The Cell Wall-Related Gene Families of Wheat (Triticum aestivum)
Abstract
1. Introduction
2. Materials and Methods
3. Results and Discussion
4. Conclusions
Supplementary Materials
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
References
- Asseng, S.; Guarin, J.R.; Raman, M.; Monje, O.; Kiss, G.; Despommier, D.D.; Meggars, F.M.; Gauthier, P.P.G. Wheat yield potential in controlled-environment vertical farms. Proc. Natl. Acad. Sci. USA 2020, 117, 19131–19135. [Google Scholar] [CrossRef]
- Wheat Explorer. Available online: https://ipad.fas.usda.gov/cropexplorer/cropview/commodityView.aspx?cropid=0410000 (accessed on 21 August 2023).
- Tripathi, M.K.; Karim, S.A.; Chaturvedi, O.H.; Verma, D.L. Nutritional value of animal feed grade wheat as a replacement for maize in lamb feeding for mutton production. J. Sci. Food Agric. 2007, 87, 2447–2455. [Google Scholar] [CrossRef]
- Patwa, N.; Penning, B.W. Environmental impact on cereal crop grain damage from pre-harvest sprouting and late maturity alpha-amylase. In Sustainable Agriculture in the Era of Climate Change; Roychowdhury, R., Choudhury, S., Hasanuzzaman, M., Srivastava, S., Eds.; Springer Nature: Cham, Switzerland, 2020; pp. 23–41. [Google Scholar] [CrossRef]
- Farrokhi, N.; Burton, R.A.; Brownfield, L.; Hrmova, M.; Wilson, S.M.; Bacic, A.; Fincher, G.B. Plant Cell wall biosynthesis: Genetic, biochemical and functional genomics approaches to the identification of key genes. Plant Biotechnol. J. 2006, 4, 145–167. [Google Scholar] [CrossRef]
- Shrivastava, B.; Jain, K.K.; Kalra, A.; Kuhad, R.C. Bioprocessing of wheat straw into nutritionally rich and digested cattle feed. Sci. Rep. 2014, 4, 6360. [Google Scholar] [CrossRef] [PubMed]
- Tsegaye, B.; Balomajumder, C.; Roy, P. Optimization of microwave and NaOH pretreatments of wheat straw for enhancing biofuel yield. Energy Convers. Manag. 2019, 186, 82–92. [Google Scholar] [CrossRef]
- Yong, W.; Link, B.; O’Malley, R.; Tewari, J.; Hunter, C.T.; Lu, C.A.; Li, X.; Bleecker, A.B.; Koch, K.E.; McCann, M.C.; et al. Genomics of plant cell wall biogenesis. Planta 2005, 221, 747–751. [Google Scholar] [CrossRef]
- Nirmal, R.C.; Furtado, A.; Rangan, P.; Henry, R.J. Fasciclin-like arabinogalactan protein gene expression is associated with yield of flour in the milling of wheat. Sci. Rep. 2017, 7, 12539. [Google Scholar] [CrossRef]
- Penning, B.W. Gene expression differences related to pre-harvest sprouting uncovered in related wheat varieties by RNAseq analysis. Plant Gene 2023, 33, 100404. [Google Scholar] [CrossRef]
- Drula, E.; Garron, M.L.; Dogan, S.; Lombard, V.; Henrissat, B.; Terrapon, N. The carbohydrate-active enzyme database: Functions and literature. Nucl. Acids Res. 2022, 50, D571–D577. [Google Scholar] [CrossRef] [PubMed]
- Penning, B.W.; McCann, M.C.; Carpita, N.C. Evolution of the cell wall gene families of grasses. Front. Plant Sci. 2019, 10, 1205. [Google Scholar] [CrossRef]
- Penning, B.W.; Hunter, C.T.; Tayengwa, R.; Eveland, A.L.; Dugard, C.K.; Olek, A.T.; Vermerris, W.; Koch, K.E.; McCarty, D.R.; Davis, M.F.; et al. Genetic Resources for maize cell wall biology. Plant Physiol. 2009, 151, 1703–1728. [Google Scholar] [CrossRef]
- Potter, S.C.; Luciani, A.; Eddy, S.R.; Park, Y.; Lopez, R.; Finn, R.D. HMMER webserver: 2018 update. Nucl. Acids Res. 2018, 46, W200–W204. [Google Scholar] [CrossRef] [PubMed]
- Sigrist, C.J.A.; de Castro, E.; Cerutti, L.; Cuche, B.A.; Hulo, N.; Bridge, A.; Bougueleret, L.; Xenarios, I. New and continuing developments at PROSITE. Nucl. Acids Res. 2013, 41, D344–D347. [Google Scholar] [CrossRef]
- Carpita, N.C. Structure and biogenesis of the cell walls of grasses. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1996, 47, 445–476. [Google Scholar] [CrossRef]
- Penning, B.W.; Shiga, T.M.; Klimek, J.F.; SanMiguel, P.J.; Shreve, J.; Thimmapuram, J.; Sykes, R.W.; Davis, M.F.; McCann, M.C.; Carpita, N.C. Expression profiles of cell-wall related genes vary broadly between two common maize inbreds during stem development. BMC Genom. 2019, 20, 785. [Google Scholar] [CrossRef] [PubMed]
- CAZY. Available online: http://www.cazy.org/ (accessed on 10 August 2023).
- Orellana, A.; Moraga, C.; Araya, M.; Moreno, A. Overview of nucleotide sugar transporter gene family functions across multiple species. J. Mol. Biol. 2016, 428, 3150–3165. [Google Scholar] [CrossRef] [PubMed]
- Bonawitz, N.D.; Chapple, C. The genetics of lignin biosynthesis: Connecting genotype to phenotype. Annu. Rev. Genet. 2010, 44, 337–363. [Google Scholar] [CrossRef]
- Reiter, W.D.; Vanzin, G.F. Molecular genetics of nucleotide sugar interconversion pathways in plants. Plant Mol. Biol. 2001, 47, 95–113. [Google Scholar] [CrossRef]
- Yin, Y.; Huang, J.; Gu, X.; Bar-Peled, M.; Xu, Y. Evolution of plant nucleotide-sugar interconversion enzymes. PLoS ONE 2011, 6, e27995. [Google Scholar] [CrossRef]
- Favery, B.; Ryan, E.; Foreman, J.; Linstead, P.; Boudonck, K.; Steer, M.; Shaw, P.; Dolan, L. KOJAK encodes a cellulose synthase-like protein required for root hair cell morphogenesis in Arabidopsis. Genes Dev. 2001, 15, 79–89. [Google Scholar] [CrossRef]
- Holland, N.; Holland, D.; Helentjaris, T.; Dhugga, K.; Xoconostle-Cazares, B.; Delmer, D.P. A comparative analysis of the plant cellulose synthase (CesA) gene family. Plant Physiol. 2000, 123, 1313–1323. [Google Scholar] [CrossRef]
- Burton, R.A.; Wilson, S.M.; Hrmova, M.; Harvey, A.J.; Shirley, N.J.; Medhurst, A.; Stone, B.A.; Newbigin, E.J.; Bacic, A.; Fincher, G.B. Cellulose synthase-like CslF genes mediate the synthesis of cell wall (1,3;1,4)-b-D-glucans. Science 2006, 311, 1940–1942. [Google Scholar] [CrossRef]
- Cocuron, J.C.; Lerouxel, O.; Drakakaki, G.; Alonso, A.P.; Liepman, A.H.; Keegstra, K.; Raikhel, N.; Wilkerson, C.G. A gene from the cellulose synthase-like C family encodes a β-1,4 glucan synthase. Proc. Natl. Acad. Sci. USA 2007, 104, 8550–8555. [Google Scholar] [CrossRef]
- Dhugga, K.S.; Barreiro, R.; Whitten, B.; Stecca, K.; Hazebroek, J.; Randhawa, G.S.; Dolan, M.; Kinney, A.J.; Tomes, D.; Nichols, S.; et al. Guar seed β-mannan synthase is a member of the cellulose synthase super gene family. Science 2004, 303, 363–366. [Google Scholar] [CrossRef]
- Liu, X.; Zhang, H.; Zhang, W.; Xu, W.; Li, S.; Chen, X.; Chen, H. Genome-wide bioinformatics analysis of cellulose synthase gene family in common bean (Phaseolus vulgaris L.) and the expression in the pod development. BMC Genom. Data 2022, 23, 9. [Google Scholar] [CrossRef] [PubMed]
- Wang, B.; Andargie, M.; Fang, R. The function and biosynthesis of callose in high plants. Heliyon 2022, 8, e09248. [Google Scholar] [CrossRef]
- Anders, N.; Wilkinson, M.D.; Lovegrove, A.; Freeman, J.; Tryfona, T.; Pellny, T.K.; Weimar, T.; Mortimer, J.C.; Stott, K.; Baker, J.M.; et al. Glycosyl transferases in family 61 mediate arabinofuranosyl transfer onto xylan in grasses. Proc. Natl. Acad. Sci. USA 2012, 109, 989–993. [Google Scholar] [CrossRef] [PubMed]
- Chiniquy, D.; Sharma, V.; Schultink, A.; Baidoo, E.E.; Rautengarten, C.; Cheng, K.; Carroll, A.; Ulvskov, P.; Harholt, J.; Keasling, J.D.; et al. XAX1 from glycosyltransferase family 61 mediates xylosyl transfer to rice xylan. Proc. Natl. Acad. Sci. USA 2012, 109, 17117–17122. [Google Scholar] [CrossRef] [PubMed]
- Feijao, C.; Morreel, K.; Anders, A.; Tryfona, T.; Busse-Wicher, M.; Kotake, T.; Boerjan, W.; Dupree, P. Hydroxycinnamic acid-modified xylan side chains and their cross-linking products in rice cell walls are reduced in the Xylosyl arabinosyl substitution of xylan 1 mutant. Plant J. 2022, 109, 1152–1167. [Google Scholar] [CrossRef]
- Egelund, J.; Obel, N.; Ulvskov, P.; Geshi, N.; Pauly, M.; Bacic, A.; Petersen, B.L. Molecular characterization of two Arabidopsis thaliana glycosyltransferase mutants, rra1 and rra2, which have a reduced residual arabinose content in a polymer tightly associated with the cellulosic wall residue. Plant Mol. Biol. 2007, 64, 439–451. [Google Scholar] [CrossRef] [PubMed]
- Scheller, H.V.; Jensen, J.K.; Sørensen, S.O.; Harholt, J.; Geshi, N. Biosynthesis of pectin. Physiol. Plant. 2007, 129, 283–295. [Google Scholar] [CrossRef]
- Sterling, J.D.; Atmodjo, M.A.; Inwood, S.E.; Kolli, V.S.K.; Quigley, H.F.; Hahn, M.G.; Mohnen, D. Functional identification of an Arabidopsis pectin biosynthesic homogalacturonan galacturonosyltransferase. Proc. Natl. Acad. Sci. USA 2006, 103, 5236–5241. [Google Scholar] [CrossRef]
- Lee, C.; Zhong, R.; Richardson, E.A.; Himmelsbach, D.S.; McPhail, B.T.; Ye, Z.H. The PARVUS gene is expressed in cells undergoing secondary wall thickening and is essential for glucuronoxylan biosynthesis. Plant Cell Physiol. 2007, 48, 1659–1672. [Google Scholar] [CrossRef] [PubMed]
- Qu, Y.; Egelund, J.; Gilson, P.R.; Houghton, F.; Gleeson, P.A.; Schultz, C.J.; Bacic, A. Identification of a novel group of putative Arabidopsis thaliana β-(1,3)-galactosyltransferases. Plant Mol. Biol. 2008, 68, 43–59. [Google Scholar] [CrossRef]
- Strasser, R.; Bondili, J.S.; Vavra, U.; Schoberer, J.; Svoboda, B.; Glössl, J.; Léonard, R.; Stadlmann, J.; Altmann, F.; Steinkellner, H.; et al. A unique β1,3-galactosyltransferase is indispensable for the biosynthesis of N-glycans containing Lewis a structures in Arabidopsis thaliana. Plant Cell 2007, 19, 2278–2292. [Google Scholar] [CrossRef]
- Cavalier, D.M.; Lerouxel, O.; Neumetzler, L.; Yamauchi, K.; Reinecke, A.; Freshour, G.; Zabotina, O.A.; Hahn, M.G.; Burgert, I.; Pauly, M.; et al. Disrupting two Arabidopsis thaliana xylosyltransferase genes results in plants deficient in xyloglucan, a major primary cell wall component. Plant Cell 2008, 20, 1519–1537. [Google Scholar] [CrossRef] [PubMed]
- Sarria, R.; Wagner, T.A.; O’Neill, M.A.; Faik, A.; Wilkerson, C.G.; Keegstra, K.; Raikhel, N.V. Characterization of a family of Arabidopsis genes related to xyloglucan fucosyltransferase1. Plant Physiol. 2001, 127, 1595–1606. [Google Scholar] [CrossRef] [PubMed]
- Vanzin, G.F.; Madson, M.; Carpita, N.C.; Raikhel, N.V.; Keegstra, K.; Reiter, W.D. The mur2 mutant of Arabidopsis thaliana lacks fucosylated xyloglucan because of a lesion in fucosyltransferase AtFUT1. Proc. Natl. Acad. Sci. USA 2002, 99, 3340–3345. [Google Scholar] [CrossRef] [PubMed]
- Harholt, J.; Jensen, J.K.; Sørensen, S.O.; Orfila, C.; Pauly, M.; Scheller, H.V. ARABINAN DEFICIENT1 is a putative arabinosyltransferase involved in biosynthesis of pectic arabinan in Arabidopsis. Plant Physiol. 2006, 140, 49–58. [Google Scholar] [CrossRef]
- Jensen, J.K.; Sørensen, S.O.; Harholt, J.; Geshi, N.; Sakuragi, Y.; Møller, I.; Zandleven, J.; Bernal, A.J.; Jensen, N.B.; Sørensen, C.; et al. Identification of a xylogalacturonan xylosyltransferase involved in pectin biosynthesis in Arabidopsis. Plant Cell 2008, 20, 1289–1302. [Google Scholar] [CrossRef]
- Madson, M.; Dunand, C.; Li, X.; Verma, R.; Vanzin, G.F.; Caplan, J.; Shoue, D.A.; Carpita, N.C.; Reiter, W.-D. The MUR3 gene of Arabidopsis encodes a xyloglucan galactosyltransferase that is evolutionarily related to animal exostosins. Plant Cell 2003, 15, 1662–1670. [Google Scholar] [CrossRef]
- Peña, M.J.; Zhong, R.; Zhou, G.K.; Richardson, E.A.; O’Neill, M.A.; Darvill, A.G.; York, W.S.; Ye, Z.H. Arabidopsis irregular xylem8 and irregular xylem9: Implications for the complexity of glucuronoxylan biosynthesis. Plant Cell 2007, 19, 549–563. [Google Scholar] [CrossRef] [PubMed]
- Zhong, R.; Peña, M.J.; Zhou, G.K.; Nairn, C.J.; Wood-Jones, A.; Richardson, E.A.; Morrison, W.H.; Darvill, A.G.; York, W.S.; Ye, Z.-H. Arabidopsis fragile fiber8, which encodes a putative glucuronosyltransferase, is essential for normal secondary wall synthesis. Plant Cell 2005, 17, 3390–3408. [Google Scholar] [CrossRef] [PubMed]
- Brown, D.M.; Zhang, Z.; Stephens, E.; Dupree, P.; Turner, S.R. Characterization of IRX10 and IRX10-like reveals an essential role in glucuronoxylan biosynthesis in Arabidopsis. Plant J. 2009, 57, 732–746. [Google Scholar] [CrossRef]
- Cosgrove, D.J. Loosening of plant cell walls by expansins. Nature 2000, 407, 321–326. [Google Scholar] [CrossRef] [PubMed]
- Rose, J.K.C.; Braam, J.; Fry, S.C.; Nishitani, K. The XTH family of enzymes involved in xyloglucan endotransglucosylation and endohydrolysis: Current perspectives and a new unifying nomenclature. Plant Cell Physiol. 2002, 43, 1421–1435. [Google Scholar] [CrossRef]
- Akiyama, T.; Pillai, M.A.; Sentoku, N. Cloning, characterization and expression of OsGLN2, a rice endo-1,3-beta-glucanase gene regulated developmentally in flowers and hormonally in germinating seeds. Planta 2004, 220, 129–139. [Google Scholar] [CrossRef]
- Liu, B.; Lu, Y.; Xin, Z.; Zhang, Z. Identification and antifungal assay of a wheat B-1,3-glucanase. Biotechnol. Lett. 2009, 31, 1005–1010. [Google Scholar] [CrossRef]
- Minic, Z. Physiological roles of plant glycoside hydrolases. Planta 2008, 227, 723–740. [Google Scholar] [CrossRef]
- Nawaz, M.A.; Rehman, H.M.; Imtiaz, M.; Baloch, F.S.; Lee, J.D.; Yang, S.H.; Lee, S.I.; Chung, G. Systems identification and characterization of cell wall reassembly and degradation related genes in Glycine max (L.) Merill, a bioenergy legume. Sci. Rep. 2017, 7, 10862. [Google Scholar] [CrossRef]
- Schlumbaum, A.; Mauch, F.; Vögeli, U.; Boller, T. Plant chitinases are potent inhibitors of fungal growth. Nature 1986, 324, 365–367. [Google Scholar] [CrossRef]
- Jindou, S.; Xu, Q.; Kenig, R.; Shulman, M.; Shoham, Y.; Bayer, E.A.; Lamed, R. Novel architecture of family-9 glycoside hydrolases identified in cellulosal enzymes of Acetivibrio cellulyticus and Clostridium thermocellum. FEMS Microbiol. Lett. 2006, 254, 308–316. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Jenkins, E.S.; Paul, W.; Craze, M.; Whitelaw, A.; Weigand, A.; Roberts, J.A. Dehiscence-related expression of an Arabidopsis thaliana gene encoding a polygalacturonase in transgenic plants of Brassica napus. Plant Cell Environ. 1999, 22, 159–167. [Google Scholar] [CrossRef]
- Sampedro, J.; Gianzo, C.; Iglesias, N.; Guitián, E.; Revilla, G.; Zarra, I. AtBGAL10 is the main xyloglucan β-galactosidase in Arabidopsis, and its absence results in unusual xyloglucan subunits and growth defects. Plant Physiol. 2012, 158, 1146–1157. [Google Scholar] [CrossRef] [PubMed]
- Gou, Y.Y.; Miller, L.M.; Hou, G.; Yu, X.H.; Chen, X.Y.; Liu, C.J. Acetylesterase-mediated deacetylation of pectin impairs cell elongation, pollen germination, and plant reproduction. Plant Cell 2012, 24, 50–65. [Google Scholar] [CrossRef]
- Philippe, F.; Pelloux, J.; Rayon, C. Plant pectin acetylesterase structure and function: New insights from bioinformatic analysis. BMG Genom. 2017, 18, 456. [Google Scholar] [CrossRef]
- Louvet, R.; Cavel, E.; Gutierrez, L.; Guénin, S.; Roger, D.; Gillet, F.; Guerineau, F.; Pelloux, J. Comprehensive expression profiling of the pectin methylesterase gene family during silique development in Arabidopsis thaliana. Planta 2006, 224, 782–791. [Google Scholar] [CrossRef]
- Domingo, C.; Roberts, K.; Stacey, N.J.; Connerton, I.; Ruíz-Teran, F.; McCann, M.C. A pectate lyase from Zinnia elegans is auxin inducible. Plant J. 1998, 13, 17–28. [Google Scholar] [CrossRef]
- Ochoa-Jiménez, V.A.; Berumen-Varela, G.; Burgara-Estrella, A.; Orozco-Avitia, J.A.; Ojeda-Contreras, A.J.; Trillo-Hernández, E.A.; Rivera-Domínguez, M.; Troncoso-Rojas, R.; Báez-Sañudo, R.; Datsenka, T.; et al. Functional analysis of tomato rhamnogalacturonan lyase gene Solyc11g011300 during fruit development and ripening. J. Plant Physiol. 2018, 231, 31–40. [Google Scholar] [CrossRef]
- Canut, H.; Albenne, C.; Jamet, E. Post-translational modifications of plant cell wall proteins and peptides: A survey from a proteomics point of view. Biochim. Biophys. Acta–Proteins Proteom. 2016, 1864, 983–990. [Google Scholar] [CrossRef]
- Sterjiades, R.; Dean, J.F.D.; Gamble, G.; Himmelsbach, D.S.; Eriksson, K.L. Extracellular laccases and peroxidases from sycamore maple (Acer pseudoplatanus) cell-suspension cultures. Planta 1993, 190, 75–83. [Google Scholar] [CrossRef]
- Brown, D.M.; Zeef, L.A.H.; Ellis, J.; Goodacre, R.; Turner, S.R. Identification of novel genes in Arabidopsis involved in secondary cell wall formation using expression profiling and reverse genetics. Plant Cell 2005, 17, 2281–2295. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Qian, Q.; Zhou, Y.; Yan, M.; Sun, L.; Zhang, M.; Fu, Z.; Wang, Y.; Han, B.; Pang, X.; et al. BRITTLE CULM1, which encodes a COBRA-like protein, affects the mechanical properties of rice plants. Plant Cell 2003, 15, 2020–2031. [Google Scholar] [CrossRef] [PubMed]
- Velasquez, S.M.; Ricardi, M.M.; Poulsen, C.P.; Oikawa, A.; Dilokpimol, A.; Halim, A.; Mangano, S.; Juarez, S.P.D.; Marzol, E.; Salter, J.D.S.; et al. Complex regulation of prolyl-5-hydroxylases impacts root hair expansion. Mol. Plant 2015, 8, 734–746. [Google Scholar] [CrossRef]
- Califar, B.; Sng, N.J.; Zupanska, A.; Paul, A.L.; Ferl, R.J. Root skewing-associated genes impact the spaceflight response of Arabidopsis thaliana. Front. Plant Sci. 2020, 11, 239. [Google Scholar] [CrossRef]
- Sedbrook, J.C.; Carroll, K.L.; Hung, K.F.; Masson, P.H.; Somerville, C.R. The Arabidopsis SKU5 gene encodes an extracellular glycosyl phosphatidylinositol-anchored glycoprotein involved in directional root growth. Plant Cell 2002, 14, 1635–1648. [Google Scholar] [CrossRef]
- Zhou, C.; Dong, Z.; Zhang, T.; Wu, J.; Yu, S.; Zeng, Q.; Han, D.; Tong, W. Genome-scale analysis of homologous genes among subgenomes of bread wheat (Triticum aestivum L.). Int. J. Mol. Sci. 2020, 21, 3015. [Google Scholar] [CrossRef]
- Johnson, D.A.; Thomas, M.A. The monosaccharide transporter gene family in Arabidopsis and rice: A history of duplications, adaptive evolution, and functional divergence. Mol. Biol. Evol. 2007, 24, 2412–2423. [Google Scholar] [CrossRef]
- Xu, A.F.; Molinuevo, R.; Fazzari, E.; Tom, H.; Zhang, Z.; Menendez, J.; Casey, K.M. Subfunctionalized expression drives evolutionary retention of ribosomal protein paralogs Rps27 and Rps27l in vertebrates. eLife 2023, 12, e78695. [Google Scholar] [CrossRef]
- Zhu, T.; Wang, L.; Rimbert, H.; Rodriguez, J.C.; Deal, K.R.; De Oliveira, R.; Choulet, F.; Keeble-Gagnère, G.; Tibbits, J.; Rogers, J.; et al. Optical maps refine the bread wheat Triticum aestivum cv Chinese Spring genome assembly. Plant J. 2021, 107, 303–314. [Google Scholar] [CrossRef]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef] [PubMed]
- International Wheat Genome Sequencing Consortium. Available online: https://www.wheatgenome.org/ (accessed on 21 August 2023).
- MaizeGDB. Available online: https://www.maizegdb.org/ (accessed on 21 August 2023).
- Rice Genome Annotation Project. Available online: http://rice.uga.edu/ (accessed on 21 August 2023).
- The Arabidopsis Information Reseource. Available online: https://www.arabidopsis.org/ (accessed on 21 August 2023).
- Chenna, R.; Sugawara, H.; Koike, T.; Lopez, R.; Gibson, T.J.; Higgins, D.G.; Thompson, J.D. Multiple sequence alignment with the Clustal series of programs. Nucl. Acids Res. 2003, 31, 3497–3500. [Google Scholar] [CrossRef]
- Saitou, N.; Nei, M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987, 4, 406–425. [Google Scholar] [CrossRef]
- Thompson, J.D.; Higgins, D.G.; Gibson, T.J. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position specific gap penalties and weight matrix choice. Nucl. Acids Res. 1994, 22, 4673–4680. [Google Scholar] [CrossRef] [PubMed]
- Chevenet, F.; Brun, C.; Banuls, A.L.; Jacq, B.; Christen, R. TreeDyn: Towards dynamic graphics and annotations for analyses of trees. BMC Bioinform. 2006, 7, 439. [Google Scholar] [CrossRef]
- HMMER. Available online: https://www.ebi.ac.uk/Tools/hmmer/ (accessed on 11 August 2023).
- Prosite. Available online: https://prosite.expasy.org (accessed on 11 August 2023).
- Clustal Omega. Available online: https://www.ebi.ac.uk/Tools/msa/clustalo/ (accessed on 11 August 2023).
- Madeira, F.; Pearce, M.; Tivey, A.R.; Basutkar, P.; Lee, J.; Edbali, O.; Madhusoodanan, N.; Kolesnikov, A.; Lopez, R. Search and sequence analysis tools services from EMBL-EBI in 2022. Nucl. Acids Res. 2022, 50, W276–W279. [Google Scholar] [CrossRef]
- Pellny, T.K.; Patil, A.; Wood, A.J.; Freeman, J.; Halsey, K.; Plummer, A.; Kosik, O.; Temple, H.; Collins, J.D.; Dupree, P.; et al. Loss of TaIRX9b gene function in wheat decreases chain length and amount of arabinoxylan in grain but increases cross-linking. Plant Biotech. J. 2020, 18, 2316–2327. [Google Scholar] [CrossRef]
- Gille, S.; Hänsel, U.; Ziemann, M.; Pauly, M. Identification of plant cell wall mutants by means of a forward chemical genetic approach using hydrolases. Proc. Natl. Acad. Sci. USA 2009, 25, 14699–14704. [Google Scholar] [CrossRef]
- Knoch, E.; Dilokpimol, A.; Tryfona, T.; Poulsen, C.P.; Xiong, G.; Harholt, J.; Petersen, B.L.; Ulvskov, P.; Hadi, M.Z.; Kotake, T.; et al. A β-glucuronosyltransferase from Arabidopsis thaliana involved in biosynthesis of type II arabinogalactan has a role in cell elongation during seedling growth. Plant J. 2013, 76, 1016–1029. [Google Scholar] [CrossRef]
- Sampedro, J.; Cosgrove, D.J. The expansin superfamily. Genome Biol. 2005, 6, 242. [Google Scholar] [CrossRef][Green Version]
- Tenhaken, R. Cell wall remodeling under abiotic stress. Front. Plant Sci. 2015, 5, 771. [Google Scholar] [CrossRef]
- Chono, M.; Honda, I.; Shinoda, S.; Kushiro, T.; Kamiya, Y.; Nambara, E.; Kawakami, N.; Kaneko, S.; Watanabe, Y. Field studies on the regulation of abscisic acid content and germinability during grain development of barley: Molecular and chemical analysis of pre-harvest sprouting. J. Exp. Bot. 2006, 57, 2421–2434. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Wang, C.; Zhu, M.; Yu, Y.; Zhang, Y.; Wei, Z. Generation and characterization of transgenic poplar plants overexpressing a cotton laccase gene. Plant Cell Tissue Organ Cult. 2008, 93, 303–310. [Google Scholar] [CrossRef]
- Shigeto, J.; Tsutsumi, Y. Diverse functions and reactions of class III peroxidases. New Phytol. 2016, 209, 1395–1402. [Google Scholar] [CrossRef]
- Zipor, G.; Duarte, P.; Carqueijeiro, I.; Shahar, L.; Ovadia, R.; Teper-Bamnolker, P.; Eshel, D.; Levin, Y.; Doron-Faigenboim, A.; Sottomayor, M.; et al. In planta anthocyanin degradation by a vacuolar class III peroxidase in Brunfelsia calycina flowers. New Phytol. 2015, 205, 653–665. [Google Scholar] [CrossRef]
- Martinez, M.; Gómez-Cabellos, S.; Giménez, M.J.; Barro, F.; Diaz, I.; Diaz-Mendoza, M. Plant Proteases: From key enzymes in germination to allies for fighting human gluten-related disorders. Front. Plant Sci. 2019, 10, 721. [Google Scholar] [CrossRef]
- Berthet, S.; Thevin, J.; Baratiny, D.; Demont-Caulet, N.; Debeaujon, I.; Bidzinski, P.; Leple, J.; Huis, R.; Hawkins, S.; Gomez, L.D.; et al. Chapter 5—Role of plant laccases in lignin polymerization. In Advances in Botanical Research; Jouanin, J., Lapierre, C., Eds.; Academic Press: London, UK, 2012; Volume 61, pp. 145–172. [Google Scholar] [CrossRef]
- Pant, S.; Huang, Y. Genome-wide studies of PAL genes in sorghum and their responses to aphid infestation. Sci. Rep. 2022, 12, 22537. [Google Scholar] [CrossRef]
- Riaz, M.W.; Yousaf, M.I.; Hussain, Q.; Yasir, M.; Sajjad, M.; Shah, L. Role of lignin in wheat plant for the enhancement of resistance against lodging and biotic and abiotic stresses. Stresses 2023, 3, 434–453. [Google Scholar] [CrossRef]
- Pellny, T.K.; Lovegrove, A.; Freeman, J.; Tosi, P.; Love, C.G.; Knox, J.P.; Shewry, P.R.; Mitchell, R.A.C. Cell walls of developing wheat starchy endosperm: Comparison of composition and RNA-seq transcriptome. Plant Phyisol. 2012, 158, 612–627. [Google Scholar] [CrossRef]

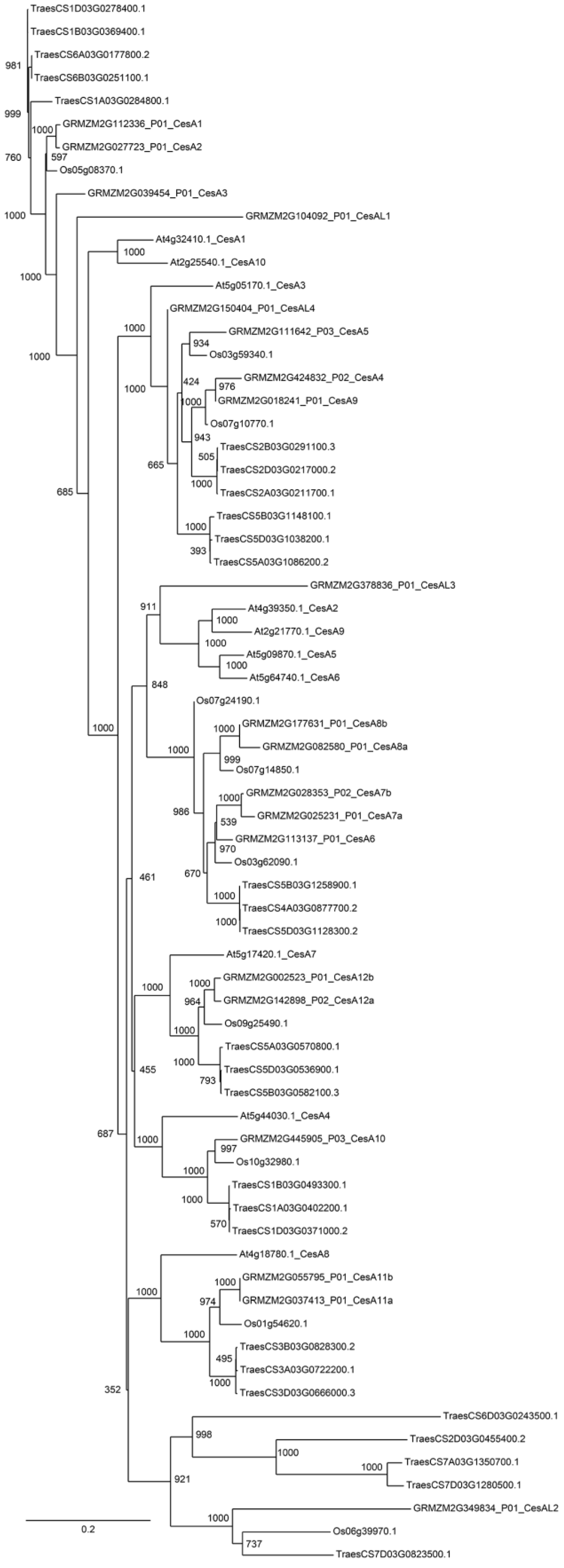
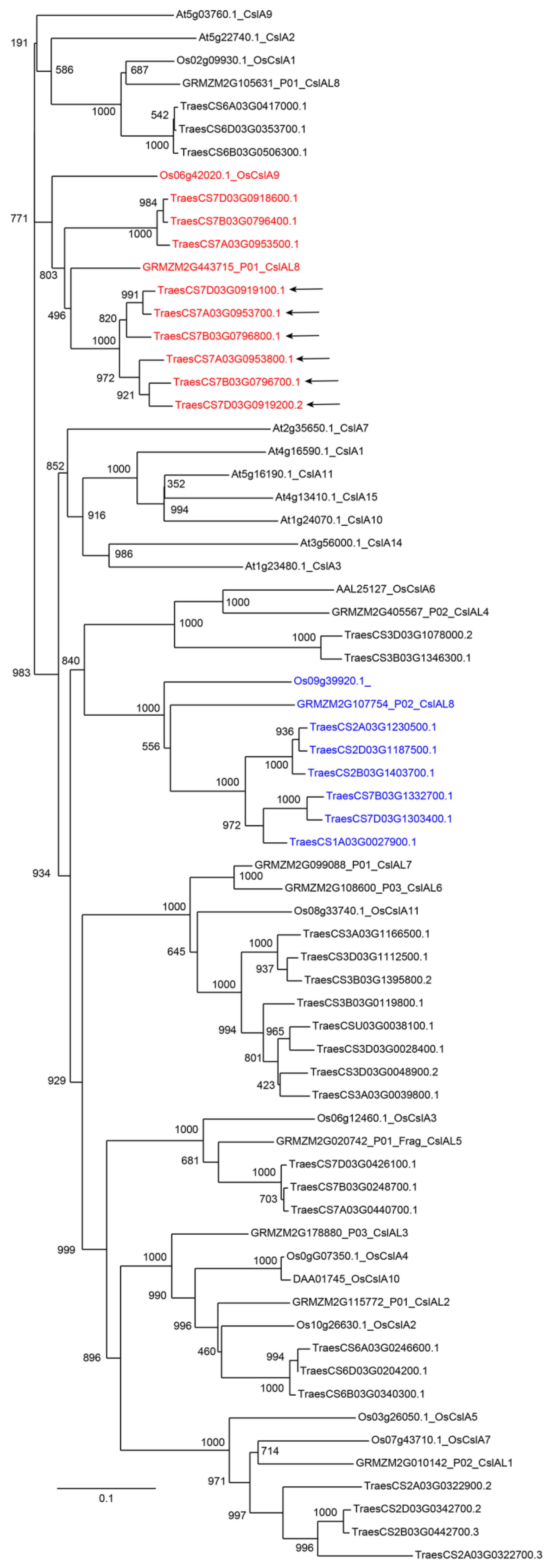
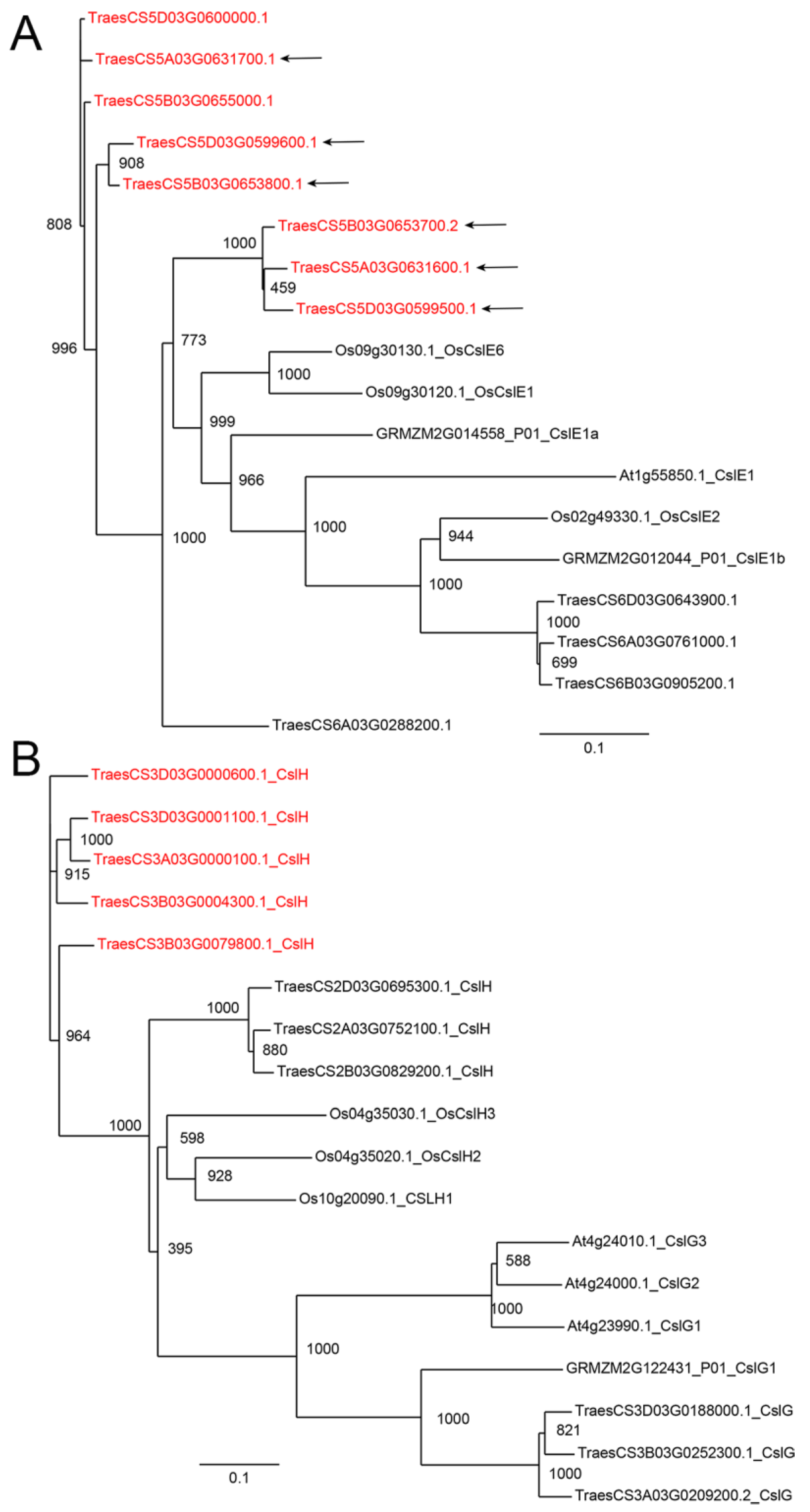
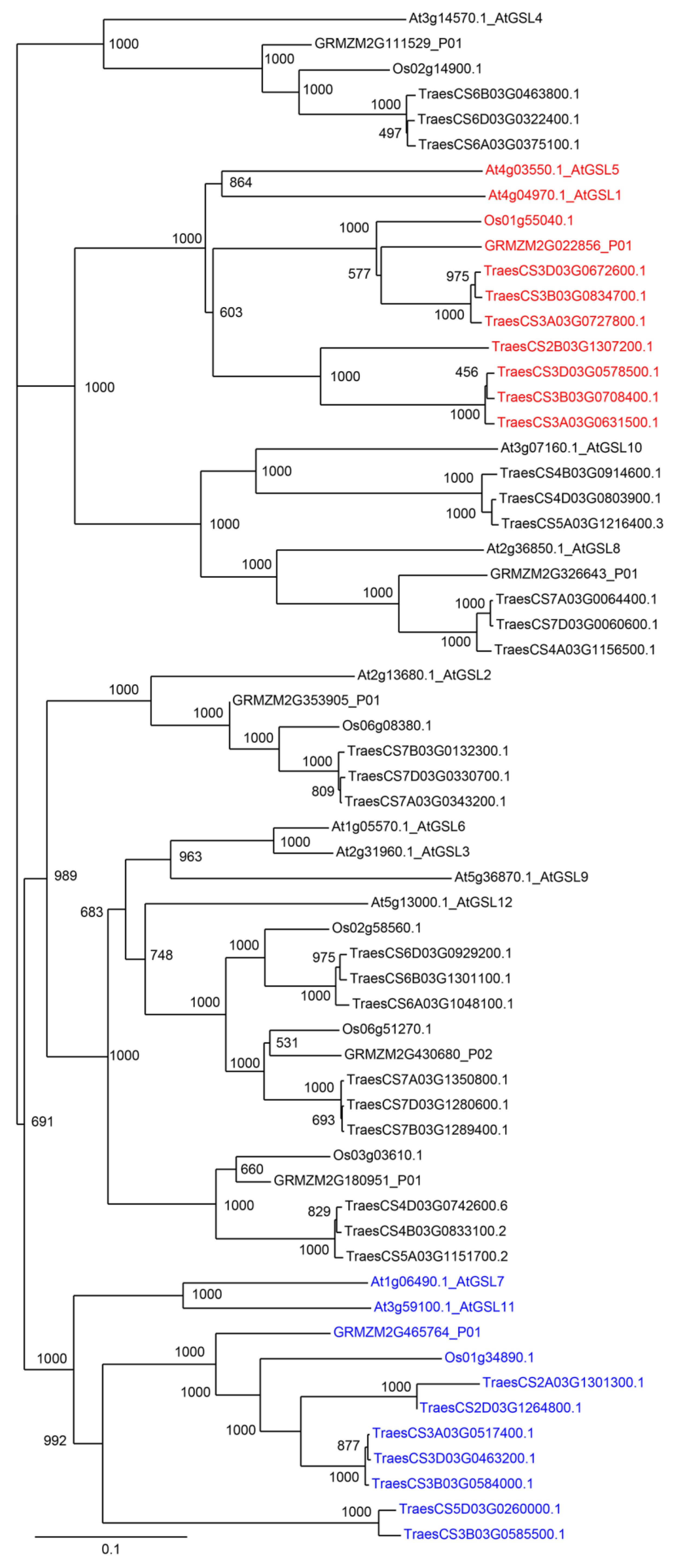
| Number of Genes per Family | ||||
|---|---|---|---|---|
| Cell Wall Family | Wheat | Maize | Rice | Arabidopsis |
| Nucleotide Sugar Transferases | 133 | 63 | 50 | 51 |
| PP 4CL | 45 | 10 | 14 | 13 |
| PP C3H_C4H_F5H | 50 | 8 | 10 | 6 |
| PP CAD | 48 | 4 | 12 | 9 |
| PP CCoAOMT | 21 | 6 | 6 | 7 |
| PP COMT | 33 | 3 | 7 | 16 |
| PP CCR | 121 | 18 | 24 | 7 |
| PP HCT | 149 | 38 | 5 | 1 |
| PP PAL | 47 | 10 | 9 | 4 |
| GT1–NSI–AUD_SUD | 15 | 9 | 6 | 6 |
| GT1–NSI–AXS | 3 | 1 | 1 | 2 |
| GT1–NSI–GAE | 15 | 9 | 4 | 6 |
| GT1–NSI–GER | 3 | 1 | 1 | 2 |
| GT1–NSI–GMD | 6 | 1 | 1 | 2 |
| GT1–NSI–GME | 6 | 2 | 2 | 1 |
| GT1–NSI–RHM_UER | 8 | 4 | 3 | 4 |
| GT1–NSI–UGD | 12 | 3 | 5 | 4 |
| GT1–NSI–UGE | 9 | 3 | 4 | 5 |
| GT1–NSI–UXE | 9 | 4 | 3 | 4 |
| GT2–CesA | 28 | 20 | 10 | 10 |
| GT2–CSL A | 38 | 10 | 11 | 9 |
| GT2–CSL C | 15 | 8 | 6 | 5 |
| GT2–CSL D | 15 | 5 | 5 | 5 |
| GT2–CSL E | 12 | 2 | 3 | 1 |
| GT2–CSL F | 18 | 7 | 8 | 0 |
| GT2–CSL G & H | 11 | 1 | 3 | 3 |
| GT4–Sucrose Synthases | 21 | 5 | 7 | 6 |
| GT8–A | 15 | 7 | 4 | 5 |
| GT8–B | 13 | 2 | 2 | 8 |
| GT8–C | 21 | 10 | 8 | 10 |
| GT8–D | 52 | 23 | 22 | 15 |
| GT8–E | 12 | 5 | 5 | 3 |
| GT10 | 6 | 3 | 3 | 3 |
| GT13 | 6 | 1 | 1 | 1 |
| GT14 | 44 | 10 | 11 | 9 |
| GT16 | 3 | 1 | 1 | 1 |
| GT17 | 11 | 4 | 4 | 6 |
| GT21 | 3 | 1 | 1 | 1 |
| GT22 | 9 | 6 | 4 | 3 |
| GT24 | 3 | 2 | 1 | 1 |
| GT29 | 12 | 8 | 5 | 3 |
| GT30 | 3 | 1 | 1 | 1 |
| GT31 | 107 | 40 | 39 | 33 |
| GT32 | 6 | 2 | 2 | 6 |
| GT33 | 6 | 2 | 2 | 1 |
| GT34 | 24 | 17 | 7 | 6 |
| GT37 | 53 | 17 | 18 | 10 |
| GT41 | 6 | 2 | 3 | 2 |
| GT43 | 27 | 16 | 10 | 4 |
| GT47 | 126 | 50 | 38 | 39 |
| GT48–Callose Synthases | 35 | 7 | 7 | 12 |
| GT50 | 2 | 1 | 1 | 1 |
| GT57 | 3 | 1 | 1 | 1 |
| GT58 | 3 | 1 | 1 | 1 |
| GT59 | 3 | 1 | 1 | 1 |
| GT61 | 117 | 33 | 24 | 8 |
| GT64 | 6 | 3 | 3 | 3 |
| GT66 | 9 | 4 | 2 | 2 |
| GT68 | 3 | 1 | 1 | 3 |
| GT75–RGP | 12 | 9 | 3 | 5 |
| GT76 | 3 | 2 | 1 | 1 |
| GT77 | 74 | 23 | 16 | 18 |
| GT92 | 9 | 3 | 3 | 3 |
| GT96 | 3 | 2 | 1 | 1 |
| GH9 | 82 | 22 | 25 | 25 |
| GH16–XTHs | 142 | 31 | 28 | 33 |
| GH17 | 209 | 51 | 60 | 47 |
| GH18–Yieldins | 60 | 10 | 9 | 10 |
| GH28–Pgases–A | 29 | 12 | 11 | 13 |
| GH28–Pgases–B | 18 | 0 | 1 | 10 |
| GH28–Pgases–C | 11 | 8 | 8 | 9 |
| GH28–Pgases–D | 34 | 7 | 9 | 6 |
| GH28–Pgases–E | 15 | 3 | 3 | 8 |
| GH28–Pgases–F | 14 | 16 | 6 | 10 |
| GH28–Pgases–G | 13 | 1 | 4 | 8 |
| GH35–Beta Galactosidases | 48 | 16 | 10 | 17 |
| Expansins | 248 | 55 | 61 | 34 |
| CE8–PMEs | 120 | 34 | 41 | 66 |
| CE13–PAEs | 53 | 11 | 10 | 11 |
| PL1–Pectin Lyases | 31 | 12 | 12 | 26 |
| PL4–RG Lyases | 15 | 4 | 3 | 7 |
| P4H | 28 | 10 | 12 | 11 |
| Laccases | 113 | 24 | 27 | 17 |
| Peroxidases | 643 | 124 | 135 | 73 |
| SKU | 34 | 12 | 4 | 3 |
| COBRA-like | 38 | 9 | 11 | 11 |
| GDPlike | 15 | 7 | 6 | 7 |
| Proteases | 296 | 49 | 50 | 18 |
| HIP-like | 11 | 3 | 3 | 3 |
| AGPs | 22 | 9 | 8 | 39 |
| HRGPs | 3 | 2 | 2 | 13 |
| Total | 4086 | 1118 | 1036 | 955 |
| Gene Family | Location (bp) | Distance Apart (kb) | ||
|---|---|---|---|---|
| Wheat Gene | Start | Stop | ||
| CAD | TraesCS2A03G0131200 | 34,450,307 | 34,451,760 | 19.9 |
| TraesCS2A03G0131300 | 34,471,637 | 34,473,153 | ||
| CAD | TraesCS2B03G0178200 | 50,262,749 | 50,264,076 | 78.9 |
| TraesCS2B03G0178300 | 50,342,947 | 50,344,421 | ||
| CAD | TraesCS2B03G0191100 | 54,229,490 | 54,230,578 | 33.8 |
| TraesCS2B03G0191300 | 54,264,332 | 54,266,016 | ||
| CAD | TraesCS3A03G1041000 | 688,689,431 | 688,692,028 | 34.1 |
| TraesCS3A03G1041600 | 688,726,119 | 688,729,557 | ||
| CAD | TraesCS3B03G1199000 | 745,631,611 | 745,634,419 | 321.7 |
| TraesCS3B03G1200700 | 745,956,098 | 745,958,925 | ||
| CAD | TraesCS3D03G0968100 | 552,033,322 | 552,035,463 | 116.5 |
| TraesCS3D03G0968500 | 552,151,988 | 552,155,191 | ||
| CAD | TraesCS6A03G0939400 | 597,869,237 | 597,875,251 | 17.6 |
| TraesCS6A03G0939600LC | 597,892,856 | 597,894,201 | 21.3 | |
| TraesCS6A03G0939900 | 597,915,465 | 597,921,915 | ||
| CAD | TraesCS6B03G1146700 | 690,063,231 | 690,067,770 | 26.8 |
| TraesCS6B03G1147100 | 690,094,598 | 690,099,654 | 43.3 | |
| TraesCS6B03G1147200 | 690,142,991 | 690,147,748 | ||
| CAD | TraesCS6D03G0816500 | 470,593,540 | 470,597,691 | 26.6 |
| TraesCS6D03G0816600 | 470,624,314 | 470,629,321 | 26.0 | |
| TraesCS6D03G0817100 | 470,655,366 | 470,661,613 | ||
| PAL | TraesCS1A03G0089500 | 22,809,328 | 22,817,055 | 2.8 |
| TraesCS1A03G0089700 | 22,819,864 | 22,822,522 | ||
| PAL | TraesCS1B03G0108100 | 30,156,990 | 30,159,840 | 13.2 |
| TraesCS1B03G0108300 | 30,173,031 | 30,175,842 | 48.2 | |
| TraesCS1B03G0108400 | 30,224,080 | 30,226,937 | 67.0 | |
| TraesCS1B03G0108600 | 30,293,907 | 30,296,751 | 37.8 | |
| TraesCS1B03G0108700 | 30,334,559 | 30,337,342 | ||
| PAL | TraesCS1D03G0078900 | 20,552,689 | 20,561,537 | 1.4 |
| TraesCS1D03G0079200 | 20,562,959 | 20,565,612 | ||
| PAL | TraesCS2A03G0922600 | 628,113,634 | 628,116,620 | 32.0 |
| TraesCS2A03G0922700 | 628,148,609 | 628,151,525 | 49.4 | |
| TraesCS2A03G0922800 | 628,200,915 | 628,203,659 | 8.4 | |
| TraesCS2A03G0922900 | 628,212,066 | 628,214,916 | ||
| PAL | TraesCS2B03G1014000 | 572,920,722 | 572,923,789 | 145.0 |
| TraesCS2B03G1015000 | 573,068,831 | 573,071,748 | 140.2 | |
| TraesCS2B03G1015300 | 573,211,969 | 573,214,748 | ||
| PAL | TraesCS2D03G0862600 | 483,823,970 | 483,826,687 | 84.6 |
| TraesCS2D03G0863100 | 483,911,277 | 483,914,370 | 268.0 | |
| TraesCS2D03G0863200 | 484,182,377 | 484,185,488 | 23.8 | |
| TraesCS2D03G0863300 | 484,209,252 | 484,211,934 | ||
| CslA | TraesCS7A03G0953700 | 576,059,990 | 576,064,129 | 49.7 |
| TraesCS7A03G0953800 | 576,113,787 | 576,117,206 | ||
| CslA | TraesCS7B03G0796700 | 537,109,006 | 537,112,692 | 3.1 |
| TraesCS7B03G0796800 | 537,115,750 | 537,132,129 | ||
| CslA | TraesCS7D03G0919100 | 506,334,490 | 506,337,868 | 9.5 |
| TraesCS7D03G0919200 | 506,347,416 | 506,352,132 | ||
| CslE | TraesCS5A03G0631600 | 469,521,391 | 469,527,274 | 2.4 |
| TraesCS5A03G0631700 | 469,529,676 | 469,535,899 | ||
| CslE | TraesCS5B03G0653700 | 437,270,468 | 437,275,686 | 2.7 |
| TraesCS5B03G0653800 | 437,278,393 | 437,284,101 | ||
| CslE | TraesCS5D03G0599500 | 370,151,008 | 370,156,445 | 4.7 |
| TraesCS5D03G0599600 | 370,161,140 | 370,165,637 | ||
| GH18 | TraesCS3A03G0882000 | 623,487,813 | 623,487,813 | 6.4 |
| TraesCS3A03G0882100 | 623,494,204 | 623,495,097 | 2.5 | |
| TraesCS3A03G0882200 | 623,497,609 | 623,498,502 | 137.5 | |
| TraesCS3A03G0882500 | 623,636,002 | 623,636,895 | 13.2 | |
| TraesCS3A03G0882600 | 623,650,108 | 623,651,001 | 6.9 | |
| TraesCS3A03G0882700 | 623,657,857 | 623,658,750 | 26.5 | |
| TraesCS3A03G0882800 | 623,685,242 | 623,686,135 | 214.1 | |
| TraesCS3A03G0883100 | 623,900,253 | 623,901,602 | ||
| GH18 | TraesCS3B03G1008300 | 655,681,956 | 655,682,987 | 4.1 |
| TraesCS3B03G1008400 | 655,687,134 | 655,688,027 | 27.7 | |
| TraesCS3B03G1008600 | 655,715,751 | 655,716,644 | 6.3 | |
| TraesCS3B03G1008700 | 655,722,943 | 655,723,836 | 6.7 | |
| TraesCS3B03G1009100 | 655,730,571 | 655,731,464 | 122.3 | |
| TraesCS3B03G1009200 | 655,853,730 | 655,854,731 | ||
| GH18 | TraesCS3D03G0810000 | 480,870,250 | 480,871,131 | 3.9 |
| TraesCS3D03G0810100 | 480,875,009 | 480,875,902 | 31.7 | |
| TraesCS3D03G0810300LC | 480,907,624 | 480,908,268 | 199.2 | |
| TraesCS3D03G0810400 | 481,107,472 | 481,108,365 | 16.6 | |
| TraesCS3D03G0810500 | 481,124,934 | 481,125,827 | 457.3 | |
| TraesCS3D03G0810900 | 481,583,081 | 481,584,377 | ||
| PAE | TraesCS3A03G1263600 | 748,276,487 | 748,280,357 | 53.1 |
| TraesCS3A03G1264000 | 748,333,460 | 748,335,357 | 20.7 | |
| TraesCS3A03G1264100 | 748,356,041 | 748,358,835 | 3.3 | |
| TraesCS3A03G1264200 | 748,362,146 | 748,365,282 | 8.5 | |
| TraesCS3A03G1264400 | 748,373,743 | 748,377,054 | ||
| PAE | TraesCS3B03G1508100 | 844,439,077 | 844,442,856 | 27.3 |
| TraesCS3B03G1508200 | 844,470,116 | 844,472,978 | 28.1 | |
| TraesCS3B03G1508300 | 844,501,107 | 844,509,112 | 13.6 | |
| TraesCS3B03G1508500 | 844,522,685 | 844,524,017 | 22.3 | |
| TraesCS3B03G1508700 | 844,546,309 | 844,549,502 | ||
| PAE | TraesCS3D03G1188000 | 615,245,943 | 615,249,493 | 29.9 |
| TraesCS3D03G1188400 | 615,279,382 | 615,282,166 | 9.9 | |
| TraesCS3D03G1188500 | 615,292,106 | 615,295,886 | 2.7 | |
| TraesCS3D03G1188600 | 615,298,614 | 615,301,615 | 31.6 | |
| TraesCS3D03G1188700 | 615,333,190 | 615,336,310 | 40.9 | |
| TraesCS3D03G1188900 | 615,377,258 | 615,380,183 | ||
| PAE | TraesCS5A03G1140500 | 658,736,919 | 658,740,227 | 72.9 |
| TraesCS5A03G1140600 | 658,813,140 | 658,817,069 | ||
| PAE | TraesCS5B03G1211700 | 666,788,941 | 666,792,805 | 127.0 |
| TraesCS5B03G1212500 | 666,919,777 | 666,924,347 | ||
| PAE | TraesCS5D03G1094700 | 532,276,790 | 532,280,796 | 29.9 |
| TraesCS5D03G1094800 | 532,310,668 | 532,314,929 | ||
| ProteinV1 | Description from Penning 2023 | ProteinV2_1 | Gene Family |
|---|---|---|---|
| TraesCS5A01G405700.1 | Cellulose synthase-like protein | TraesCS5A03G0962500.1 | GT2–CSL C |
| TraesCS5B01G410400.1 | Cellulose synthase-like protein | TraesCS5B03G1012800.1 | GT2–CSL C |
| TraesCS5D01G415700.1 | Cellulose synthase-like protein | TraesCS5D03G0916900.2 | GT2–CSL C |
| TraesCS7A01G298600.1 | Cellulose synthase-like protein | TraesCS7A03G0733400.1 | GT2–CSL F |
| TraesCS7B01G188400.2 | Cellulose synthase-like protein | TraesCS7B03G0538200.1 | GT2–CSL F |
| TraesCS1A01G152500.1 | UDP-glucose:glycoprotein glucosyltransferase | TraesCS1A03G0409400.1 | GT24 |
| TraesCS1D01G149400.1 | UDP-glucose:glycoprotein glucosyltransferase | TraesCS1D03G0377700.1 | GT24 |
| TraesCS6A01G102100.1 | Glycosyltransferase | TraesCS6A03G0237900.1 | GT61 |
| TraesCS3A01G440800.1 | glycosyltransferase family exostosin protein | TraesCS3A03G1025100.1 | GT47 E |
| TraesCS3D01G433400.1 | glycosyltransferase family exostosin protein | TraesCS3D03G0953500.2 | GT47 E |
| TraesCS2A01G051600.2 | O-methyltransferase | TraesCS2A03G0099800.2 | 1.3 COMT |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Penning, B.W. The Cell Wall-Related Gene Families of Wheat (Triticum aestivum). Diversity 2023, 15, 1135. https://doi.org/10.3390/d15111135
Penning BW. The Cell Wall-Related Gene Families of Wheat (Triticum aestivum). Diversity. 2023; 15(11):1135. https://doi.org/10.3390/d15111135
Chicago/Turabian StylePenning, Bryan W. 2023. "The Cell Wall-Related Gene Families of Wheat (Triticum aestivum)" Diversity 15, no. 11: 1135. https://doi.org/10.3390/d15111135
APA StylePenning, B. W. (2023). The Cell Wall-Related Gene Families of Wheat (Triticum aestivum). Diversity, 15(11), 1135. https://doi.org/10.3390/d15111135






