Emerging Technologies for the Discovery of Novel Diversity in Cyanobacteria and Algae and the Elucidation of Their Valuable Metabolites
Abstract
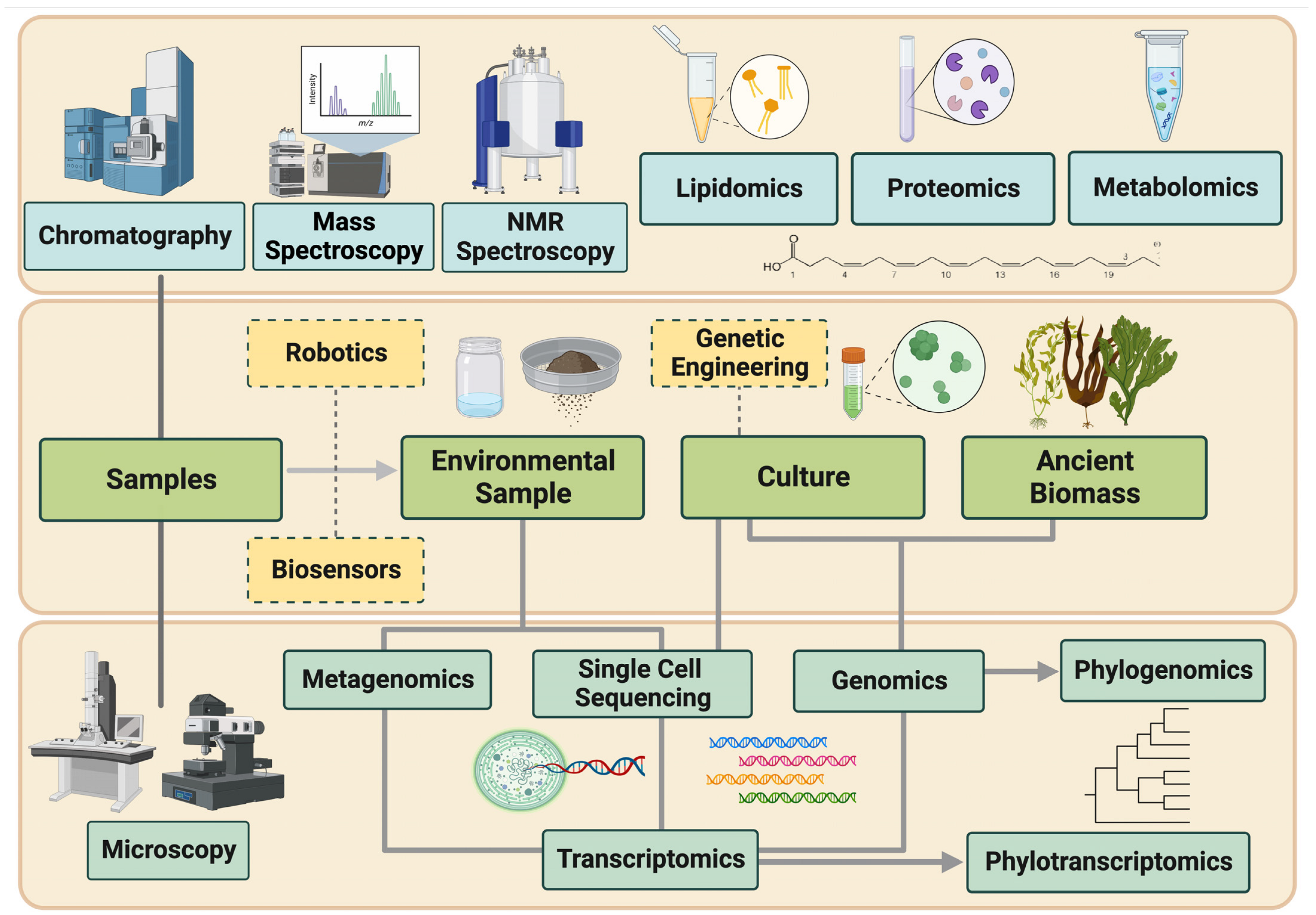
:1. Introduction
2. Sampling and Robotics
3. Microscopy
3.1. Atomic Force Microscopy
3.2. Cryo-Electron Microscopy
4. Metagenomics
4.1. Environmental DNA
4.2. Microbiomes
5. Single-Cell Sequencing
5.1. Single-Cell Genome Sequencing
5.2. Single-Cell RNA Sequencing
6. Phylogenomics and Phylotranscriptomics
7. Ancient Cyanobacteria and Algae
7.1. Fossils and Archaeological Remains
7.2. Ancient DNA
8. Metabolomics, Metabolic Profiling and Functional Genomics
8.1. Metabolite Target Analysis
8.2. Metabolite Profiling
8.3. Metabolite Fingerprinting
8.4. Metabolite Footprinting
8.5. Flux Analysis
9. The OMICS Approach
9.1. Proteomics
9.2. Integrated Omics (Multi-Omics)
10. Genetic Engineering
CRISPR-Cas
11. HiTES and Chemical Biology
12. Microsensors
12.1. Oxygen Sensors
12.2. Temperature Sensors
12.3. pH Sensors
12.4. Biosensors
13. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
References
- Zammit, G.; Kaštovský, J.; Albertano, P. A first cytomorphological and molecular characterisation of a new Stigonematalean cyanobacterial morphotype isolated from Maltese catacombs. Algol. Stud. 2010, 135, 1–14. [Google Scholar] [CrossRef]
- Zammit, G.; Billi, D.; Shubert, E.; Kaštovsky, J.; Albertano, P. The biodiversity of subaerophytic phototrophic biofilms from Maltese hypogea. Fottea 2011, 11, 187–201. [Google Scholar] [CrossRef]
- Zammit, G. Phototrophic biofilm communities and adaptation to growth on ancient archaeological surfaces. Ann. Microbiol. 2019, 69, 1047–1058. [Google Scholar] [CrossRef]
- Schembri, S.; Zammit, G. The biodiversity of epilithic microalgal communities colonising a central Mediterranean coastline. J. Coast. Res. 2022, 38, 249–260. [Google Scholar] [CrossRef]
- Zammit, G.; Schembri, S.; Fenech, M. Phototrophic biofilms and microbial mats from the marine littoral of the central Mediterranean. Acta Bot. Croat. 2021, 80, 112–116. [Google Scholar] [CrossRef]
- Zammit, G.; Agius, M. Ecophysiology and metabolites of biofilm-forming strains of the microalgal genus Jenufa (Chlorophyceae). Phycologia 2022, 61, 409–418. [Google Scholar] [CrossRef]
- Zammit, G.; Billi, D.; Albertano, P. The subaerophytic cyanobacterium Oculatella subterranea (Oscillatoriales, Cyanophyceae) gen. et sp. nov: A cytomorphological and molecular description. Eur. J. Phycol. 2012, 47, 341–354. [Google Scholar] [CrossRef]
- Zammit, G. Systematics and biogeography of sciophilous cyanobacteria: An ecological and molecular description of Albertania skiophila (Leptolyngbyaceae) gen et sp. nov. Phycologia 2018, 57, 481–491. [Google Scholar] [CrossRef]
- Bartolo, A.; Zammit, G.; Peters, A.F.; Kuepper, F.C. The current state of DNA Barcoding of Macroalgae in the Mediterranean Sea: Presently lacking but urgently required. Bot. Mar. 2020, 63, 253–272. [Google Scholar] [CrossRef]
- Bartolo, A.G.; Zammit, G.; Russell, H.; Peters, A.F.; Küpper, F.C. DNA barcoding of marine algae from Malta: New records from the central Mediterranean. Acta Bot. Croat. 2021, 80, 176–183. [Google Scholar] [CrossRef]
- Bartolo, A.G.; Zammit, G.; Küpper, F.C. Germling culture and molecular analysis of evasive micro-filamentous green algae growing in the Maltese islands (central Mediterranean). Bot. Mar. 2022, 65, 243–254. [Google Scholar] [CrossRef]
- Bartolo, A.G.; Zammit, G.; Küpper, F.C. New records of Palisada tenerrima and Hincksia mitchelliae from the Maltese Islands revealed by molecular analysis. Mediterr. Mar. Sci. 2022, 23, 766–776. [Google Scholar] [CrossRef]
- Bartolo, A.G.; Zammit, G.; Küpper, F.C. Ulva biodiversity in the central Mediterranean Sea: Cryptic species and new records. Cryptogam. Algol. 2022, 43, 215–225. [Google Scholar] [CrossRef]
- Bartolo, A.G.; Zammit, G.; Kytinou, E.; Küpper, F.C. Laurencia mediterranea sp. nov., (Ceramiales, Rhodophyta) from the central Mediterranean Sea. Bot. Mar. 2023. [Google Scholar] [CrossRef]
- Crognale, S.; Venturi, S.; Tassi, F.; Rossetti, S.; Rashed, H.; Cabassi, J.; Capecchiacci, F.; Nisi, B.; Vaselli, O.; Morrison, H.G.; et al. Microbiome profiling in extremely acidic soils affected by hydrothermal fluids: The case of the Solfatara Crater (Campi Flegrei, southern Italy). FEMS Microbiol. Ecol. 2018, 94, fiy90. [Google Scholar] [CrossRef] [PubMed]
- Sánchez-García, L.; Fernández-Martínez, M.A.; Moreno-Paz, M.; Carrizo, D.; García-Villadangos, M.; Manchado, J.M.; Stoker, C.R.; Glass, B.; Parro, V. Simulating Mars Drilling Mission for Searching for Life: Ground-Truthing Lipids and Other Complex Microbial Biomarkers in the Iron-Sulfur Rich Río Tinto Analog. Astrobiology 2020, 20, 1029–1047. [Google Scholar] [CrossRef] [PubMed]
- Gusmão, A.C.B.; Peres, F.V.; Paula, F.S.; Pellizari, V.H.; Kolm, H.E.; Signori, C.N. Microbial communities in the deep-sea sediments of the South São Paulo Plateau, Southwestern Atlantic Ocean. Int. Microbiol. 2023, 26, 1041–1051. [Google Scholar] [CrossRef] [PubMed]
- Garel, M.; Bonin, P.; Martini, S.; Guasco, S.; Roumagnac, M.; Bhairy, N.; Armougom, F.; Tamburini, C. Pressure-Retaining Sampler and High-Pressure Systems to Study Deep-Sea Microbes Under in situ Conditions. Front. Microbiol. 2019, 10, 453. [Google Scholar] [CrossRef]
- Salman, I.; Karapetyan, N.; Venkatachari, A.; Li, A.Q.; Bourbonnais, A.; Rekleitis, I. Multi-Modal Lake Sampling for Detecting Harmful Algal Blooms. In Proceedings of the OCEANS 2022, Hampton Roads, VA, USA, 17–20 October 2022; pp. 1–9. [Google Scholar]
- Moore, S.K.; Mickett, J.B.; Doucette, G.J.; Adams, N.G.; Mikulski, C.M.; Birch, J.M.; Roman, B.; Michel-Hart, N.; Newton, J.A. An Autonomous Platform for Near Real-Time Surveillance of Harmful Algae and Their Toxins in Dynamic Coastal Shelf Environments. J. Mar. Sci. Eng. 2021, 9, 336. [Google Scholar] [CrossRef]
- Sellanes, J.; Gorny, M.; Zapata-Hernández, G.; Alvarez, G.; Muñoz, P.; Tala, F. A new threat to local marine biodiversity: Filamentous mats proliferating at mesophotic depths off Rapa Nui. PeerJ 2021, 9, e12052. [Google Scholar] [CrossRef]
- Mišić Radić, T.; Vukosav, P.; Ačković, A.; Dulebo, A. Insights into the Morphology and Surface Properties of Microalgae at the Nanoscale by Atomic Force Microscopy: A Review. Water 2023, 15, 1983. [Google Scholar] [CrossRef]
- Müller, D.J.; Dufrêne, Y.F. Atomic force microscopy as a multifunctional molecular toolbox in nanobiotechnology. Nat. Nanotechnol. 2008, 3, 261–269. [Google Scholar] [CrossRef] [PubMed]
- Novosel, N.; Mišić Radić, T.; Zemla, J.; Lekka, M.; Čačković, A.; Kasum, D.; Legović, T.; Žutinić, P.; Gligora Udovič, M.; Ivošević DeNardis, N. Temperature-induced response in algal cell surface properties and behaviour: An experimental approach. J. Appl. Phycol. 2022, 34, 243–259. [Google Scholar] [CrossRef]
- Demir, I.; Lüchtefeld, I.; Lemen, C.; Dague, E.; Guiraud, P.; Zambelli, T.; Formosa-Dague, C. Probing the interactions between air bubbles and (bio)interfaces at the nanoscale using FluidFM technology. J. Colloid. Interface Sci. 2021, 604, 785–797. [Google Scholar] [CrossRef] [PubMed]
- Mišić Radić, T.; Vukosav, P.; Komazec, B.; Formosa-Dague, C.; Domazet Jurašin, D.; Peharec Štefanić, P.; Čačković, A.; Juraić, K.; Ivošević DeNardis, N. Nanoplastic-Induced Nanostructural, Nanomechanical, and Antioxidant Response of Marine Diatom Cylindrotheca closterium. Water 2022, 14, 2163. [Google Scholar] [CrossRef]
- Deniset-Besseau, A.; Coat, R.; Moutel, B.; Rebois, R.; Mathurin, J.; Grizeau, D.; Dazzi, A.; Gonçalves, O. Revealing Lipid Body Formation and its Subcellular Reorganization in Oleaginous Microalgae Using Correlative Optical Microscopy and Infrared Nanospectroscopy. Appl. Spectrosc. 2021, 75, 1538–1547. [Google Scholar] [CrossRef] [PubMed]
- Yin, C. Structural biology revolution led by technical breakthroughs in cryo-electron microscopy. Chin. Phys. B 2018, 27, 58703. [Google Scholar] [CrossRef]
- Benjin, X.; Ling, L. Developments, applications, and prospects of cryo-electron microscopy. Protein Sci. 2020, 29, 872–882. [Google Scholar] [CrossRef]
- Semchonok, D.A.; Mondal, J.; Cooper, C.J.; Schlum, K.; Li, M.; Amin, M.; Sorzano, C.O.S.; Ramírez-Aportela, E.; Kastritis, P.L.; Boekema, E.J.; et al. Cryo-EM structure of a tetrameric photosystem I from Chroococcidiopsis TS-821, a thermophilic, unicellular, non-heterocyst-forming cyanobacterium. Plant Commun. 2022, 3, 100248. [Google Scholar] [CrossRef]
- Kawakami, K.; Hamaguchi, T.; Hirose, Y.; Kosumi, D.; Miyata, M.; Kamiya, N.; Yonekura, K. Core and rod structures of a thermophilic cyanobacterial light-harvesting phycobilisome. Nat. Commun. 2022, 13, 3389. [Google Scholar] [CrossRef]
- Zheng, L.; Zhang, Z.; Wang, H.; Zheng, Z.; Wang, J.; Liu, H.; Chen, H.; Dong, C.; Wang, G.; Weng, Y.; et al. Cryo-EM and femtosecond spectroscopic studies provide mechanistic insight into the energy transfer in CpcL-phycobilisomes. Nat. Commun. 2023, 14, 3961. [Google Scholar] [CrossRef] [PubMed]
- Danelius, E.; Patel, K.; Gonzalez, B.; Gonen, T. MicroED in drug discovery. Curr. Opin. Struct. Biol. 2023, 79, 102549. [Google Scholar] [CrossRef] [PubMed]
- Park, J.; Li, Y.; Moon, K.; Han, E.J.; Lee, S.R.; Seyedsayamdost, M.R. Structural Elucidation of Cryptic Algaecides in Marine Algal-Bacterial Symbioses by NMR Spectroscopy and MicroED. Angew. Chem. 2022, 61, e202114022-n/a. [Google Scholar] [CrossRef] [PubMed]
- Pessi, I.S.; Popin, R.V.; Durieu, B.; Lara, Y.; Tytgat, B.; Savaglia, V.; Roncero-Ramos, B.; Hultman, J.; Verleyen, E.; Vyverman, W.; et al. Novel diversity of polar Cyanobacteria revealed by genome-resolved metagenomics. Microb. Genom. 2023, 9, 001056. [Google Scholar] [CrossRef] [PubMed]
- Singh, G. Linking Lichen Metabolites to Genes: Emerging Concepts and Lessons from Molecular Biology and Metagenomics. J. Fungi 2023, 9, 160. [Google Scholar] [CrossRef] [PubMed]
- Dextro, R.B.; Delbaje, E.; Cotta, S.R.; Zehr, J.P.; Fiore, M.F.; Mock, T. Trends in Free-access Genomic Data Accelerate Advances in Cyanobacteria Taxonomy. J. Phycol. 2021, 57, 1392–1402. [Google Scholar] [CrossRef] [PubMed]
- Kottuparambil, S.; Ashok, A.; Barozzi, A.; Michoud, G.; Cai, C.; Daffonchio, D.; Duarte, C.M.; Agusti, S. Tracking the early signals of crude oil in seawater and plankton after a major oil spill in the Red Sea. Environ. Sci. Pollut. Res. 2023, 30, 69150–69164. [Google Scholar] [CrossRef]
- Linz, D.M.; Sienkiewicz, N.; Struewing, I.; Stelzer, E.A.; Graham, J.L.; Lu, J. Metagenomic mapping of cyanobacteria and potential cyanotoxin producing taxa in large rivers of the United States. Sci. Rep. 2023, 13, 2806. [Google Scholar] [CrossRef]
- Dong, X.; Lan, H.; Huang, L.; Zhang, H.; Lin, X.; Weng, S.; Peng, Y.; Lin, J.; Wang, J.; Peng, J.; et al. Metagenomic Views of Microbial Communities in Sand Sediments Associated with Coral Reefs. Microb. Ecol. 2023, 85, 465–477. [Google Scholar] [CrossRef]
- Meyer, J.L.; Gunasekera, S.P.; Brown, A.L.; Ding, Y.; Miller, S.; Teplitski, M.; Paul, V.J. Cryptic Diversity of Black Band Disease Cyanobacteria in Siderastrea siderea Corals Revealed by Chemical Ecology and Comparative Genome-Resolved Metagenomics. Mar. Drugs 2023, 21, 76. [Google Scholar] [CrossRef]
- Anggoro, A.W. Comparative Metagenomics of Coral Reef Associated Marine Biodiversity Across a Pollution Gradient in Western Indonesia. Doctor of Philosophy Dissertation in Biology. Ph.D. Thesis, University of California, Los Angeles, CA, USA, 2022. [Google Scholar]
- Palladino, G.; Caroselli, E.; Tavella, T.; D’Amico, F.; Prada, F.; Mancuso, A.; Franzellitti, S.; Rampelli, S.; Candela, M.; Goffredo, S.; et al. Metagenomic shifts in mucus, tissue and skeleton of the coral Balanophyllia europaea living along a natural CO2 gradient. ISME Commun. 2022, 2, 65. [Google Scholar] [CrossRef]
- Smith, H.B.; Dal Grande, F.; Muggia, L.; Keuler, R.; Divakar, P.K.; Grewe, F.; Schmitt, I.; Lumbsch, H.T.; Leavitt, S.D. Metagenomic data reveal diverse fungal and algal communities associated with the lichen symbiosis. Symbiosis 2020, 82, 133–147. [Google Scholar] [CrossRef]
- Ghosh, S.; Pramanik, S. Structural diversity, functional aspects and future therapeutic applications of human gut microbiome. Arch. Microbiol. 2021, 203, 5281–5308. [Google Scholar] [CrossRef]
- Nayfach, S.; Shi, Z.J.; Seshadri, R.; Pollard, K.S.; Kyrpides, N.C. New insights from uncultivated genomes of the global human gut microbiome. Nature 2019, 568, 505–510. [Google Scholar] [CrossRef] [PubMed]
- Brenner, S.R. Blue-green algae or cyanobacteria in the intestinal micro-flora may produce neurotoxins such as Beta-N-Methylamino-l-Alanine (BMAA) which may be related to development of amyotrophic lateral sclerosis, Alzheimer’s disease and Parkinson-Dementia-Complex in humans and Equine Motor Neuron Disease in Horses. Med. Hypotheses 2013, 80, 103. [Google Scholar] [CrossRef]
- Ghaisas, S.; Maher, J.; Kanthasamy, A. Gut microbiome in health and disease: Linking the microbiome–gut–brain axis and environmental factors in the pathogenesis of systemic and neurodegenerative diseases. Pharmacol. Ther. 2016, 158, 52–62. [Google Scholar] [CrossRef] [PubMed]
- Hernandez, B.Y.; Zhu, X.; Risch, H.A.; Lu, L.; Ma, X.; Irwin, M.L.; Lim, J.K.; Taddei, T.H.; Pawlish, K.S.; Stroup, A.M.; et al. Oral Cyanobacteria and Hepatocellular Carcinoma. Cancer Epidemiol. Biomark. Prev. 2022, 31, 221–229. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Chen, L.; Chen, Z.; Zhang, W. RNA-seq based transcriptomic analysis of single bacterial cells. Integr. Biol. 2015, 7, 1466–1476. [Google Scholar] [CrossRef]
- Davison, M.; Hall, E.; Zare, R.; Bhaya, D. Challenges of metagenomics and single-cell genomics approaches for exploring cyanobacterial diversity. Photosynth. Res. 2015, 126, 135–146. [Google Scholar] [CrossRef]
- Nakayama, T.; Nomura, M.; Takano, Y.; Tanifuji, G.; Shiba, K.; Inaba, K.; Inagaki, Y.; Kawata, M. Single-cell genomics unveiled a cryptic cyanobacterial lineage with a worldwide distribution hidden by a dinoflagellate host. Proc. Natl. Acad. Sci. USA 2019, 116, 15973–15978. [Google Scholar] [CrossRef]
- Liu, Y.; Jeraldo, P.; Herbert, W.; McDonough, S.; Eckloff, B.; Schulze-Makuch, D.; de Vera, J.; Cockell, C.; Leya, T.; Baqué, M.; et al. Whole genome sequencing of cyanobacterium Nostoc sp. CCCryo 231-06 using microfluidic single cell technology. iScience 2022, 25, 104291. [Google Scholar] [CrossRef] [PubMed]
- Schön, M.E.; Zlatogursky, V.V.; Singh, R.P.; Poirier, C.; Wilken, S.; Mathur, V.; Strassert, J.F.H.; Pinhassi, J.; Worden, A.Z.; Keeling, P.J.; et al. Single cell genomics reveals plastid-lacking Picozoa are close relatives of red algae. Nat. Commun. 2021, 12, 6651. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Gao, J.; Huang, Y.; Wang, J. Recent Developments in Single-Cell RNA-Seq of Microorganisms. Biophys. J. 2018, 115, 173–180. [Google Scholar] [CrossRef] [PubMed]
- Homberger, C.; Barquist, L.; Vogel, J. Ushering in a new era of single-cell transcriptomics in bacteria. microLife 2022, 3, uqac020. [Google Scholar] [CrossRef] [PubMed]
- Imdahl, F.; Vafadarnejad, E.; Homberger, C.; Saliba, A.; Vogel, J. Single-cell RNA-sequencing reports growth-condition-specific global transcriptomes of individual bacteria. Nat. Microbiol. 2020, 5, 1202–1206. [Google Scholar] [CrossRef] [PubMed]
- Nishimura, M.; Takeyama, H.; Hosokawa, M. Enhancing the sensitivity of bacterial single-cell RNA sequencing using RamDA-seq and Cas9-based rRNA depletion. J. Biosci. Bioeng. 2023, 136, 152–158. [Google Scholar] [CrossRef] [PubMed]
- Ma, F.; Salomé, P.A.; Merchant, S.S.; Pellegrini, M. Single-cell RNA sequencing of batch Chlamydomonas cultures reveals heterogeneity in their diurnal cycle phase. Plant Cell 2021, 33, 1042–1057. [Google Scholar] [CrossRef] [PubMed]
- Cheon, S.; Zhang, J.; Park, C. Is phylotranscriptomics as reliable as phylogenomics? Mol. Biol. Evol. 2020, 37, 3672–3683. [Google Scholar] [CrossRef]
- Komárek, J.; Kastovsky, J.; Mares, J.; Johansen, J.R. Taxonomic classification of cyanoprokaryotes (cyanobacterial genera) (2014), using a polyphasic approach. Preslia 2014, 86, 295–335. [Google Scholar]
- Strunecký, O.; Ivanova, A.P.; Mareš, J. An updated classification of cyanobacterial orders and families based on phylogenomic and polyphasic analysis. J. Phycol. 2023, 59, 12–51. [Google Scholar] [CrossRef] [PubMed]
- Boo, G.H.; Hughey, J.R. Phylogenomics and multigene phylogenies decipher two new cryptic marine algae from California, Gelidium gabrielsonii and G. kathyanniae (Gelidiales, Rhodophyta). J. Phycol. 2019, 55, 160–172. [Google Scholar] [CrossRef]
- Díaz-Tapia, P.; Pasella, M.M.; Verbruggen, H.; Maggs, C.A. Morphological evolution and classification of the red algal order Ceramiales inferred using plastid phylogenomics. Mol. Phylogeny Evol. 2019, 137, 76–85. [Google Scholar] [CrossRef]
- Rehmanji, M.; Kumar, A.; Nesamma, A.A.; Khan, N.J.; Fatma, T.; Jutur, P.P. Elucidation of Functional Genes Associated with Long Chain-Polyunsaturated Fatty Acids (LC-PUFAs) Metabolism in Oleaginous Diatom Phaeodactylum tricornutum. Hydrobiology 2022, 1, 451–468. [Google Scholar] [CrossRef]
- Goh, F.Q.Y.; Jeyakani, J.; Tipthara, P.; Cazenave-Gassiot, A.; Ghosh, R.; Bogard, N.; Yeo, Z.; Wong, G.K.S.; Melkonian, M.; Wenk, M.R.; et al. Gains and losses of metabolic function inferred from a phylotranscriptomic analysis of algae. Sci. Rep. 2019, 9, 10482. [Google Scholar] [CrossRef] [PubMed]
- Liu, B.; Chen, Y.; Zhu, H.; Liu, G. Phylotranscriptomic and Evolutionary Analyses of the Green Algal Order Chaetophorales (Chlorophyceae, Chlorophyta). Genes 2022, 13, 1389. [Google Scholar] [CrossRef] [PubMed]
- Cheon, S.; Lee, S.G.; Hong, H.H.; Lee, H.G.; Kim, K.Y.; Park, C.; Cheon, S.; Lee, S.G.; Hong, H.H.; Lee, H.G.; et al. A guide to phylotranscriptomic analysis for phycologists. Algae 2021, 36, 333–340. [Google Scholar] [CrossRef]
- Sellam, Y.; Gruchola, S.; Reghizzi, M.; Lugli, S.; Riedo, A.; Wurz, P. Chemical identification of fossil filament entrapped in Messinian gypsum using space Laser Mass Spectrometry, application for Mars astrobiology. In Proceedings of the EGU General Assembly, Vienna, Austria, 23–28 April 2023. [Google Scholar]
- Guillemin, M.; Valero, M.; Mauger, S.; Faugeron, S.; Destombe, C.; Utge, J.; Verdu, P.; Bon, C.; Corre, E.; Le Corguillé, G.; et al. Sequencing of seaweeds ancient DNA from Monte Verde, one of the oldest archeological sites from South America. In Proceedings of the 8th European Phycological Congress, EPC2023, Brest, France, 20–26 August 2023. [Google Scholar]
- Picard, M. Investigating Drivers of Cyanobacterial Blooms in Aotearoa—New Zealand Lakes Using Sedimentary Ancient DNA. Ph.D. Thesis, The University of Waikato, Hamilton, New Zealand, 2023. [Google Scholar]
- Nwosu, E.C.; Brauer, A.; Monchamp, M.; Pinkerneil, S.; Bartholomäus, A.; Theuerkauf, M.; Schmidt, J.; Stoof-Leichsenring, K.R.; Wietelmann, T.; Kaiser, J.; et al. Early human impact on lake cyanobacteria revealed by a Holocene record of sedimentary ancient DNA. Commun. Biol. 2023, 6, 72. [Google Scholar] [CrossRef] [PubMed]
- Hobbs, W.O.; Dreher, T.W.; Davis, E.W.; Vinebrooke, R.D.; Wong, S.; Weissman, T.; Dawson, M. Using a lake sediment record to infer the long-term history of cyanobacteria and the recent rise of an anatoxin producing Dolichospermum sp. Harmful Algae 2021, 101, 101971. [Google Scholar] [CrossRef] [PubMed]
- Schwarz, D.; Orf, I.; Kopka, J.; Hagemann, M. Recent applications of metabolomics toward cyanobacteria. Metabolites 2013, 3, 72–100. [Google Scholar] [CrossRef] [PubMed]
- Fernie, A.R.; Aharoni, A.; Willmitzer, L.; Stitt, M.; Tohge, T.; Kopka, J.; Carroll, A.J.; Saito, K.; Fraser, P.D.; DeLuca, V. Recommendations for reporting metabolite data. Plant. Cell. 2011, 23, 2477–2482. [Google Scholar] [CrossRef]
- Ferrinho, S. Gifts from Nature: Genomic and Metabolomic Approaches to Natural Product Discovery from Cyanobacteria and Actinomycetes. Doctor of Philosophy Dissertation in Chemistry. Ph.D. Thesis, The University of St Andrews, St Andrews, UK, 2023. [Google Scholar]
- Strieth, D.; Lenz, S.; Ulber, R. In vivo and in silico screening for antimicrobial compounds from cyanobacteria. MicrobiologyOpen 2022, 11, e1268. [Google Scholar] [CrossRef]
- Hiller, S.; Krock, B.; Cembella, A.; Luckas, B. Rapid detection of cyanobacterial toxins in precursor ion mode by liquid chromatography tandem mass spectrometry. J. Mass. Spectrom. 2007, 42, 1238–1250. [Google Scholar] [CrossRef] [PubMed]
- Gugger, M.; Lyra, C.; Suominen, I.; Tsitko, I.; Humbert, J.F.; Salkinoja-Salonen, M.S.; Sivonen, K. Cellular fatty acids as chemotaxonomic markers of the genera Anabaena, Aphanizomenon, Microcystis, Nostoc and Planktothrix (cyanobacteria). Int. J. Syst. Evol. Microbiol. 2002, 52, 1007–1015. [Google Scholar] [PubMed]
- Young, J.D.; Shastri, A.A.; Stephanopoulos, G.; Morgan, J.A. Mapping photoautotrophic metabolism with isotopically non stationary 13C flux analysis. Metab. Eng. 2011, 13, 656–665. [Google Scholar] [CrossRef] [PubMed]
- Kleigrewe, K.; Almaliti, J.; Tian, I.Y.; Kinnel, R.B.; Korobeynikov, A.; Monroe, E.A.; Duggan, B.M.; Di Marzo, V.; Sherman, D.H.; Dorrestein, P.C.; et al. Combining mass spectrometric metabolic profiling with genomic analysis: A powerful approach for discovering natural products from cyanobacteria. J. Nat. Prod. 2015, 78, 1671–1682. [Google Scholar] [CrossRef] [PubMed]
- Eisenhut, M.; Huege, J.; Schwarz, D.; Bauwe, H.; Kopka, J.; Hagemann, M. Metabolome phenotyping of inorganic carbon limitation in cells of wild type and photorespiratory mutants of the cyanobacterium Synechocystis sp. strain PCC 6803. Plant. Physiol. 2008, 148, 2109–2120. [Google Scholar] [CrossRef] [PubMed]
- Baran, R.; Bowen, B.P.; Northen, T.R. Untargeted metabolic footprinting reveals a surprising breadth of metabolite uptake and release by Synechococcus. sp. PCC 7002. Mol. Biosyst. 2011, 7, 3200–3206. [Google Scholar] [CrossRef]
- Huege, J.; Goetze, J.; Schwarz, D.; Bauwe, H.; Hagemann, M.; Kopka, J. Modulation of the major paths of carbon in photorespiratory mutants of Synechocystis. PLoS ONE 2011, 6, e16278. [Google Scholar] [CrossRef]
- Maghembe, R.; Damian, D.; Makaranga, A.; Nyandoro, S.S.; Lyantagaye, S.L.; Kusari, S.; Hatti-Kaul, R. Omics for bioprospecting and drug discovery from bacteria and microalgae. Antibiotics 2020, 9, 229. [Google Scholar] [CrossRef]
- Hassan, S.; Meenatchi, R.; Pachillu, K.; Bansal, S.; Brindangnanam, P.; Arockiaraj, J.; Kiran, G.S.; Selvin, J. Identification and characterization of the novel bioactive compounds from microalgae and cyanobacteria for pharmaceutical and nutraceutical applications. J. Basic. Microbiol. 2022, 62, 999–1029. [Google Scholar] [CrossRef]
- Srivastava, A.; Shukla, P. Emerging tools and strategies in cyanobacterial omics. Trends Biotechnol. 2022, 40, 4–7. [Google Scholar] [CrossRef] [PubMed]
- Shi, Y.; Che, Y.; Wang, Y.; Luan, S.; Hou, X. Loss of mature D1 leads to compromised CP43 assembly in Arabidopsis thaliana. BMC Plant Biol. 2021, 21, 106. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Chen, L.; Zhang, W. Proteomic and metabolomic analyses reveal metabolic responses to 3-hydroxypropionic acid synthesized internally in cyanobacterium Synechocystis sp. PCC 6803. Biotechnol. Biofuels 2016, 9, 209. [Google Scholar] [CrossRef] [PubMed]
- Pathania, R.; Srivastava, A.; Srivastava, S.; Shukla, P. Metabolic systems biology and multi-omics of cyanobacteria: Perspectives and future directions. Bioresour. Technol. 2022, 343, 126007. [Google Scholar] [CrossRef]
- Babele, P.K.; Kumar, J.; Chaturvedi, V. Proteomic De-Regulation in Cyanobacteria in Response to Abiotic Stresses. Front. Microbiol. 2019, 10, 1315. [Google Scholar] [CrossRef]
- Mo, R.; Yang, M.; Chen, Z.; Cheng, Z.; Yi, X.; Li, C.; He, C.; Xiong, Q.; Chen, H.; Wang, Q.; et al. Acetylome analysis reveals the involvement of lysine acetylation in photosynthesis and carbon metabolism in the model Cyanobacterium Synechocystis sp. PCC 6803. J. Proteome Res. 2015, 14, 1275–1286. [Google Scholar] [CrossRef]
- Yang, M.K.; Qiao, Z.X.; Zhang, W.Y.; Xiong, Q.; Zhang, J.; Li, T.; Ge, F.; Zhao, J.D. Global phosphoproteomic analysis reveals diverse functions of serine/ threonine/tyrosine phosphorylation in the model cyanobacterium Synechococcus sp. strain PCC 7002. J. Proteome Res. 2013, 12, 1909–1923. [Google Scholar] [CrossRef]
- Amer, B.; Baidoo, E.E.K. Omics-driven biotechnology for industrial applications. Front. Bioeng. Biotechnol. 2021, 9, 613307. [Google Scholar] [CrossRef]
- Yang, Y.; Feng, J.; Li, T.; Ge, F.; Zhao, J. CyanOmics: An integrated database of omics for the model cyanobacterium Synechococcus sp. PCC 7002. Database 2015, 2015, bau127. [Google Scholar] [CrossRef]
- Brunk, E.; Mih, N.; Monk, J.; Zhang, Z.; O’Brien, E.J.; Bliven, S.E.; Chen, K.; Chang, R.L.; Bourne, P.E.; Palsson, B.O. Systems biology of the structural proteome. BMC Syst. Biol. 2016, 10, 26. [Google Scholar] [CrossRef]
- Gu, C.; Kim, G.B.; Kim, W.J.; Kim, H.U.; Lee, S.Y. Current status and applications of genome-scale metabolic models. Genome Biol. 2019, 20, 121. [Google Scholar] [CrossRef] [PubMed]
- Chen, M.; Hernandez-Prieto, M.A.; Loughlin, P.C.; Li, Y.; Willows, R.D. Genome and proteome of the chlorophyll f-producing cyanobacterium Halomicronema hongdechloris: Adaptative proteomic shifts under different light conditions. BMC Genom. 2019, 20, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Kleessen, S.; Irgang, S.; Klie, S.; Giavalisco, P.; Nikoloski, Z. Integration of transcriptomics and metabolomics data specifies the metabolic response of Chlamydomonas to rapamycin treatment. Plant J. 2015, 81, 822–835. [Google Scholar] [CrossRef] [PubMed]
- Sanchez, B.J.; Zhang, C.; Nilsson, A.; Lahtvee, P.J.; Kerkhoven, E.J.; Nielsen, J. Improving the phenotype predictions of a yeast genome-scale metabolic model by incorporating enzymatic constraints. Mol. Syst. Biol. 2017, 13, 935. [Google Scholar] [CrossRef] [PubMed]
- Pandey, V.; Hadadi, N.; Hatzimanikatis, V.; Patil, K.R. Enhanced flux prediction by integrating relative expression and relative metabolite abundance into thermodynamically consistent metabolic models. PLoS Comput. Biol. 2019, 15, e1007036. [Google Scholar] [CrossRef] [PubMed]
- Merwin, N.J.; Mousa, W.K.; Dejong, C.A.; Skinnider, M.A.; Cannon, M.J.; Li, H.; Dial, K.; Gunabalasingam, M.; Johnston, C.; Magarvey, N.A. DeepRiPP integrates multiomics data to automate discovery of novel ribosomally synthesized natural products. Proc. Natl. Acad. Sci. USA 2020, 117, 371–380. [Google Scholar] [CrossRef] [PubMed]
- Gaj, T.; Gersbach, C.A.; Barbas, C.F., 3rd. ZFN, TALEN, and CRISPR/Cas-based methods for genome engineering. Trends Biotechnol. 2013, 31, 397–405. [Google Scholar] [CrossRef]
- Ghribi, M.; Nouemssi, S.B.; Meddeb-Mouelhi, F.; Desgagné-Penix, I. Genome Editing by CRISPR-Cas: A Game Change in the Genetic Manipulation of Chlamydomonas. Life 2020, 10, 295. [Google Scholar] [CrossRef]
- Jiang, W.; Brueggeman, A.J.; Horken, K.M.; Plucinak, T.M.; Weeks, D.P. Successful transient expression of Cas9 and single guide RNA genes in Chlamydomonas reinhardtii. Eukaryot. Cell. 2014, 13, 1465–1469. [Google Scholar] [CrossRef]
- Baek, K.; Kim, D.H.; Jeong, J.; Sim, S.J.; Melis, A.; Kim, J.S.; Jin, E.; Bae, S. DNA-free two-gene knockout in Chlamydomonas reinhardtii via CRISPR-Cas9 ribonucleoproteins. Sci. Rep. 2016, 6, 30620. [Google Scholar] [CrossRef]
- Greiner, A.; Kelterborn, S.; Evers, H.; Kreimer, G.; Sizova, I.; Hegemann, P. Targeting of Photoreceptor Genes in Chlamydomonas reinhardtii via Zinc-Finger Nucleases and CRISPR/Cas9. Plant Cell 2017, 29, 2498–2518. [Google Scholar] [CrossRef] [PubMed]
- Poliner, E.; Takeuchi, T.; Du, Z.Y.; Benning, C.; Farré, E.M. Nontransgenic Marker-Free Gene Disruption by an Episomal CRISPR System in the Oleaginous Microalga, Nannochloropsis oceanica CCMP1779. ACS Synth. Biol. 2018, 7, 962–968. [Google Scholar] [CrossRef] [PubMed]
- Stukenberg, D.; Zauner, S.; Dell’Aquila, G.; Maier, U.G. Optimizing CRISPR/cas9 for the diatom Phaeodactylum tricornutum. Front. Plant Sci. 2018, 9, 740. [Google Scholar] [CrossRef] [PubMed]
- Hopes, A.; Nekrasov, V.; Kamoun, S.; Mock, T. Editing of the urease gene by CRISPR-Cas in the diatom Thalassiosira pseudonana. Plant Methods 2016, 12, 49. [Google Scholar] [CrossRef] [PubMed]
- Lin, W.R.; Ng, I.S. Development of CRISPR/Cas9 system in Chlorella vulgaris FSP-E to enhance lipid accumulation. Enzym. Microb. Technol. 2020, 133, 109458. [Google Scholar] [CrossRef] [PubMed]
- Yao, L.; Shabestary, K.; Björk, S.M.; Asplund-Samuelsson, J.; Joensson, H.N.; Jahn, M.; Hudson, E.P. Pooled CRISPRi screening of the cyanobacterium Synechocystis sp PCC 6803 for enhanced industrial phenotypes. Nat. Commun. 2020, 11, 1666. [Google Scholar] [CrossRef] [PubMed]
- Welkie, D.G.; Rubin, B.E.; Chang, Y.G.; Diamond, S.; Rifkin, S.A.; LiWang, A.; Golden, S.S. Genome-wide fitness assessment during diurnal growth reveals an expanded role of the cyanobacterial circadian clock protein KaiA. Proc. Natl. Acad. Sci. USA 2018, 115, E7174–E7183. [Google Scholar] [CrossRef] [PubMed]
- Hsieh, S.C.; Peters, J.E. Discovery and characterization of novel type ID CRISPR-guided transposons identified among diverse Tn7-like elements in cyanobacteria. Nucleic Acids Res. 2023, 51, 765–782. [Google Scholar] [CrossRef]
- Okada, B.K.; Seyedsayamdost, M.R. Antibiotic dialogues: Induction of silent biosynthetic gene clusters by exogenous small molecules. FEMS Microbiol. Rev. 2017, 41, 19–33. [Google Scholar] [CrossRef]
- Mao, D.; Okada, B.K.; Wu, Y.; Xu, F.; Seyedsayamdost, M.R. Recent advances in activating silent biosynthetic gene clusters in bacteria. Curr. Opin. Microbiol. 2018, 45, 156–163. [Google Scholar] [CrossRef]
- Moon, K.; Xu, F.; Zhang, C.; Seyedsayamdost, M.R. Bioactivity-HiTES unveils cryptic antibiotics encoded in actinomycete bacteria. ACS Chem. Biol. 2019, 14, 767–774. [Google Scholar] [CrossRef] [PubMed]
- Xu, F.; Wu, Y.; Zhang, C.; Davis, K.M.; Moon, K.; Bushin, L.B.; Seyedsayamdost, M.R. A genetics-free method for high-throughput discovery of cryptic microbial metabolites. Nat. Chem. Biol. 2019, 15, 161–168. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez, D.J.; Haste, N.M.; Hollands, A.; Fleming, T.C.; Hamby, M.; Pogliano, K.; Nizet, V.; Dorrestein, P.C. Microbial competition between Bacillus subtilis and Staphylococcus aureus monitored by imaging mass spectrometry. Microbiology 2011, 157, 2485–2492. [Google Scholar] [CrossRef] [PubMed]
- Grindberg, R.V.; Ishoey, T.; Brinza, D.; Esquenazi, E.; Coates, R.C.; Liu, W.T.; Gerwick, L.; Dorrestein, P.C.; Pevzner, P.; Lasken, R.; et al. Single cell genome amplification accelerates identification of the apratoxin biosynthetic pathway from a complex microbial assemblage. PLoS ONE 2011, 6, e18565. [Google Scholar] [CrossRef] [PubMed]
- Gerwick, W.H.; Moore, B.S. Lessons from the past and charting the future of marine natural products drug discovery and chemical biology. Chem. Biol. 2012, 19, 85–98. [Google Scholar] [CrossRef] [PubMed]
- Molinski, T.F. NMR of natural products at the “nanomole-scale”. Nat. Prod. Rep. 2010, 27, 321. [Google Scholar] [CrossRef] [PubMed]
- Mohimani, H.; Liu, W.T.; Mylne, J.S.; Poth, A.G.; Colgrave, M.L.; Tran, D.; Selsted, M.E.; Dorrestein, P.C.; Pevzner, P.A. Cycloquest: Identification of cyclopeptides via database search of their mass spectra against genome databases. J. Proteome Res. 2011, 10, 4505–4512. [Google Scholar] [CrossRef] [PubMed]
- Saccomano, S.C.; Jewell, M.P.; Cash, K.J. A review of chemosensors and biosensors for monitoring biofilm dynamics. Sens. Actuators Rep. 2021, 3, 100043. [Google Scholar] [CrossRef]
- Cheah, Y.T.; Chan, D.J.C. A methodological review on the characterization of microalgal biofilm and its extracellular polymeric substances. J. Appl. Microbiol. 2022, 132, 3490–3514. [Google Scholar] [CrossRef]
- Kliphuis, A.M.J. Modeling of Microalgal Metabolism. Ph.D. Thesis, Wageningen University, Wageningen, The Netherlands, 2010. [Google Scholar]
- López-Gejo, J.; Haigh, D.; Orellana, G. Relationship between the microscopic and macroscopic world in optical oxygen sensing: A luminescence lifetime microscopy study. Langmuir 2010, 26, 2144–2150. [Google Scholar] [CrossRef] [PubMed]
- Zhong, N.; Wu, Y.; Wang, Z.; Chang, H.; Zhong, D.; Xu, Y.; Hu, X.; Huang, L. Monitoring Microalgal Biofilm Growth and Phenol Degradation with Fiber-Optic Sensors. Anal. Chem. 2019, 91, 15155–15162. [Google Scholar] [CrossRef] [PubMed]
- Wolfbeis, O.S. Luminescent sensing and imaging of oxygen: Fierce competition to the Clark electrode. BioEssays 2015, 37, 921–928. [Google Scholar] [CrossRef] [PubMed]
- Rincon, S.M.; Urrego, N.F.; Avila, K.J.; Romero, H.M.; Beyenal, H. Photosynthetic activity assessment in mixotrophically cultured Chlorella vulgaris biofilms at various developmental stages. Algal Res. 2019, 38, 101408. [Google Scholar] [CrossRef]
- Chen, M.; Xin, X.; Liu, H.; Wu, Y.; Zhong, N.; Chang, H. Monitoring biohydrogen production and metabolic heat in biofilms by fiber Bragg grating sensors. Anal. Chem. 2019, 91, 7842–7849. [Google Scholar] [CrossRef] [PubMed]
- Boukazia, Y.; Delaplace, G.; Cadé, M.; Bellouard, F.; Fillaudeau, L. On-line biofouling monitoring and qualification based on local thermal and periodic excitation with MEMS sensor. Food Bioprod. Process. 2021, 126, 12–22. [Google Scholar] [CrossRef]
- Babauta, J.T.; Nguyen, H.D.; Harrington, T.D.; Renslow, R.; Beyenal, H. pH, redox potential and local biofilm potential microenvironments within geobacter sulfurreducens biofilms and their roles in electron transfer. Biotechnol. Bioeng. 2012, 109, 2651–2662. [Google Scholar] [CrossRef] [PubMed]
- Kawamura, A.; Miyata, T. Biosensors. In Biomaterials Nanoarchitectonics; Ebara, M., Ed.; Elsevier: Amsterdam, The Netherlands, 2016; Chapter 1; pp. 157–176. [Google Scholar]
- Vaz, R.; Valpradinhos, B.; Frasco, M.F.; Sales, M.G.F. Emerging optical materials in sensing and discovery of bioactive compounds. Sensors 2021, 21, 5784. [Google Scholar] [CrossRef]
- Páscoa, I.; Biltes, R.; Sousa, J.; Correia Preto, M.A.; Vasconcelos, V.; Castro, L.F.; Ruivo, R.; Cunha, I. A Multiplex Molecular Cell-Based Sensor to Detect Ligands of PPARs: An Optimized Tool for Drug Discovery in Cyanobacteria. Sensors 2023, 23, 1338. [Google Scholar] [CrossRef]
- Li, M.; Paidi, S.K.; Sakowski, E.; Preheim, S.; Barman, I. Ultrasensitive Detection of Hepatotoxic Microcystin Production from Cyanobacteria Using Surface-Enhanced Raman Scattering Immunosensor. ACS Sens. 2019, 4, 1203–1210. [Google Scholar] [CrossRef]
- Thomas, A.M.; Segata, N. Multiple levels of the unknown in microbiome research. BMC Biol. 2019, 17, 48. [Google Scholar] [CrossRef]
- Cheng, J.; Zhang, K.; Hou, Y. The current situations and limitations of genetic engineering in cyanobacteria: A mini review. Mol. Biol. Rep. 2023, 50, 5481–5487. [Google Scholar] [CrossRef]
- Shailaja, V.L.; John, C.M.; Kalaivani, M.K. Genetic engineering of algae material: Merits and demerits. In Algae Materials: Applications Benefitting Health; Arunkumar, K., Arun, A., Raja, R., Palaniappan, R., Eds.; Elsevier Inc.: London, UK, 2023; pp. 355–382. [Google Scholar]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zammit, G.; Zammit, M.G.; Buttigieg, K.G. Emerging Technologies for the Discovery of Novel Diversity in Cyanobacteria and Algae and the Elucidation of Their Valuable Metabolites. Diversity 2023, 15, 1142. https://doi.org/10.3390/d15111142
Zammit G, Zammit MG, Buttigieg KG. Emerging Technologies for the Discovery of Novel Diversity in Cyanobacteria and Algae and the Elucidation of Their Valuable Metabolites. Diversity. 2023; 15(11):1142. https://doi.org/10.3390/d15111142
Chicago/Turabian StyleZammit, Gabrielle, Maria G. Zammit, and Kyle G. Buttigieg. 2023. "Emerging Technologies for the Discovery of Novel Diversity in Cyanobacteria and Algae and the Elucidation of Their Valuable Metabolites" Diversity 15, no. 11: 1142. https://doi.org/10.3390/d15111142
APA StyleZammit, G., Zammit, M. G., & Buttigieg, K. G. (2023). Emerging Technologies for the Discovery of Novel Diversity in Cyanobacteria and Algae and the Elucidation of Their Valuable Metabolites. Diversity, 15(11), 1142. https://doi.org/10.3390/d15111142





