea basin (the Northern Dvina and Onega rivers), the Black Sea basin (the Upper Dnieper and lower reaches of the Don River), the Caspian Sea basin (the Volga River basin) | [25,48] |
| Lota lota | 5 | the Baltic region | [49] |
| Gasterosteus aculeatus | 6–7 | the north-eastern part of the Atlantic Ocean, the Phȏne River, the White and Barents Seas, the Black Sea | [23,50] |
| Pungitius sp. | 3 | from the north of France (the North Sea, English Channel, Seine, Meuse, and Rhine drainages) to the Netherlands | [24,51] |
| 2 | the Baltic Sea, Skagerrak/Kattegat | [52] | |
| Perca fluviatilis | 3–9 | the Oslofjord (Norway), the eastern part of the Baltic region, the North Sea basin, the Danube River, southern Europe, Lake Constance | [53,54,55] |
| Cottus gobio | 7 | Northern Scandinavia (the northern Baltic), the Lower Rhine | [56,57,58] |
| Lineages | Level of Clades Subdivison | N | S | k | τ | Theta Obs. | Theta0 | Ri | Tajima’s D | Fs | Time Since Expansion |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Cisco species | |||||||||||
| All haplotypes | 2–1 | 240 | 137 | 6.706 | 1.770 | 24.770 | 4.936 | 0.008 | −2.259 ** | −24.611 *** | 91 425–45 712 |
| IC | 1–1 | 59 | 53 | 4.414 | 4.414 | 11.838 | 0 | 0.018 | −2.130 * | −25.751 *** | 227 052–113 529 |
| IIC | 1–2 | 167 | 93 | 3.967 | 3.591 | 17.392 | 0.396 | 0.030 | −2.421 ** | −25.691 *** | 184 912–92 456 |
| IIIC | 1–3 | 7 | 3 | 1.238 | 0.566 | 1.224 | 0.673 | 0.277 | 0.050 NS | 0.406 NS | – |
| IVC | 1–4 | 7 | 5 | 2.095 | 2.095 | 2.041 | 0 | 0.091 | 0.132 NS | −1.447 NS | – |
| Whitefish | |||||||||||
| All haplotypes | 3–1 | 562 | 175 | 6.058 | 4.967 | 27.939 | 1.091 | 0.006 | −2.313 ** | −24.406 *** | 255 242–127 621 |
| IW | 2–1 | 330 | 123 | 5.072 | 4.280 | 20.550 | 0.791 | 0.010 | −2.272 ** | −24.898 *** | 219 938–109 969 |
| IIW | 2–2 | 232 | 99 | 3.526 | 3.526 | 17.437 | 0 | 0.026 | −2.447 ** | −25.798 *** | 180 820–90 410 |
| IW1 | 1–1 | 91 | 12 | 1.878 | 0.998 | 2.361 | 0.880 | 0.027 | −0.550 NS | −1.242 NS | – |
| IW2 | 1–2 | 239 | 113 | 4.646 | 4.202 | 19.830 | 0.444 | 0.018 | −2.355 ** | −25.211 *** | 215 930–107 965 |
| CA | CS | CF | CL | COx | CPr | CBt | CAn | CFl | CCl | CMg | CCd | CUs | CN | PC | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CA | 0–3.3 0.1 | ||||||||||||||
| CS | 0–3.4 0.1 | 0–1.6 0.1 | |||||||||||||
| CF | 0–3.1 0.6 | 0.03–0.1 0.06 | 0.02–0.04 0.03 | ||||||||||||
| CL | 3.1–4.5 3.6 | 2.8–4.3 3.5 | 3.0–4.1 3.5 | 0–1.4 0.1 | |||||||||||
| COx | 3.3–4.0 3.5 | 2.9–3.8 3.4 | 3.4–3.6 3.5 | 0–1.2 0.06 | 0–0.1 0.02 | ||||||||||
| CPr | 2.9–4.7 3.7 | 2.8–4.2 3.5 | 3.4–3.8 3.6 | 0.01–1.4 0.06 | 0.02–0.1 0.06 | 0.01–0.1 0.03 | |||||||||
| CBt | 3.3–3.6 3.5 | 3.0–3.8 3.4 | 3.1–3.6 3.5 | 0–1.1 0.06 | 0.03–0.1 0.05 | 0.03–0.1 0.06 | 0–0.05 0.02 | ||||||||
| CAn | 4.0–4.7 4.1 | 3.9–4.2 4.0 | 3.9–4.4 4.1 | 2.9–3.4 3.1 | 2.5–3.6 3.1 | 2.5–3.4 3.1 | 2.6–3.5 3.0 | 0.01–0.1 0.04 | |||||||
| CFl | 3.5–4.1 3.6 | 3.2–3.9 3.5 | 3.5–3.7 3.6 | 1.4–2.0 1.7 | 0.02–2.0 1.7 | 1.4–2.0 1.7 | 1.1–1.9 1.5 | 2.0–2.9 2.5 | 0.02–0.1 0.04 | ||||||
| CCl | 2.9–3.6 3.1 | 2.7–3.4 3.1 | 3.1–3.3 3.15 | 1.6–2.2 1.9 | 1.8–2.1 1.9 | 1.6–2.2 1.9 | 1.7–2.1 1.8 | 2.5–3.4 2.8 | 1.7–2.2 1.9 | 0–0.03 0.01 | |||||
| CMg | 3.5–4.3 3.7 | 3.3–4.3 3.6 | 3.5–4.1 3.7 | 1.8–2.4 2.1 | 1.9–2.5 2.1 | 1.7–2.6 2.1 | 0–2.5 1.7 | 2.7–3.8 3.2 | 1.1–1.9 1.5 | 1.8–2.5 2.0 | 0.01–0.1 0.03 | ||||
| CCd | 4.5–5.3 4.7 | 4.5–5.1 4.6 | 4.4–4.9 4.7 | 2.6–3.3 2.9 | 2.8–3.2 3.0 | 2.8–3.3 3.1 | 2.4–3.1 2.8 | 3.2–4.0 3.6 | 2.3–2.8 2.6 | 2.9–3.1 2.95 | 2.4–2.8 2.6 | 0.04 | |||
| CUs | 4.4–5.2 4.6 | 4.1–4.8 4.5 | 4.3–4.8 4.6 | 2.7–3.2 2.9 | 2.9–3.3 3.0 | 2.6–3.3 3.0 | 2.3–3.2 2.8 | 3.1–4.0 3.5 | 2.2–2.8 2.5 | 2.7–3.1 2.8 | 2.3–2.8 2.5 | 0.04–0.1 0.05 | 0–0.04 0.02 | ||
| CN | 3.8–4.3 4.0 | 3.8–4.6 3.9 | 4.1–4.4 4.3 | 2.4–3.0 2.6 | 2.9–3.2 3.0 | 2.7–3.2 2.9 | 2.6–3.2 2.9 | 3.4–4.1 3.7 | 2.8–3.2 0.3 | 2.7–3.0 2.8 | 3.2–3.6 3.4 | 4.2–4.3 4.25 | 4.0–4.3 4.2 | nc | |
| PC | 17.1–17.5 17.2 | 17.0–17.6 17.3 | 17.4–17.7 17.6 | 16.9–17.5 17.3 | 17.7–17.9 17.8 | 17.5–18.0 17.7 | 17.7–18.0 17.9 | 16.6–17.0 16.8 | 17.0–17.1 17.05 | 17.9–18.0 17.95 | 17.9–18.4 18.1 | 17.5–17.8 17.7 | 17.5–17.9 17.7 | 17.9 | nc |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Borovikova, E.; Nikulina, Y. The Contact Zone of Phylogenetic Lineages of Freshwater Fish in Arctic Eurasia: Genetic Polymorphism of Coregonid Populations. Diversity 2023, 15, 163. https://doi.org/10.3390/d15020163
Borovikova E, Nikulina Y. The Contact Zone of Phylogenetic Lineages of Freshwater Fish in Arctic Eurasia: Genetic Polymorphism of Coregonid Populations. Diversity. 2023; 15(2):163. https://doi.org/10.3390/d15020163
Chicago/Turabian StyleBorovikova, Elena, and Yulia Nikulina. 2023. "The Contact Zone of Phylogenetic Lineages of Freshwater Fish in Arctic Eurasia: Genetic Polymorphism of Coregonid Populations" Diversity 15, no. 2: 163. https://doi.org/10.3390/d15020163
APA StyleBorovikova, E., & Nikulina, Y. (2023). The Contact Zone of Phylogenetic Lineages of Freshwater Fish in Arctic Eurasia: Genetic Polymorphism of Coregonid Populations. Diversity, 15(2), 163. https://doi.org/10.3390/d15020163






