Diversity and Recruitment Strategies of Rhizosphere Microbial Communities by Camellia fascicularis, a Plant Species with Extremely Small Populations in China: Plant Recruits Special Microorganisms to Get Benefit out of Them
Abstract
1. Introduction
2. Materials and Methods
2.1. Site Description and Sample Processing
2.2. Soil and Root Chemical Properties
2.3. DNA Isolation and Sequencing
2.4. Bioinformatic Analysis
2.5. Data and Statistical Analyses
3. Results
3.1. Soil Chemical Properties
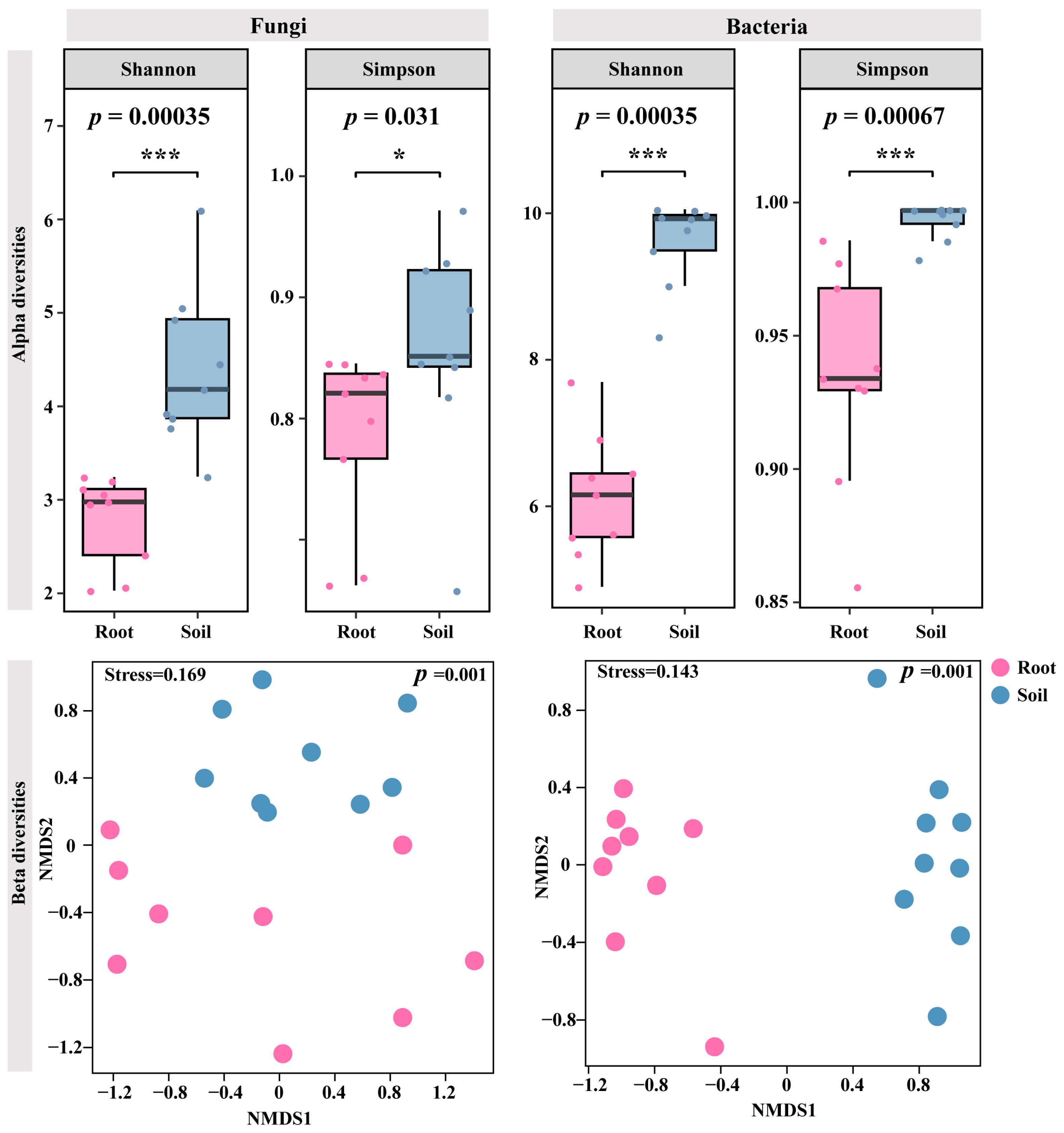
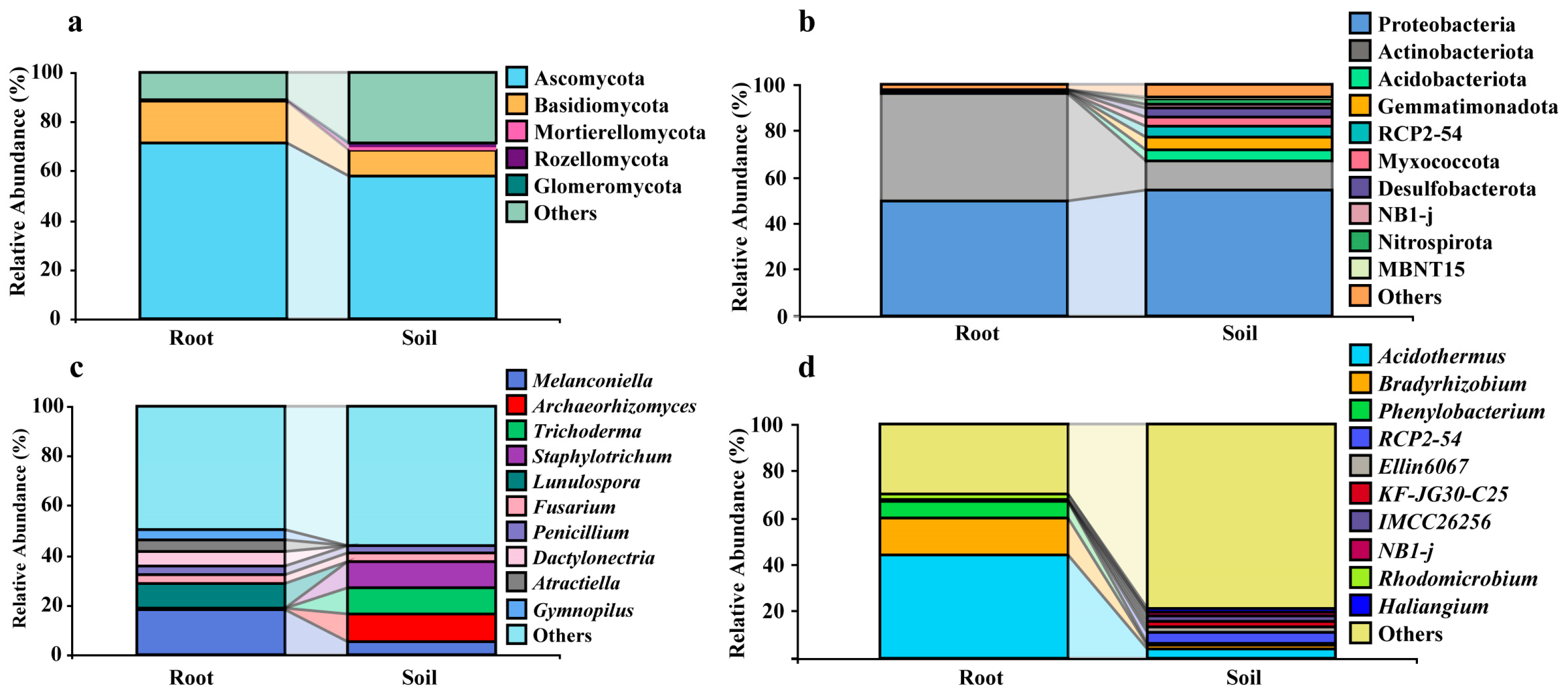
3.2. Differentiation of C. fascicularis Endophytes and Rhizosphere Soil Microbial Communities
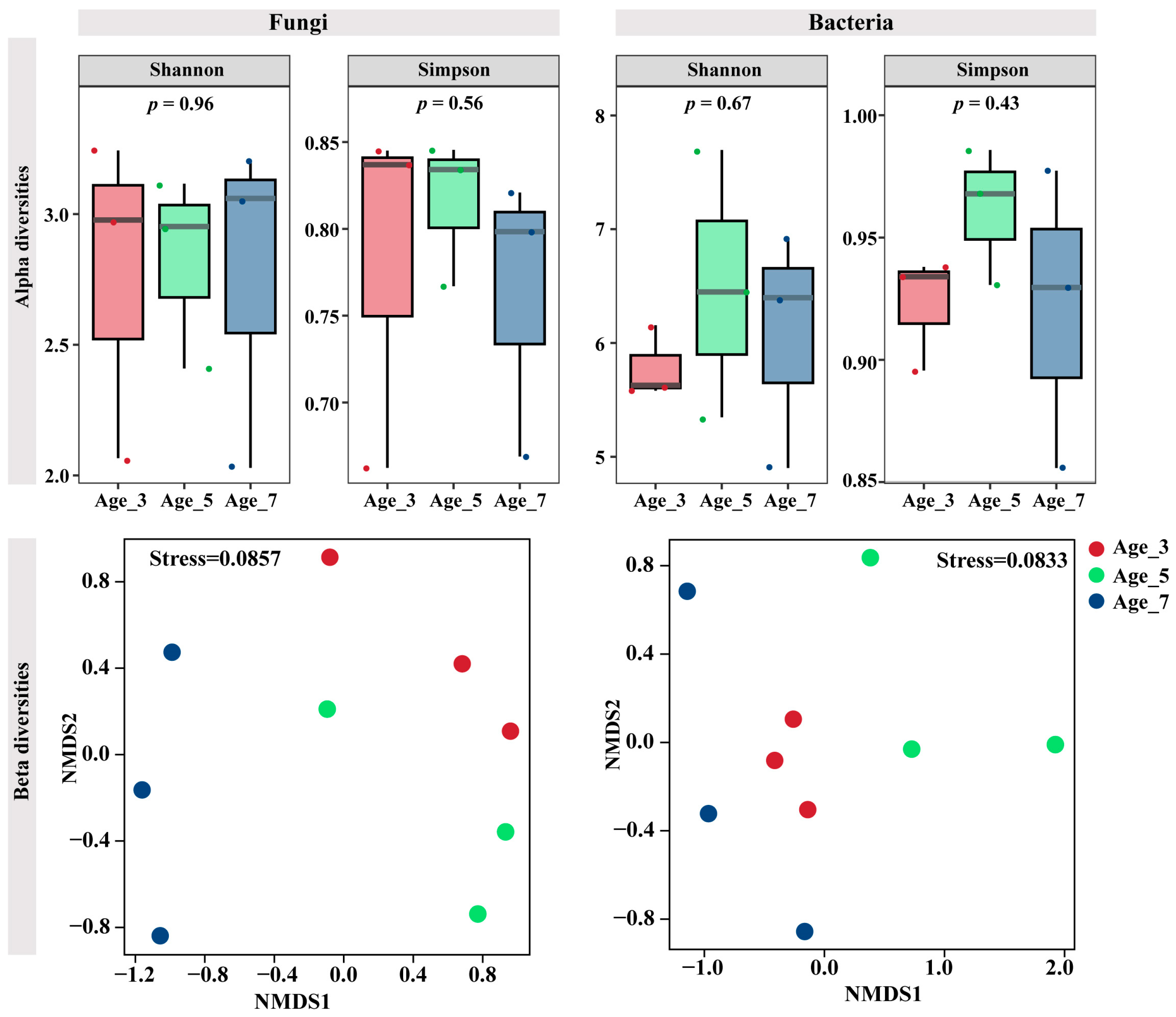
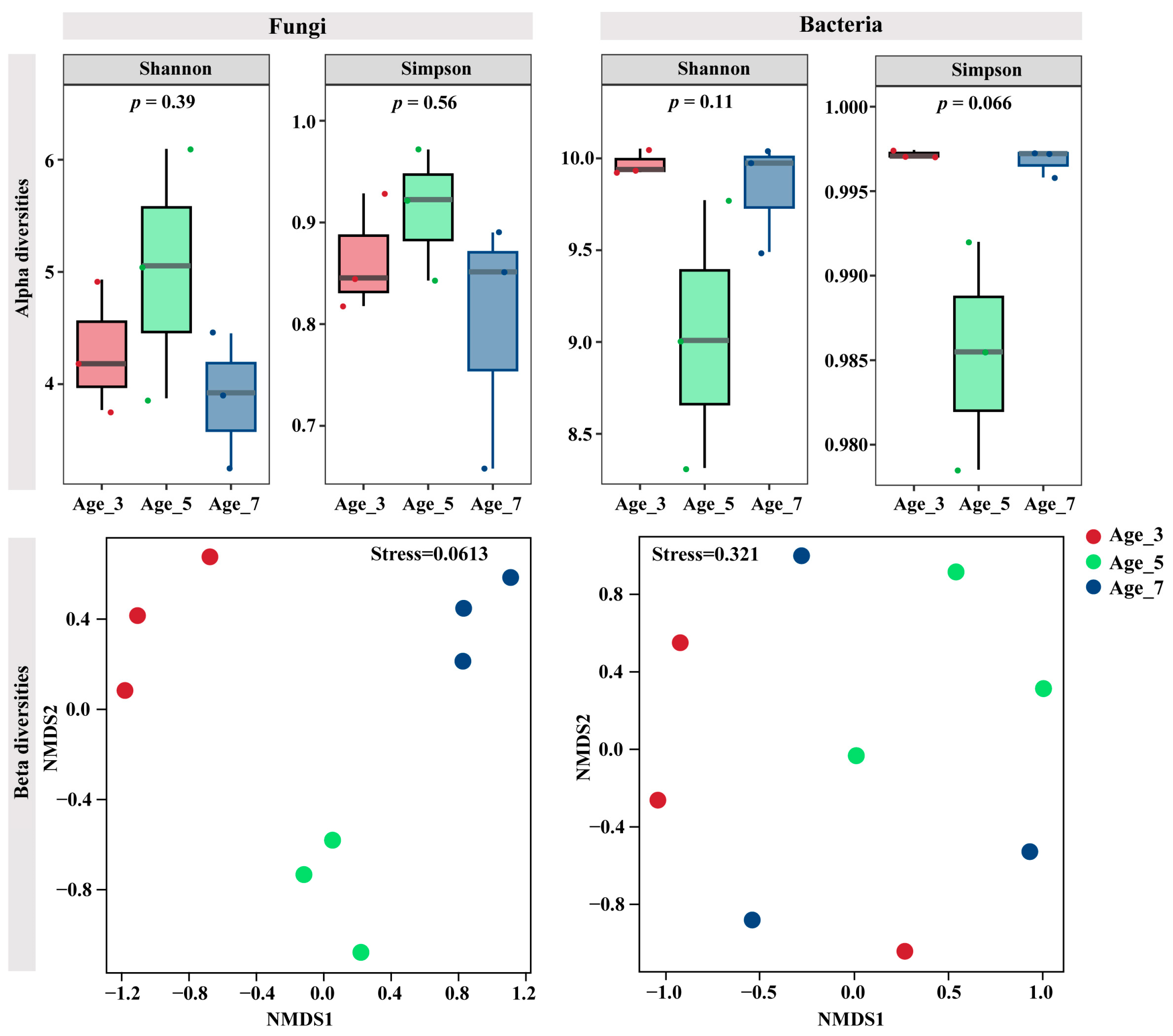
3.3. Variation in Microbial Communities in Rhizosphere Soil and Roots of Camellia for Different Cultivation Years
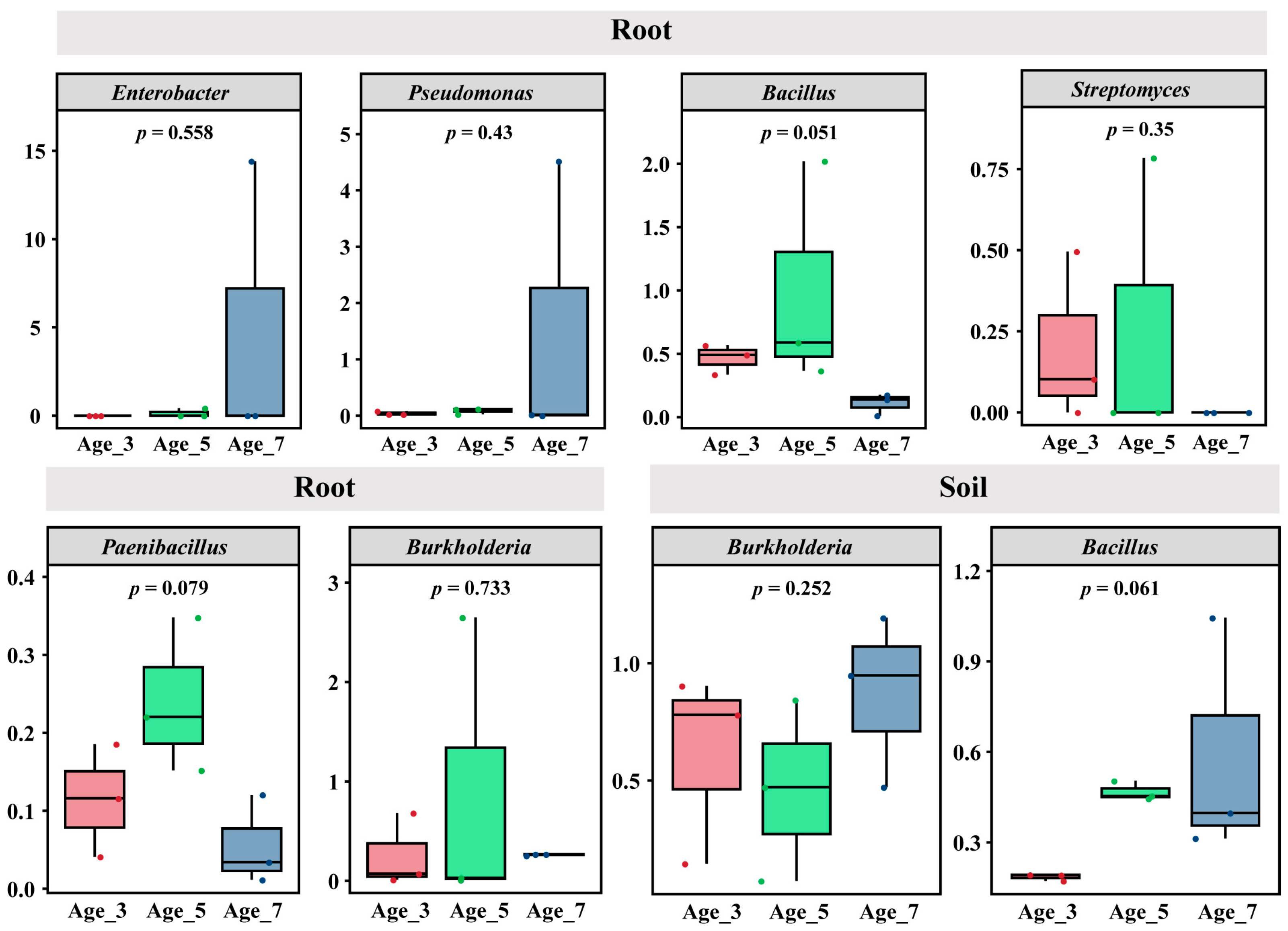
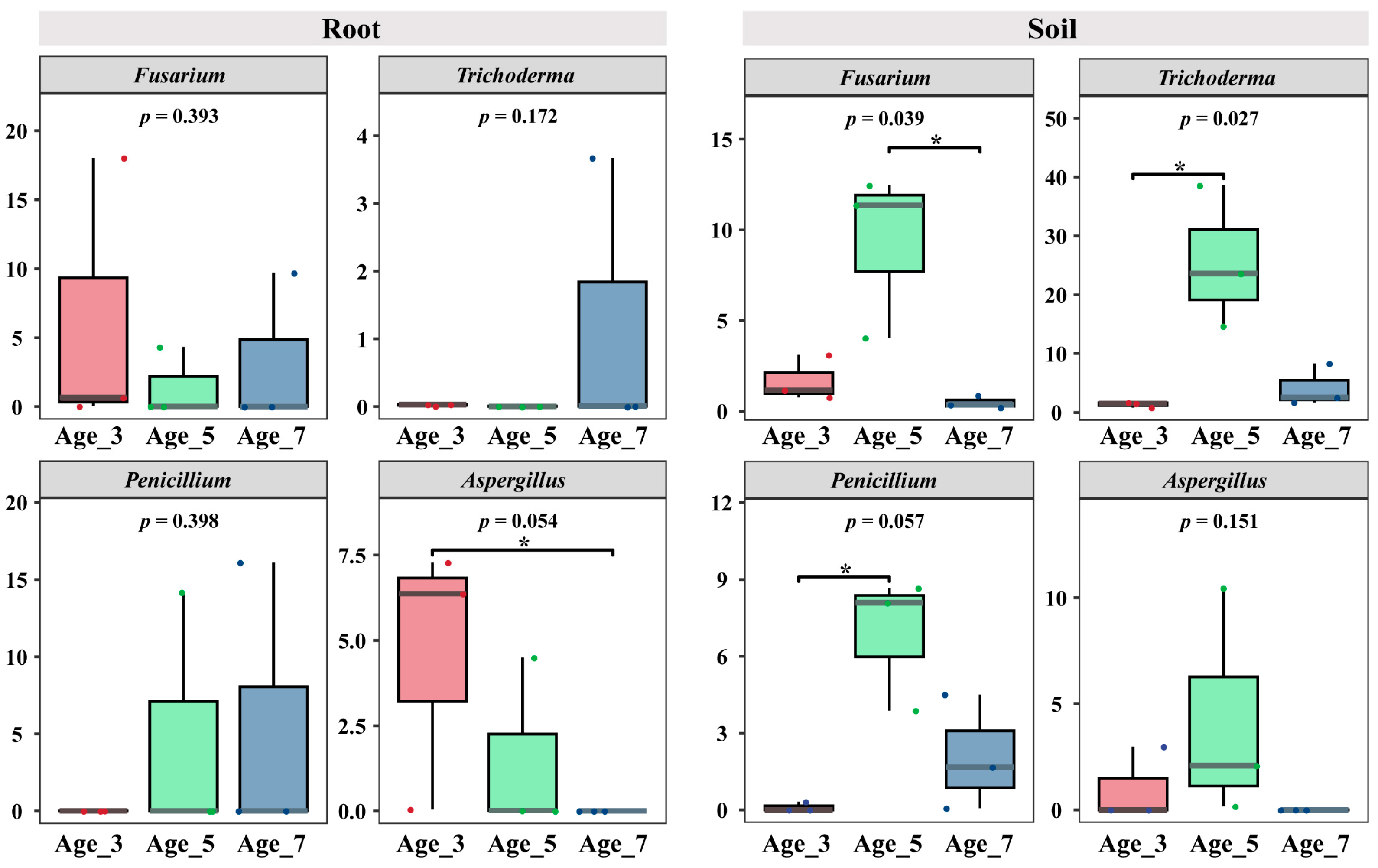
3.4. Variation in Main Pathogenic and Biocontrol Agents in Camellia with Different Cultivation Years
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
References
- Liu, Y.; Luo, X.L.; Lan, Z.Q.; Tang, J.R.; Kan, H. Ultrasonic-assisted extraction and antioxidant capacities of flavonoids from Camellia fascicularis leaves. CyTA-J. Food 2018, 16, 105–112. [Google Scholar] [CrossRef]
- Peng, X.W.; He, X.H.; Tang, J.R.; Xiang, J.Y.; Deng, J.; Kan, H.; Zhang, Y.J.; Zhang, G.L.; Zhao, P.; Liu, Y. Evaluation of the in vitro antioxidant and antitumor activity of extracts from Camellia fascicularis leaves. Front. Chem. 2022, 10, 1035949. [Google Scholar] [CrossRef] [PubMed]
- Gao, M.Z.; Peng, X.W.; Tang, J.R.; Deng, J.; Wang, F.; Zhang, Y.J.; Zhao, P.; Kan, H.; Liu, Y. Anti-Inflammatory Effects of Camellia fascicularis Polyphenols via Attenuation of NF-κB and MAPK Pathways in LPS-Induced THP-1 Macrophages. J. Inflamm. Res. 2022, 15, 851–864. [Google Scholar] [CrossRef] [PubMed]
- Eppinga, M.B.; Rietkerk, M.; Dekker, S.C.; Ruiter, P.C.D.; Putten, W.H.V.D.; Putten, W.H.V.D. Accumulation of local pathogens: A new hypothesis to explain exotic plant invasions. Oikos 2006, 114, 168–176. [Google Scholar] [CrossRef]
- Trivedi, P.; Leach, J.E.; Tringe, S.G.; Sa, T.; Singh, B.K. Plant–microbiome interactions: From community assembly to plant health. Nat. Rev. Microbiol. 2020, 18, 607–621. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Fang, K.; Zhou, J.; Yang, Z.P.; Dong, X.F.; Dai, G.H.; Zhang, H.B. Enrichment of soil rare bacteria in root by an invasive plant Ageratina adenophora. Sci. Total Environ. 2019, 683, 202–209. [Google Scholar] [CrossRef] [PubMed]
- Hassani, M.A.; Duran, P.; Hacquard, S. Microbial interactions within the plant holobiont. Microbiome 2018, 6, 58. [Google Scholar] [CrossRef]
- Durán, P.; Thiergart, T.; Garrido-Oter, R.; Agler, M.; Kemen, E.; Schulze-Lefert, P.; Hacquard, S. Microbial Interkingdom Interactions in Roots Promote Arabidopsis Survival. Cell 2018, 175, 973–983.e14. [Google Scholar] [CrossRef]
- Gray, E.J.; Smith, D.L. Intracellular and extracellular PGPR: Commonalities and distinctions in the plant–bacterium signaling processes. Soil Biol. Biochem. 2005, 37, 395–412. [Google Scholar] [CrossRef]
- Dobbelaere, S.; Vanderleyden, J.; Okon, Y. Plant Growth-Promoting Effects of Diazotrophs in the Rhizosphere. Crit. Rev. Plant Sci. 2003, 22, 107–149. [Google Scholar] [CrossRef]
- Leontidou, K.; Genitsaris, S.; Papadopoulou, A.; Kamou, N.; Bosmali, I.; Matsi, T.; Madesis, P.; Despoina, V.; Karamanoli, K.; Mellidou, I. Plant growth promoting rhizobacteria isolated from halophytes and drought-tolerant plants: Genomic characterisation and exploration of phyto-beneficial traits. Sci. Rep. 2020, 10, 14857. [Google Scholar] [CrossRef]
- Lugtenberg, B.; Kamilova, F. Plant-Growth-Promoting Rhizobacteria. Annu. Rev. Microbiol. 2009, 63, 541–556. [Google Scholar] [CrossRef] [PubMed]
- Bag, S.; Mondal, A.; Banik, A. Exploring tea (Camellia sinensis) microbiome: Insights into the functional characteristics and their impact on tea growth promotion. Microbiol. Res. 2022, 254, 126890. [Google Scholar] [CrossRef] [PubMed]
- Wilkinson, T.D.J.; Miranda, J.P.; Ferrari, J.; Hartley, S.E.; Hodge, A. Aphids Influence Soil Fungal Communities in Conventional Agricultural Systems. Front. Plant Sci. 2019, 10, 895. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.C.; Hao, X.Y.; Wang, L.; Xiao, B.; Wang, X.C.; Yang, Y.J. Diverse Colletotrichum species cause anthracnose of tea plants (Camellia sinensis (L.) O. Kuntze) in China. Sci. Rep. 2016, 6, 35287. [Google Scholar]
- Saha, D.; Dasgupta, S.; Saha, A. Antifungal Activity of Some Plant Extracts Against Fungal Pathogens of Tea (Camellia sinensis). Pharm. Biol. 2005, 43, 87–91. [Google Scholar] [CrossRef]
- Rabha, A.J.; Naglot, A.; Sharma, G.D.; Gogoi, H.K.; Vijay, V. In Vitro Evaluation of Antagonism of Endophytic Colletotrichum gloeosporioides Against Potent Fungal Pathogens of Camellia sinensis. Indian J. Microbiol. 2014, 54, 302–309. [Google Scholar] [CrossRef]
- Nisha, S.N.; Prabu, G.; Mandal, A.K.A. Biochemical and molecular studies on the resistance mechanisms in tea [Camellia sinensis (L.) O. Kuntze] against blister blight disease. Physiol. Mol. Biol. Plants 2018, 24, 867–880. [Google Scholar] [CrossRef]
- Jeyaraman, M.; Robert, P.S.A. Integrated management of branch canker disease (Macrophoma sp.) in tea under field level. J. Plant Dis. Protect. 2017, 124, 115–119. [Google Scholar] [CrossRef]
- Pandey, A.; Azariah, B. Diseases of tea crop and their current mitigation strategies. In Proceedings of the National Webinar on Plant Diseases in Eastern and Northeastern India: Current Dynamics and Proposed Action Plan for Their Management, Tripura, India, 24–25 June 2021; College of Agriculture: Tripura, India, 2021. [Google Scholar]
- Yeomans, J.C.; Bremner, J.M. A rapid and precise method for routine determination of organic carbon in soil. Commun. Soil Sci. Plant Anal. 1988, 19, 1467–1476. [Google Scholar] [CrossRef]
- Dong, J.L.; Gruda, N.; Li, X.; Tang, Y.; Duan, Z.Q. Impacts of elevated CO2 on nitrogen uptake of cucumber plants and nitrogen cycling in a greenhouse soil. Appl. Soil Ecol. 2020, 145, 103342. [Google Scholar] [CrossRef]
- Liang, F.; Xu, L.; Ji, L.; He, Q.Y.; Wu, L.L.; Yan, S.P. A new approach for biogas slurry disposal by adopting CO2-rich biogas slurry as the flower fertilizer of Spathiphyllum: Feasibility, cost and environmental pollution potential. Sci. Total Environ. 2021, 770, 145333. [Google Scholar] [CrossRef] [PubMed]
- Větrovský, T.; Baldrian, P. Analysis of soil fungal communities by amplicon pyrosequencing: Current approaches to data analysis and the introduction of the pipeline SEED. Biol. Fert. Soils 2013, 49, 1027–1037. [Google Scholar] [CrossRef]
- Bolyen, E.; Rideout, J.R.; Dillon, M.R.; Bokulich, N.A.; Abnet, C.C.; Al-Ghalith, G.A.; Alexander, H.; Alm, E.J.; Arumugam, M.; Asnicar, F.; et al. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 2019, 37, 852–857. [Google Scholar] [CrossRef]
- Martin, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 2011, 17, 10–12. [Google Scholar] [CrossRef]
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.A.; Dada, S.H. High-resolution sample inference from Illumina amplicon data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef]
- Katoh, K.; Misawa, K.; Kuma, K.; Miyata, T. MAFFT: A novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 2002, 30, 3059–3066. [Google Scholar] [CrossRef] [PubMed]
- Price, M.N.; Dehal, P.S.; Arkin, A.P. FastTree 2—Approximately Maximum-Likelihood Trees for Large Alignments. PLoS ONE 2010, 5, e9490. [Google Scholar] [CrossRef]
- Bokulich, N.A.; Kaehler, B.D.; Rideout, J.R.; Dillon, M.; Bolyen, E.; Knight, R.; Huttley, G.A.; Caporaso, J.G. Optimizing taxonomic classification of marker-gene amplicon sequences with QIIME 2′s q2-feature-classifier plugin. Microbiome 2018, 6, 90. [Google Scholar] [CrossRef]
- Oksanen, J.; Simpson, G.; Blanchet, F.; Kindt, R.; Legendre, P.; Minchin, P.; O’Hara, R.; Solymos, P.; Stevens, M.; Szoecs, E.; et al. vegan: Community Ecology Package. R Package Version 2.6-2. Available online: https://CRAN.R-project.org/package=vegan (accessed on 24 October 2023).
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2021; Available online: https://www.R-project.org/ (accessed on 23 September 2023).
- Volis, S. Securing a future for China’s plant biodiversity through an integrated conservation approach. Plant Divers. 2018, 40, 91–105. [Google Scholar] [CrossRef]
- Reinhold-Hurek, B.; Bünger, W.; Burbano, C.S.; Sabale, M.; Hurek, T. Roots Shaping Their Microbiome: Global Hotspots for Microbial Activity. Annu. Rev. Phytopathol. 2015, 53, 403–424. [Google Scholar] [CrossRef]
- Mu, D.; Tang, J.; Cai, N.; Chen, S.; He, Y.; Deng, Z.; Yang, Y.; Yang, D.; Xu, Y.; Chen, L. Effects of Microbial Communities on Elevational Gradient Adaptation Strategies of Pinus yunnanensis Franch. and Pinus densata Mast. in a Mixed Zone. Forests 2023, 14, 685. [Google Scholar] [CrossRef]
- Shan, W.; Zhou, Y.; Liu, H.; Yu, X. Endophytic Actinomycetes from Tea Plants (Camellia sinensis): Isolation, Abundance, Antimicrobial, and Plant-Growth-Promoting Activities. Biomed. Res. Int. 2018, 2018, 1470305. [Google Scholar] [CrossRef] [PubMed]
- Du, Z.; Fan, X.L.; Yang, Q.; Tian, C.M. Host and geographic range extensions of Melanconiella, with a new species M. cornuta in China. Phytotaxa 2017, 327, 252–260. [Google Scholar] [CrossRef][Green Version]
- Romila, T.; Dutta, B.K. Control of Black rot disease of tea {Camellia sinensis (L.) O Kuntze} with the mycoflora isolated from tea environment and phyllosphere. J. Mycopathol. Res. 2012, 50, 205–210. [Google Scholar]
- Liu, R.C.; Xiao, Z.Y.; Hashem, A.; Abd_Allah, E.F.; Wu, Q.S. Unraveling the Interaction between Arbuscular Mycorrhizal Fungi and Camellia Plants. Horticulturae 2021, 7, 322. [Google Scholar] [CrossRef]
- Chen, L.; Zhou, J.; Zeng, T.; Miao, Y.; Mei, L.; Yao, G.; Fang, K.; Dong, X.; Sha, T.; Yang, M.; et al. Quantifying the sharing of foliar fungal pathogens by the invasive plant Ageratina adenophora and its neighbours. New Phytol. 2020, 227, 1493–1504. [Google Scholar] [CrossRef]
- Ren, B.; Hu, Y.; Chen, B.; Zhang, Y.; Thiele, J.; Shi, R.; Liu, M.; Bu, R. Soil pH and plant diversity shape soil bacterial community structure in the active layer across the latitudinal gradients in continuous permafrost region of Northeastern China. Sci. Rep. 2018, 8, 5619. [Google Scholar] [CrossRef]
- Fan, D.; Subramanian, S.; Smith, D.L. Plant endophytes promote growth and alleviate salt stress in Arabidopsis thaliana. Sci. Rep. 2020, 10, 12740. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mu, D.; Chen, L.; Hua, G.; Pu, L.; Tian, Z.; Liu, Y.; Zhang, G.; Tang, J. Diversity and Recruitment Strategies of Rhizosphere Microbial Communities by Camellia fascicularis, a Plant Species with Extremely Small Populations in China: Plant Recruits Special Microorganisms to Get Benefit out of Them. Diversity 2023, 15, 1170. https://doi.org/10.3390/d15121170
Mu D, Chen L, Hua G, Pu L, Tian Z, Liu Y, Zhang G, Tang J. Diversity and Recruitment Strategies of Rhizosphere Microbial Communities by Camellia fascicularis, a Plant Species with Extremely Small Populations in China: Plant Recruits Special Microorganisms to Get Benefit out of Them. Diversity. 2023; 15(12):1170. https://doi.org/10.3390/d15121170
Chicago/Turabian StyleMu, Dejin, Lin Chen, Guoli Hua, Lei Pu, Zineng Tian, Yun Liu, Guiliang Zhang, and Junrong Tang. 2023. "Diversity and Recruitment Strategies of Rhizosphere Microbial Communities by Camellia fascicularis, a Plant Species with Extremely Small Populations in China: Plant Recruits Special Microorganisms to Get Benefit out of Them" Diversity 15, no. 12: 1170. https://doi.org/10.3390/d15121170
APA StyleMu, D., Chen, L., Hua, G., Pu, L., Tian, Z., Liu, Y., Zhang, G., & Tang, J. (2023). Diversity and Recruitment Strategies of Rhizosphere Microbial Communities by Camellia fascicularis, a Plant Species with Extremely Small Populations in China: Plant Recruits Special Microorganisms to Get Benefit out of Them. Diversity, 15(12), 1170. https://doi.org/10.3390/d15121170





