Terrestrial Species of Drouetiella (Cyanobacteria, Oculatellaceae) from the Russian Arctic and Subarctic Regions and Description of Drouetiella ramosa sp. nov.
Abstract
1. Introduction
2. Materials and Methods
2.1. Sampling
2.2. Isolation of Strains
2.3. Morphological Characterization
2.4. Specimens Sampling
2.5. Molecular Analyses
3. Results
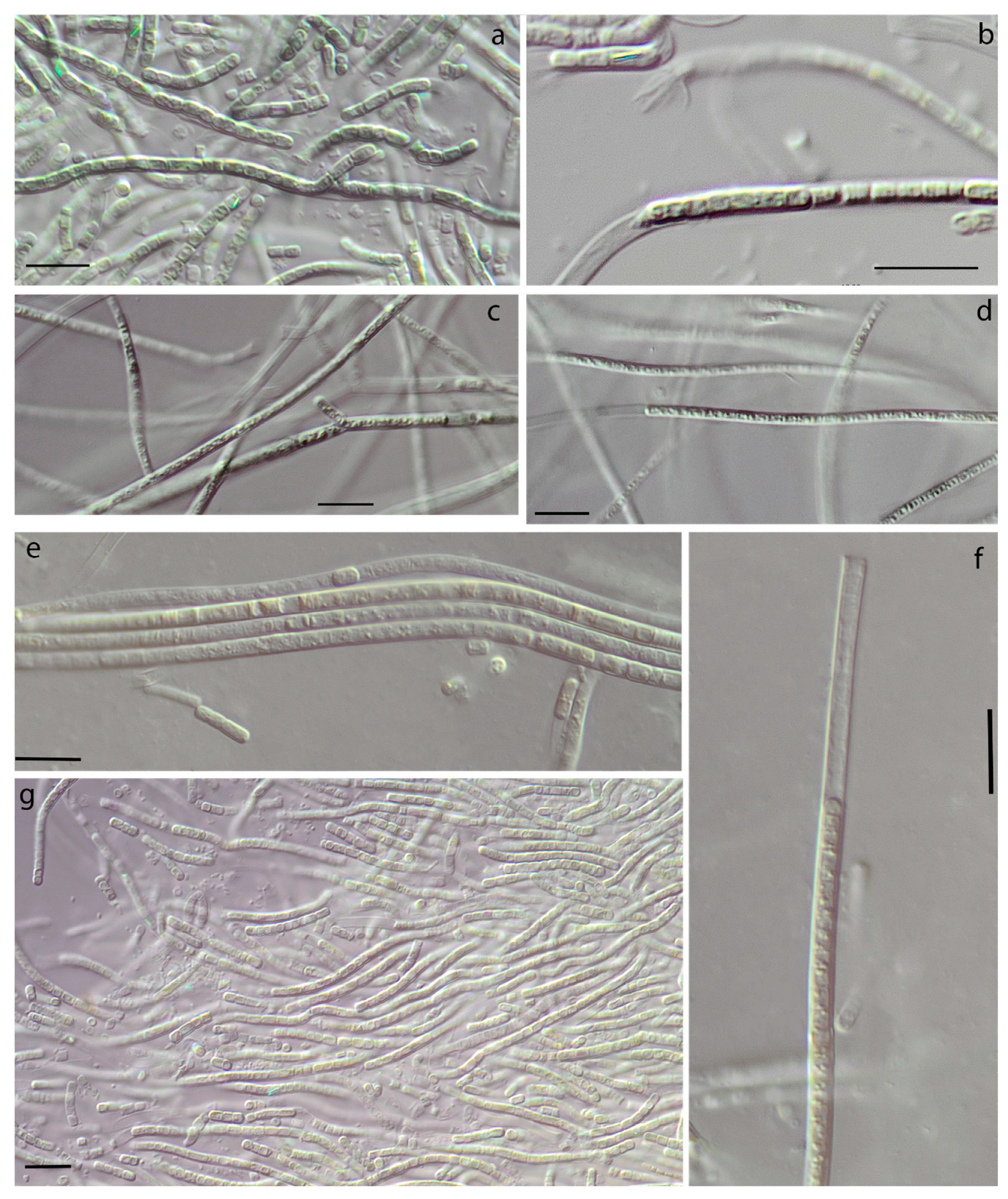
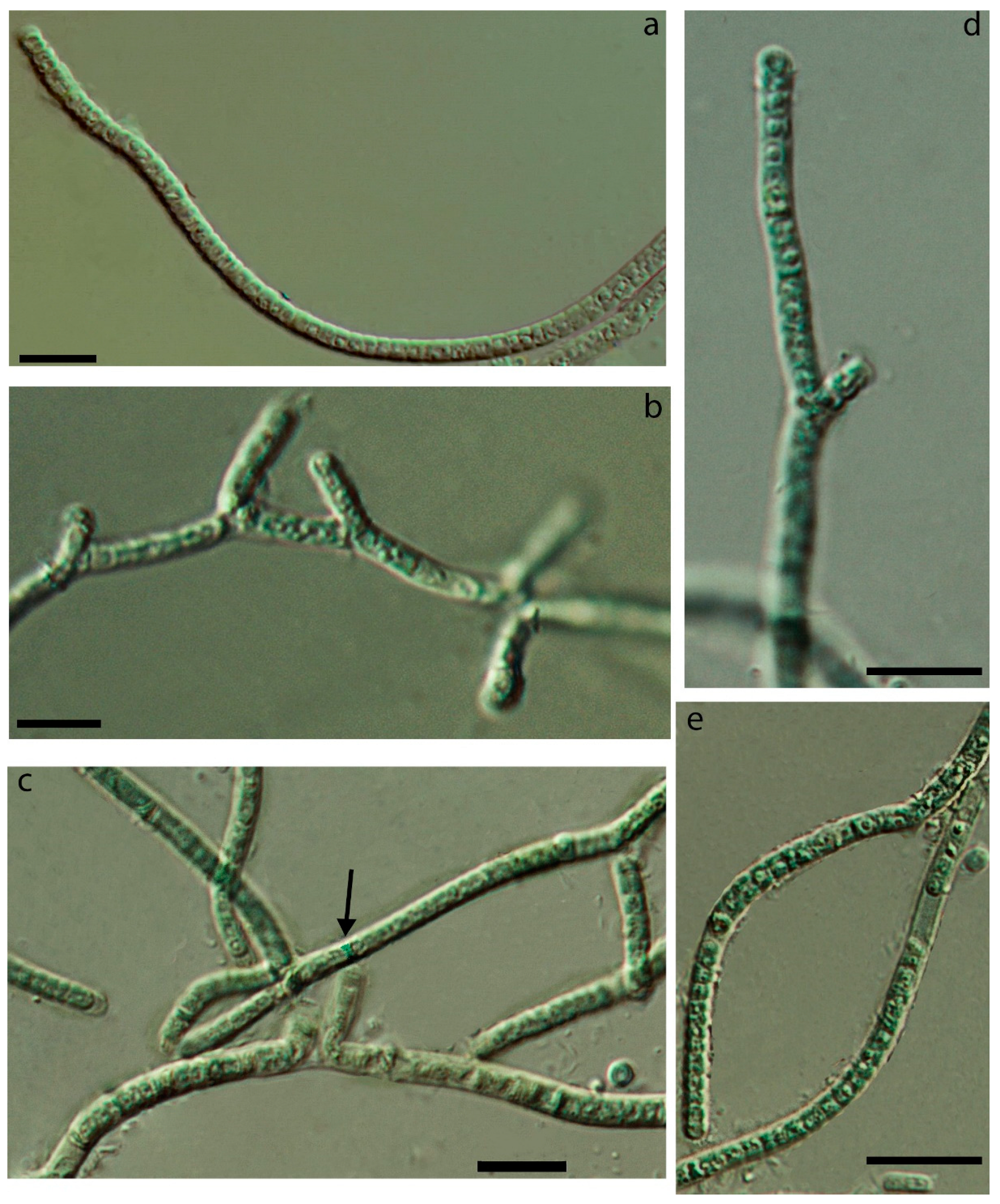
3.1. Morphology
3.2. Phylogeny
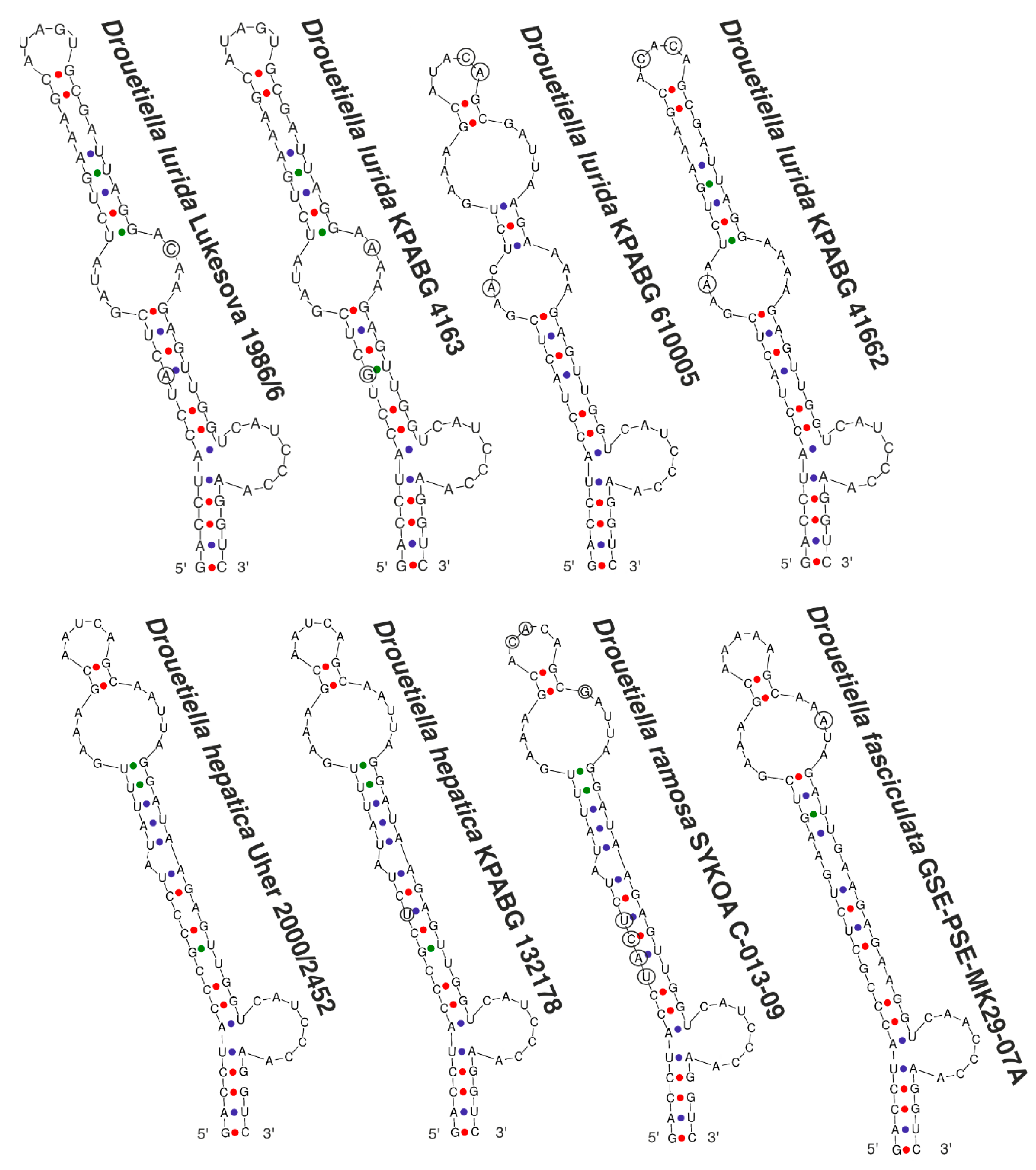
3.3. The Secondary Structure of Conserved Domains of the 16S-23S ITS rRNA
3.4. Taxonomic Description
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Whitton, B.A.; Potts, M. Introduction to the Cyanobacteria. In The Ecology of Cyanobacteria; Whitton, B.A., Potts, M., Eds.; Kluwer Academic Publishers: Dordrecht, The Netherlands, 2002; pp. 1–11. [Google Scholar] [CrossRef]
- Vincent, W.F. Cyanobacterial Dominance in the Polar Regions. In The Ecology of Cyanobacteria; Whitton, B.A., Potts, M., Eds.; Kluwer Academic Publishers: Dordrecht, The Netherlands, 2002; pp. 321–340. [Google Scholar] [CrossRef]
- Davydov, D.; Patova, E. The diversity of Cyanoprokaryota from freshwater and terrestrial habitats in the Eurasian Arctic and Hypoarctic. Hydrobiologia 2018, 811, 119–138. [Google Scholar] [CrossRef]
- Davydov, D. Diversity of the Cyanoprokaryota in polar deserts of Rijpfjorden east coast, North-East Land (Nordaustlandet) Island, Spitsbergen. Algol. Stud. 2013, 142, 29–44. [Google Scholar] [CrossRef]
- Davydov, D. Diversity of the Cyanoprokaryota of the area of settlement Pyramiden, West Spitsbergen Island, Spitsbergen archipelago. Folia Cryptogam. Est. 2014, 51, 13–23. [Google Scholar] [CrossRef]
- Davydov, D. Diversity of the Cyanoprokaryota in polar deserts of Innvika cove North-East Land (Nordaustlandet) Island, Spitsbergen. Czech Polar Rep. 2016, 6, 66–79. [Google Scholar] [CrossRef]
- Davydov, D. Cyanoprokaryotes of the west part of Oscar II Land, West Spitsbergen Island, Spitsbergen archipelago. Czech Polar Rep. 2017, 7, 94–108. [Google Scholar] [CrossRef]
- Davydov, D. Cyanobacterial diversity of Svalbard Archipelago. Polar Biol. 2021, 44, 1967–1978. [Google Scholar] [CrossRef]
- Davydov, D.; Shalygin, S.; Vilnet, A. New cyanobacterium Nodosilinea svalbardensis sp. nov. (Prochlorotrichaceae, Synechococcales) isolated from alluvium in Mimer river valley of the Svalbard archipelago. Phytotaxa 2020, 442, 61–79. [Google Scholar] [CrossRef]
- Shalygin, S.; Shalygina, R.; Johansen, J.R.; Pietrasiak, N.; Berrendero Gómez, E.; Bohunická, M.; Mareš, J.; Sheil, C.A. Cyanomargarita gen. nov. (Nostocales, Cyanobacteria): Convergent evolution resulting in a cryptic genus. J. Phycol. 2017, 53, 762–777. [Google Scholar] [CrossRef]
- Shalygin, S.; Shalygina, R.R.; Redkina, V.V.; Gargas, C.B.; Johansen, J.R. Description of Stenomitos kolaenensis and S. hiloensis sp. nov. (Leptolyngbyaceae, Cyanobacteria) with an emendation of the genus. Phytotaxa 2020, 440, 108–128. [Google Scholar] [CrossRef]
- Davydov, D.; Redkina, V. Algae and cyanoprokaryotes on naturally overgrowing ash dumps of the Apatity thermal power station (Murmansk Region). Proc. KarRC RAS 2021, 51, 51–68. [Google Scholar] [CrossRef]
- Davydov, D.; Vilnet, A. Review of the Cyanobacterial Genus Phormidesmis (Leptolyngbyaceae) with the Description of Apatinema gen. nov. Diversity 2022, 14, 731. [Google Scholar] [CrossRef]
- Rodina, O.; Davydov, D.; Vlasov, D. Lithobiotic cyanobacteria diversity of the Karelian Isthmus. Biol. Commun. 2022, 67, 97–112. [Google Scholar] [CrossRef]
- Patova, E.N.; Novakovskaya, I.V.; Deneva, S.V. Influence of edaphic and orographic factors on the diversity of algae communities of biological soil crusts on medallion spots of the Polar and Subpolar Urals. Eurasian Soil Sci. 2018, 3, 318–330. [Google Scholar] [CrossRef]
- Novakovskaya, I.V.; Patova, E.N.; Kulyugina, E.E. Changes of cyanoprokariota and algae diversity during the overgrowing of bare spots in the mountain tundra communities of the Northern Urals. Bot. Zhurnal 2019, 104, 569–586. [Google Scholar] [CrossRef]
- Novakovskaya, I.V.; Dubrovskiy, Y.A.; Patova, E.N.; Novakovskiy, A.B.; Sterlyagova, I.N. Influence of ecological factors on soil algae in different types of mountain tundra and sparse forests in the Northern Urals. Phycologia 2020, 59, 320–329. [Google Scholar] [CrossRef]
- Novakovskaya, I.V.; Patova, E.N.; Dubrovskiy, Y.A.; Novakovskiy, A.B.; Kulyugina, E.E. Distribution of algae and cyanobacteria of biological soil crusts along the elevation gradient in mountain plant communities at the Northern Urals (Russian European Northeast). J. Mt. Sci. 2022, 19, 637–646. [Google Scholar] [CrossRef]
- Davydov, D. Cyanobacterial Diversity of the Northern Polar Ural Mountains. Diversity 2021, 13, 607. [Google Scholar] [CrossRef]
- Zammit, G.; Billi, D.; Albertano, P. The subaerophytic cyanobacterium Oculatella subterranea (Oscillatoriales, Cyanophyceae) gen. et sp. nov.: A cytomorphological and molecular description. Eur. J. Phycol. 2012, 47, 341–354. [Google Scholar] [CrossRef]
- Sciuto, K.; Moschin, E.; Moro, I. Cryptic cyanobacterial diversity in the Giant Cave (Trieste, Italy): The new genus Timaviella (Leptolyngbyaceae). Cryptogam. Algol. 2017, 38, 285–323. [Google Scholar] [CrossRef]
- Mai, T.; Johansen, J.R.; Pietrasiak, N.; Bohunicka, M.; Martin, M.P. Revision of the Synechococcales (Cyanobacteria) through recognition of four families including Oculatellaceae fam. nov. and Trichocoleaceae fam. nov. and six new genera containing 14 species. Phytotaxa 2018, 365, 1–59. [Google Scholar] [CrossRef]
- Sciuto, K.; Moro, I. Detection of the new cosmopolitan genus Thermoleptolyngbya (Cyanobacteria, Leptolyngbyaceae) using the 16S rRNA gene and 16S-23S ITS region. Mol. Phylogenetics Evol. 2016, 105, 15–35. [Google Scholar] [CrossRef] [PubMed]
- Jahodářová, E.; Dvořák, P.; Hašler, P.; Holušová, K.; Poulíčková, A. Elainella gen. nov.: A new tropical cyanobacterium characterized using a complex genomic approach. Eur. J. Phycol. 2018, 53, 39–51. [Google Scholar] [CrossRef]
- Strunecky, O.; Raabova, L.; Bernardova, A.; Ivanova, A.P.; Semanova, A.; Crossley, J.; Kaftan, D. Diversity of cyanobacteria at the Alaska North Slope with description of two new genera: Gibliniella and Shackletoniella. FEMS Microbiol. Ecol. 2020, 96, fiz189. [Google Scholar] [CrossRef]
- Chakraborty, S.; Maruthanayagam, V.; Achari, A.; Pramanik, A.; Jaisankar, P.; Mukherjee, J. Aerofilum fasciculatum gen. nov., sp. nov. (Oculatellaceae) and Euryhalinema pallustris sp. nov. (Prochlorotrichaceae) isolated from an Indian mangrove forest. Phytotaxa 2021, 522, 165–186. [Google Scholar] [CrossRef]
- Pietrasiak, N.; Reeve, S.; Osorio-Santos, K.; Lipson, D.A.; Johansen, J.R. Trichotorquatus gen. nov.—A new genus of soil cyanobacteria discovered from American drylands. J. Phycol. 2021, 57, 886–902. [Google Scholar] [CrossRef] [PubMed]
- Jasser, I.; Panou, M.; Khomutovska, N.; Sandzewicz, M.; Panteris, E.; Niyatbekov, T.; Łach, Ł.; Kwiatowski, J.; Kokociński, M.; Gkelis, S. Cyanobacteria in hot pursuit: Characterization of cyanobacteria strains, including novel taxa, isolated from geothermal habitats from different ecoregions of the world. Mol. Phylogenetics Evol. 2022, 170, 107454. [Google Scholar] [CrossRef]
- Tawong, W.; Pongcharoen, P.; Nishimura, T.; Saijuntha, W. Siamcapillus rubidus gen. et sp. nov. (Oculatellaceae), a novel filamentous cyanobacterium from Thailand based on molecular and morphological analyses. Phytotaxa 2022, 558, 33–52. [Google Scholar] [CrossRef]
- Kützing, F.T. Species Algarum; F.A. Brockhaus: Lipsiae, Germany, 1849; pp. 1–922. [Google Scholar] [CrossRef]
- Andersen, R.A. Algal Culturing Techniques; Elsevier: New York, NY, USA, 2005; pp. 1–578. [Google Scholar]
- Kotai, J. Instructions for Preparation of Modified Nutrient Solution Z8 for Algae; 11/69; Norwegian Institute for Water Research: Oslo, Norway, 1972; pp. 1–5. [Google Scholar]
- Waterbury, J.B. The Cyanobacteria—Isolation, purification and identification of major groups of Cyanobacteria. In The Prokaryotes; Dworkin, M., Falkow, S., Rosenberg, E., Schleifer, K.-H., Stackebrandt, E.H., Eds.; Springer: New York, NY, USA, 2006; pp. 1053–1073. [Google Scholar] [CrossRef]
- Melekhin, A.V.; Davydov, D.A.; Borovichev, E.A.; Shalygin, S.S.; Konstantinova, N.A. CRIS—Service for input, storage and analysis of the biodiversity data of the cryptogams. Folia Cryptogam. Est. 2019, 56, 99–108. [Google Scholar] [CrossRef]
- Clark, K.; Karsch-Mizrachi, I.; Lipman, D.J.; Ostell, J.; Sayers, E.W. GenBank. Nucleic Acids Res. 2016, 44, 67–72. [Google Scholar] [CrossRef]
- Hall, T.A. BioEdit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 1999, 41, 95–98. [Google Scholar]
- Nguyen, L.-T.; Schmidt, H.A.; von Haeseler, A.; Minh, B.Q. IQ-TREE: A fast and effective stochastic algorithm for estimating maximum likelihood phylogenies. Mol. Biol. Evol. 2015, 32, 268–274. [Google Scholar] [CrossRef]
- Ronquist, F.; Teslenko, M.; Mark, P.; van der Ayres, D.L.; Darling, A.; Höhna, S.; Larget, B.; Liu, L.; Suchard, M.A.; Hülsenbeck, J.P. MrBayes 3.2: Efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 2012, 61, 539–542. [Google Scholar] [CrossRef]
- Kalyaanamoorthy, S.; Minh, B.Q.; Wong, T.K.F.; von Haeseler, A.; Jermiin, L.S. ModelFinder: Fast model selection for accurate phylogenetic estimates. Nat. Methods 2017, 14, 587–589. [Google Scholar] [CrossRef]
- Hoang, D.T.; Chernomor, O.; von Haeseler, A.; Minh, B.Q.; Vinh, L.S. UFBoot2: Improving the ultrafast bootstrap approximation. Mol. Biol. Evol. 2018, 35, 518–522. [Google Scholar] [CrossRef]
- Rambaut, A.; Drummond, A.J. Tracer v1.4. 2007. Available online: http://beast.bio.ed.ac.uk/Tracer (accessed on 10 November 2021).
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular Evolutionary Genetics Analysis version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef]
- Zuker, M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003, 31, 3406–3415. [Google Scholar] [CrossRef]
- Yarza, P.; Yilmaz, P.; Pruesse, E.; Glöckner, L.W.; Schleifer, K.H.; Witman, W.B.; Euzéby, J.; Amann, R. Rosselló-Móra Uniting the classification of cultured and uncultured bacteria and archaea using 16S rRNA gene sequences. Nat. Rev. Microbiol. 2014, 12, 635–645. [Google Scholar] [CrossRef]
- González-Resendiz, L.; Johansen, J.R.; León-Tejera, H.; Sánchez, L.; Segal-Kischinevzky, C.; Escobar-Sánchez, V.; Morales, M. A bridge too far in naming species: A total evidence approach does not support recognition of four species in Desertifilum (Cyanobacteria). J. Phycol. 2019, 55, 898–911. [Google Scholar] [CrossRef]
- Komárek, J.; Anagnostidis, K. Cyanoprokaryota 2. Teil: Oscillatoriales; Büdel, B., Gärtner, G., Krienitz, L., Schlager, M., Eds.; Spektrum Akademischer Verlag: Heidelberg, Germany, 2008; pp. 1–759. [Google Scholar]
- Voronichin, N.N. Materialy k al’goflore i rastitel’nosti mineral’nykh istochnikov gruppy K.M.V. Tr. Bal’neol.-Kavkaz. Miner. Vodakh. 1927, 5, 90–121. [Google Scholar]
- Boye Petersen, J. The fresh-water cyanophyceae of Iceland. Arb. Bot. Have København 1923, 2, 251–324. [Google Scholar]
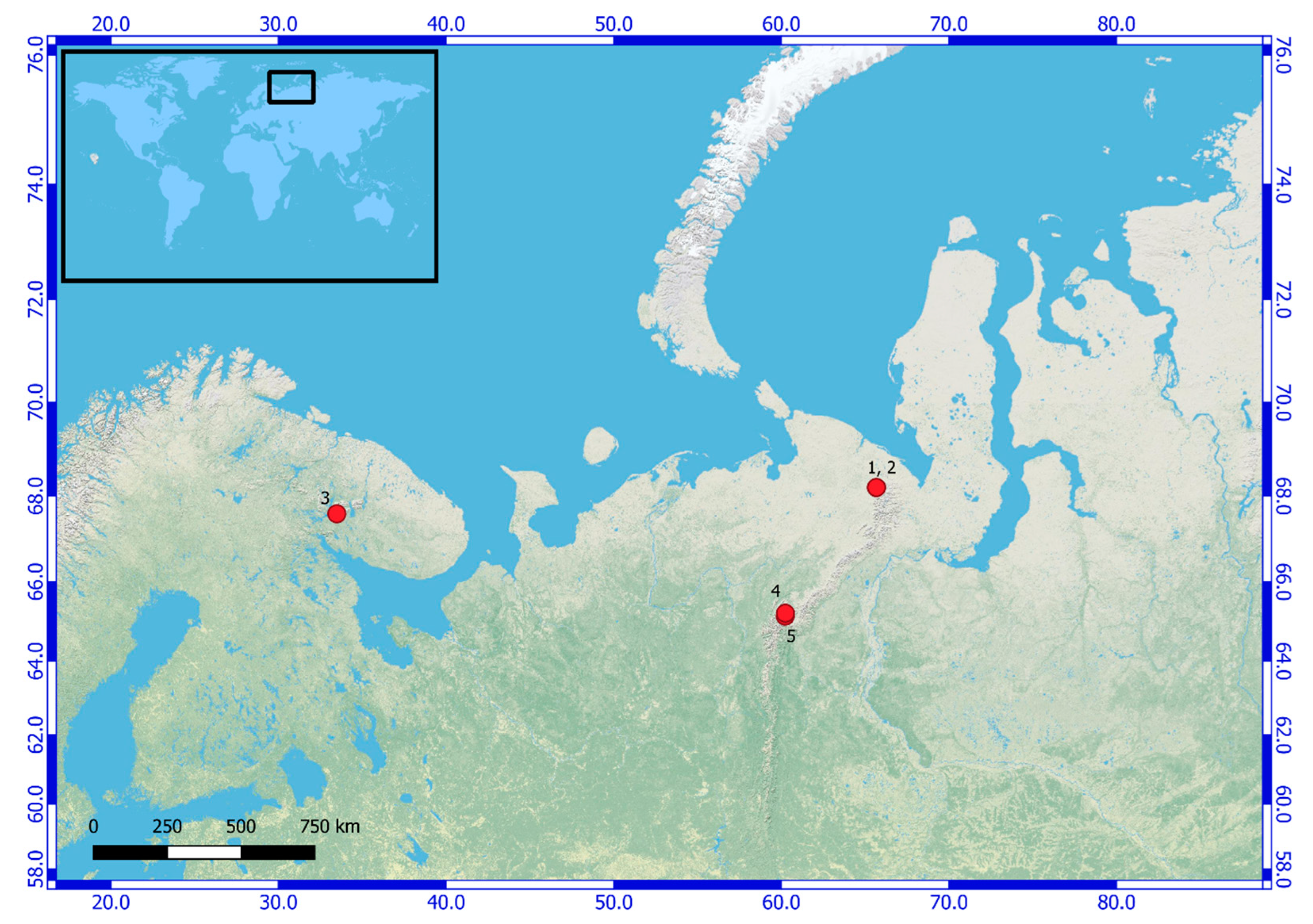


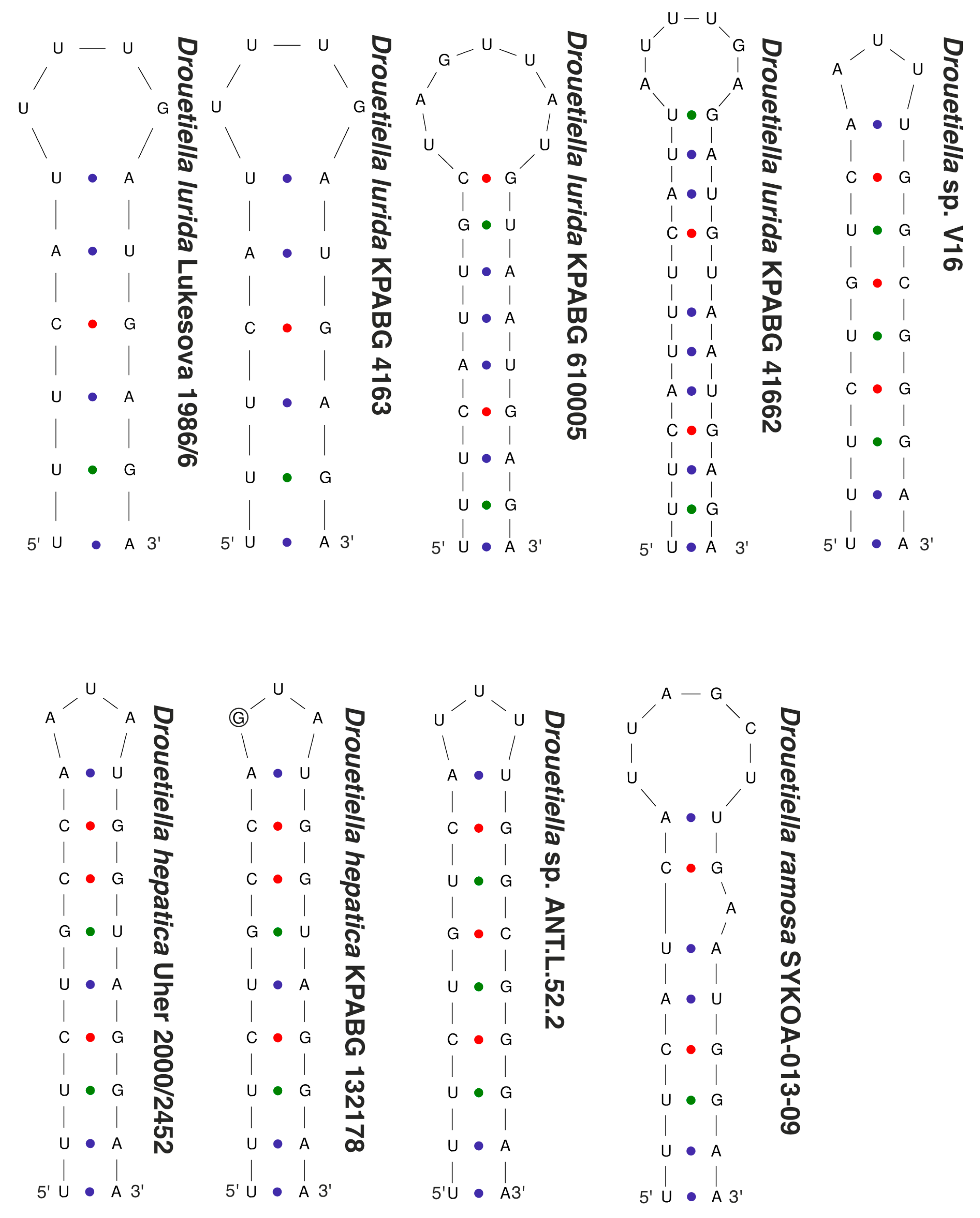
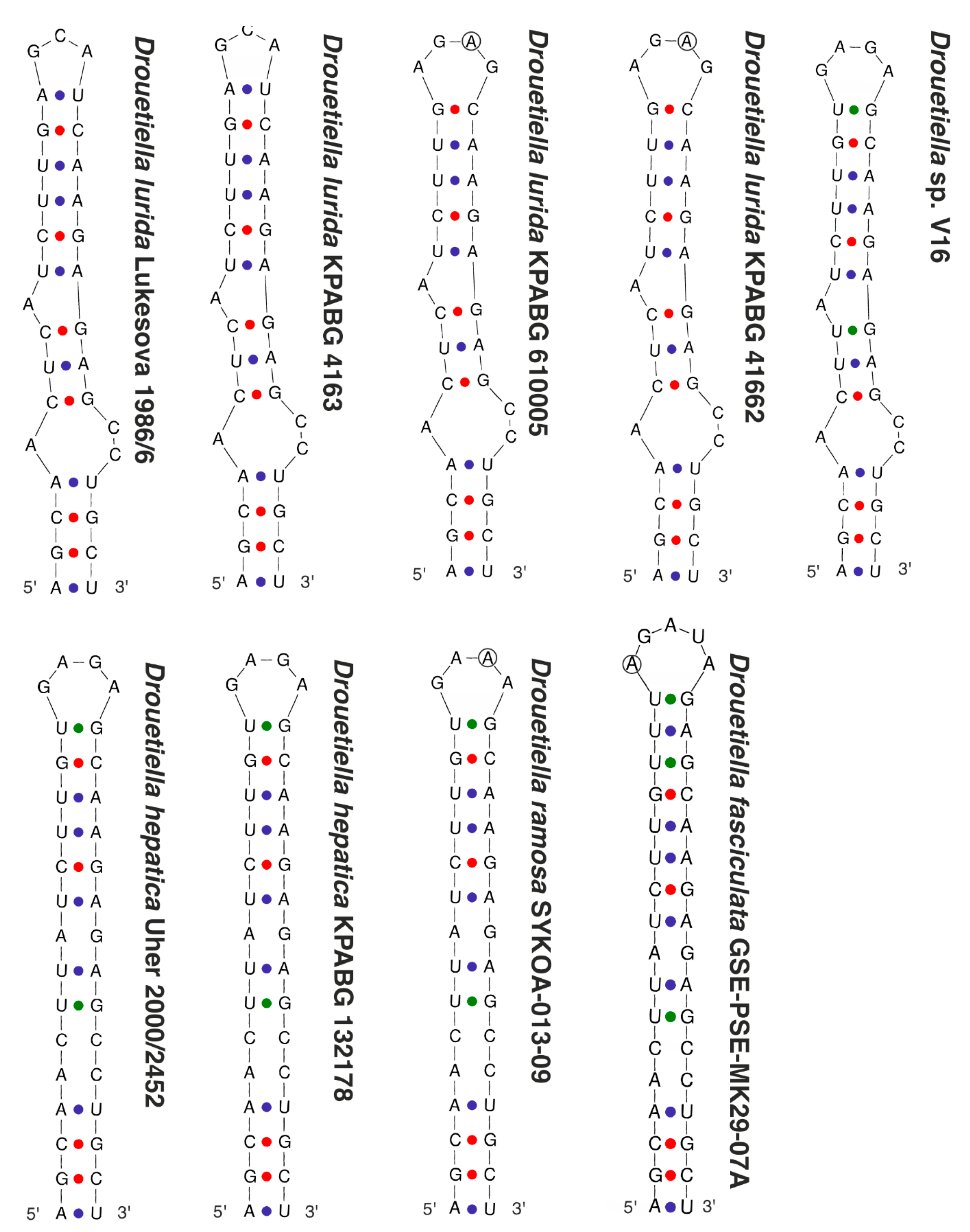
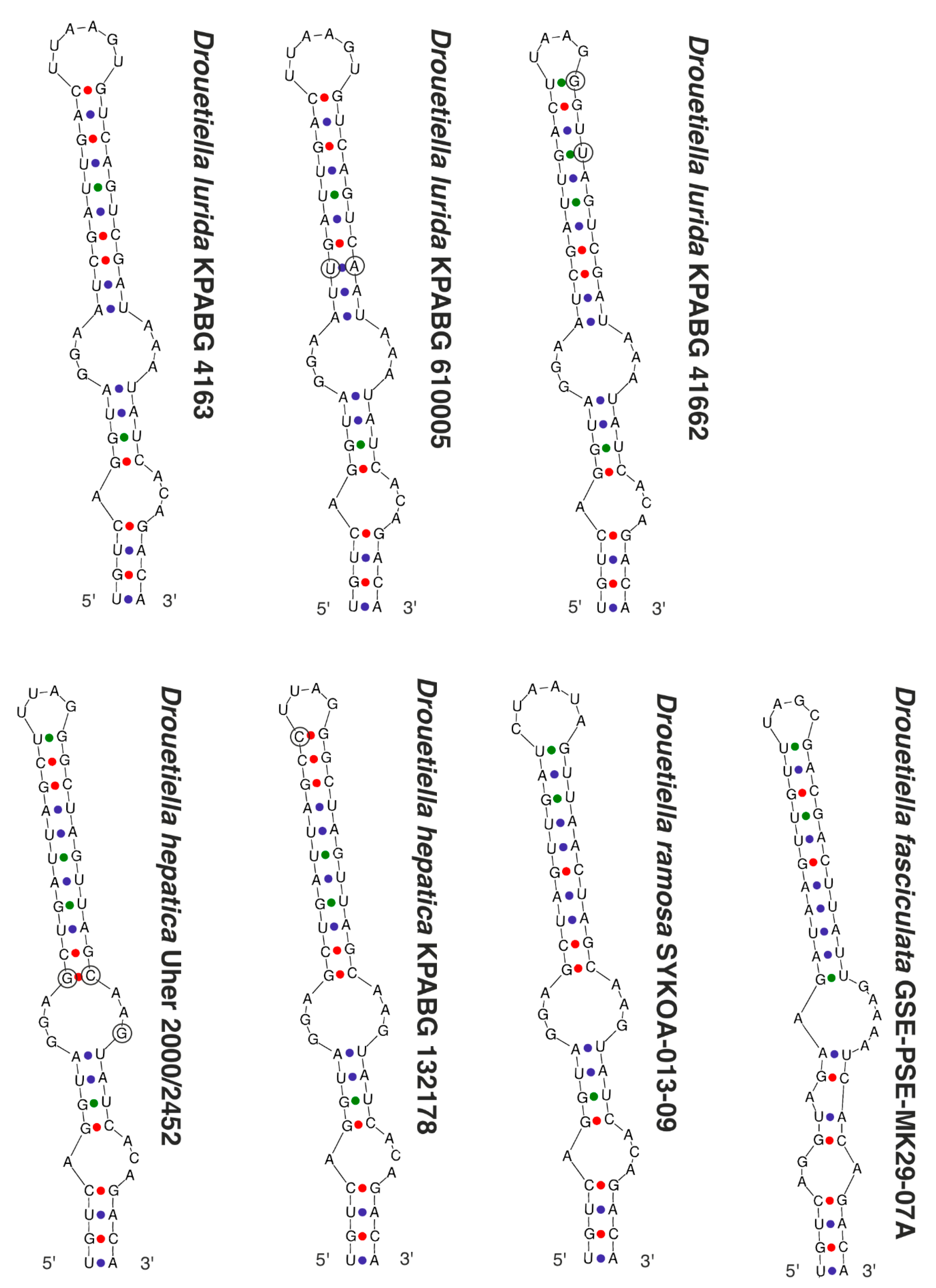
| Number of Location | Number of the Strain in Collection (GB Accession Number) | Locality | Habitats | Latitude N | Longitude E | Elev. (m) |
|---|---|---|---|---|---|---|
| 1 | KPABG 4163 (ON897679) | Yamalo-Nenets Autonomous Okrug. The Polar Urals Mountains. The Ochetyvis Valley. The left shore of the Ochetyvis River. | On the limestone rocks and boulders underwater. | 68.18995 | 65.67754 | 167 |
| 2 | KPABG 132178 (ON897678) | Yamalo-Nenets Autonomous Okrug. The Polar Urals Mountains. The Ochetyvis Valley. The left shore of the Ochetyvis River. | On a wet limestone wall of rock, near the water. | 68.19059 | 65.67815 | 154 |
| 3 | KPABG 41662 (ON897680) | Murmansk Region. Ash dumps of the Apatitsky Thermal Power Plant. | Anthropogenic habitat, crust on the sludge. | 67.599972 | 33.48128 | 202 |
| 4 | SYKOA C-013-09 (ON897677) | Komi Republic. Subpolar Urals Mountains. | Next to a quartz mine on damp quartz sand. | 65.22639 | 60.24528 | 850 |
| 5 | KPABG 610005/SYKOA C-002-10 (ON897681) | Komi Republic. Subpolar Urals Mountains. Near Maloe Balbanty Lake. | Grass–moss community next to a deer camp, in the soil. | 65.155833 | 60.231833 | 690 |
| Characteristics/Species | D. lurida | D. hepatica | D. ramosa | D. fasciculata | ||||
|---|---|---|---|---|---|---|---|---|
| Lukesova 1986/6 | KPABG 4163 | KPABG 41662 | KPABG 610005 | Uher 2000/2452 | KPABG 132178 | SYKOA C-013-09 | GSE-PSE-MK29-07A | |
| False-branching | - | + | + | - | very rare | - | common | - |
| Cells elongated | + | + | + | + | + | + | - | + |
| Constricted at cross-walls | not or slightly | distinctly | slightly | + | not or slightly | not or slightly | not or slightly | not |
| Necridia | - | + | + | - | frequent | + | + | + |
| Width of cells | 1.7–2.1 | 1.8–2.5 | 1.6–2.4 | 1.6–3.4 | 1.5–3.0 | 1.6–3.1 | 2.3–4.3 | 1.5–2.4 (3.0) |
| Length of cells | (2.1) 2.9–3.8 (5.4) | 1.2–2.6 | 1.9–3 | 1.5–3.8(4) | (2.2) 3.1–4.5 | 2.0–4.0 | 1.9–2.6 | 3.1–4.4 (5.4) |
| Meristematic zones | - | - | - | - | + | - | - | - |
| Hormogonia | - | - | - | rare | - | - | rare | rare |
| Coloration | liver-brown | olive-green | olive-green | blue-green, olive-green | brownish | olive-green | olive-green, blue-green | blue-green |
| Locality/ habitat | Czech Republic, temperate forest/aerophytic, soil | The Russian Arctic/epilithic on a boulder in a river, underwater | The Russian Subarctic/aerophytic, crust on the sludge | The Russian Subarctic/aerophytic, soil crusts | Slovakia, temperate forest/on subaerial seep wall and waterfall | The Russian Arctic/seep wall | The Russian Subarctic/aerophytic, soil | The USA, semi-arid/seep wall and waterfall |
| Taxon | Nucleotide Sequence Similarity, 16S/16S-23S ITS, % | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | |
| 1. D. lurida Lukesova 1986/6 | - | ||||||||
| 2. D. lurida KPABG 4163 | 99.90/ 99.12 | - | |||||||
| 3. D. lurida KPABG 41662 | 99.48/ 94.83 | 99.64/ 94.13 | - | ||||||
| 4. D. lurida KPABG 610005 | 99.17/ 93.53 | 99.37/ 93.72 | 99.28/ 97.42 | - | |||||
| 5. D. hepatica KPABG 132178 | 97.82/ 85.58 | 98.21/ 85.22 | 98.40/ 87.60 | 98.66/ 88.15 | - | ||||
| 6. D. hepatica Uher 2000/2452 | 97.81/ 85.58 | 97.93/ 85.14 | 98.14/ 87.88 | 98.45/ 88.41 | 100.00/ 99.40 | - | |||
| 7. Drouetiella sp. V16 | 97.81/ 87.08 | 98.21/ 87.32 | 98.40/ 88.94 | 98.66/ 89.10 | 99.82/ 95.57 | 99.80/ 95.57 | - | ||
| 8. Drouetiella sp. ANTL522 | 97.91/ 86.67 | 98.30/ 86.35 | 98.48/ 87.53 | 98.75/ 87.45 | 99.55/ 94.15 | 99.50/ 94.22 | 99.74/ 96.98 | - | |
| 9. D. ramosa SYKOA C-013-09 | 98.02/ 88.99 | 98.39/ 87.73 | 98.03/ 88.10 | 98.30/ 88.89 | 98.84/ 89.41 | 98.65/ 89.24 | 98.84/ 91.57 | 98.92/ 90.18 | - |
| 10. D. fasciculata GSE-PSE-MK29-07A | 96.42/ 79.57 | 96.51/ 78.57 | 96.17/ 80.14 | 98.43/ 79.91 | 96.52/ 80.59 | 96.54/ 80.59 | 96.82/ 83.33 | 96.90/ 80.41 | 96.60/ 80.14 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Davydov, D.; Vilnet, A.; Novakovskaya, I.; Patova, E. Terrestrial Species of Drouetiella (Cyanobacteria, Oculatellaceae) from the Russian Arctic and Subarctic Regions and Description of Drouetiella ramosa sp. nov. Diversity 2023, 15, 132. https://doi.org/10.3390/d15020132
Davydov D, Vilnet A, Novakovskaya I, Patova E. Terrestrial Species of Drouetiella (Cyanobacteria, Oculatellaceae) from the Russian Arctic and Subarctic Regions and Description of Drouetiella ramosa sp. nov. Diversity. 2023; 15(2):132. https://doi.org/10.3390/d15020132
Chicago/Turabian StyleDavydov, Denis, Anna Vilnet, Irina Novakovskaya, and Elena Patova. 2023. "Terrestrial Species of Drouetiella (Cyanobacteria, Oculatellaceae) from the Russian Arctic and Subarctic Regions and Description of Drouetiella ramosa sp. nov." Diversity 15, no. 2: 132. https://doi.org/10.3390/d15020132
APA StyleDavydov, D., Vilnet, A., Novakovskaya, I., & Patova, E. (2023). Terrestrial Species of Drouetiella (Cyanobacteria, Oculatellaceae) from the Russian Arctic and Subarctic Regions and Description of Drouetiella ramosa sp. nov. Diversity, 15(2), 132. https://doi.org/10.3390/d15020132








