Phylogenetic Study of Alternaria Potato and Tomato Pathogens in Russia
Abstract
1. Introduction
2. Materials and Methods
2.1. Isolates
2.2. PCR and Sequencing
2.3. Phylogenetic Analysis
2.4. Bioinformatic Methods
3. Results
3.1. Bioinformatic Analysis
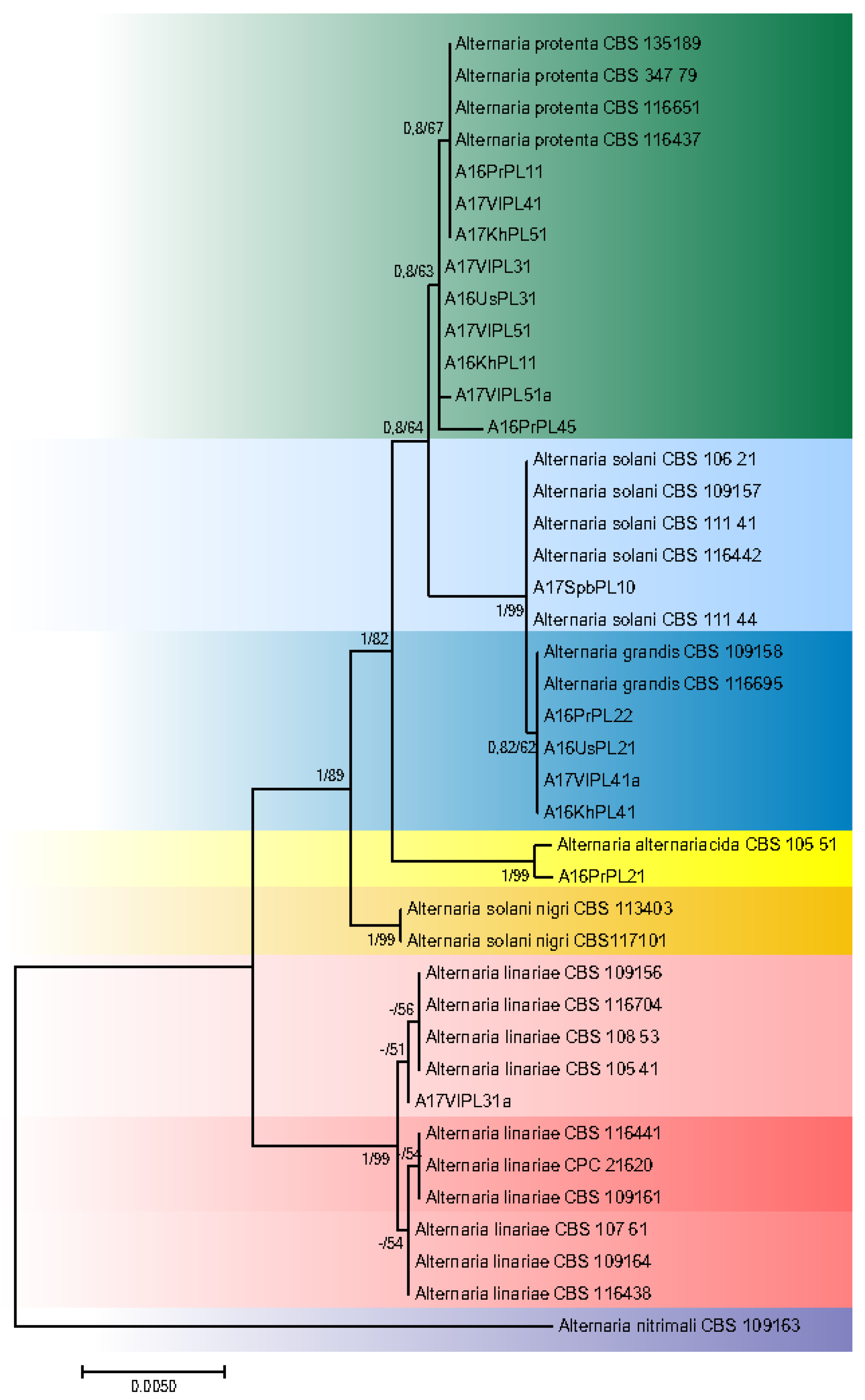
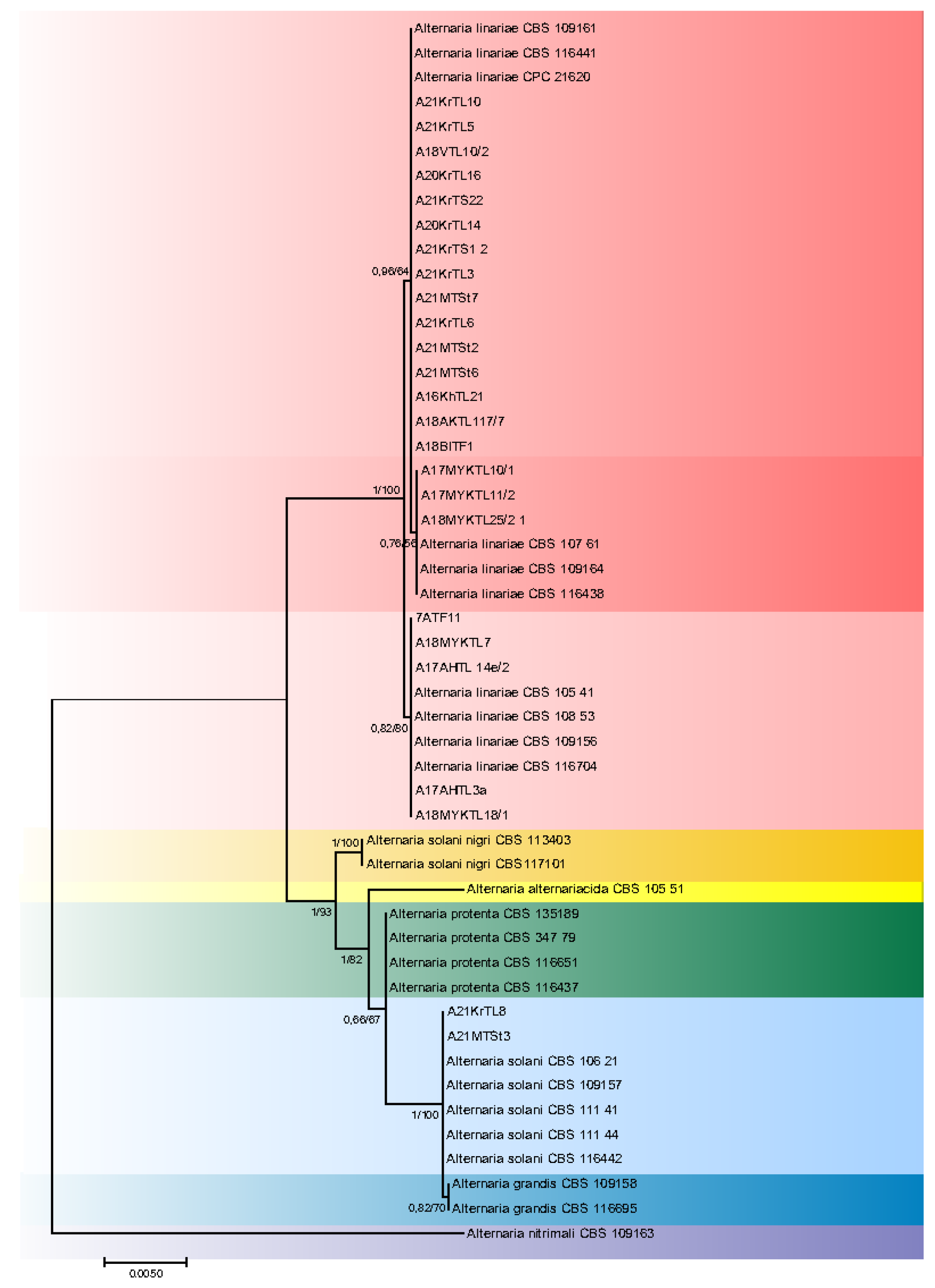
3.2. Phylogeny
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Simmons, E.G. Alternaria themes and variation (244–286) Species on Solanaceae. Mycotaxon 2000, 75, 1–115. [Google Scholar]
- Simmons, E.G. Alternaria: An Identification Manual; CBS Fungal Biodiversity Centre: Utrecht, The Netherlands, 2007. [Google Scholar]
- Woudenberg, J.H.C.; Truter, M.; Groenewald, J.Z.; Crous, P.W. Large-spored Alternaria pathogens in section Porri disentangled. Stud. Mycol. 2014, 79, 1–47. [Google Scholar] [CrossRef] [PubMed]
- Elansky, S.N.; Chudinova, E.M.; Elansky, A.S.; Kah, M.O.; Sandzhieva, D.A.; Mukabenova, B.A.; Dedov, A.G. Microorganisms in spent water-miscible metalworking fluids as a resource of strains for their disposal. J. Clean. Prod. 2022, 350, 131438. [Google Scholar] [CrossRef]
- Kutuzova, I.A.; Kokaeva, L.Y.; Pobedinskaya, M.A.; Krutyakov, Y.A.; Scolotneva, E.S.; Chudinova, E.M.; Elansky, S.N. Resistance of Helminthosporium solani strains to the fungicides applied for tuber treatment. J. Plant Pathol. 2017, 99, 635–642. [Google Scholar] [CrossRef]
- White, T.J.; Bruns, T.; Lee, S.J.W.T.; Taylor, J. Amplifcation and direct sequencing of fungal ribosomal RNA genes for phylogenetics. PCR Protoc. A Guide Methods Appl. 1990, 18, 315–322. [Google Scholar]
- Berbee, M.L.; Pirseyedi, M.; Hubbard, S. Cochliobolus phylogenetics and the origin of known, highly virulent pathogens, inferred from ITS and glyceraldehyde-3-phosphate dehydrogenase gene sequences. Mycologia 1999, 91, 964–977. [Google Scholar] [CrossRef]
- Sung, G.H.; Sung, J.M.; Hywel-Jones, N.L.; Spatafora, J.W. A multi-gene phylogeny of Clavicipitaceae (Ascomycota, Fungi): Identification of localized incongruence using a combinational bootstrap approach. Mol. Phylogenet. Evol. 2007, 44, 1204–1223. [Google Scholar] [CrossRef]
- Liu, Y.J.; Whelen, S.; Hall, B.D. Phylogenetic relationships among ascomycetes: Evidence from an RNA polymerse II subunit. Mol. Biol. Evol. 1999, 16, 1799–1808. [Google Scholar] [CrossRef]
- Carbone, I.; Kohn, L.M. A method for designing primer sets for speciation studies in filamentous ascomycetes. Mycologia 1999, 91, 553–556. [Google Scholar] [CrossRef]
- Tamura, K.; Nei, M. Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol. Biol. Evol. 1993, 10, 512–526. [Google Scholar]
- Kokaeva, L.Y.; Elansky, S.N. First report of Alternaria alternariacida causing potato leaf blight in the Far East, Russia. Plant Dis. 2022; in press. [Google Scholar]
- Ayad, D.; Aribi, D.; Hamon, B.; Kedad, A.; Simoneau, P.; Bouznad, Z. Distribution of large-spored Alternaria species associated with early blight of potato and tomato in Algeria. Phytopathol. Mediterr. 2019, 58, 139–149. [Google Scholar] [CrossRef]
- Ayad, D.; Hamon, B.; Kedad, A.; Bouznad, Z.; Simoneau, P. First Report of Early Blight Caused by Alternaria linariae on Potato in Algeria. Plant Dis. 2018, 102, 2651. [Google Scholar] [CrossRef]
- Ayad, D.; Leclerc, S.; Hamon, B.; Kedad, A.; Bouznad, Z.; Simoneau, P. First Report of Early Blight Caused by Alternaria protenta on Potato in Algeria. Plant Dis. 2017, 101, 836. [Google Scholar] [CrossRef]
- Bessadat, N.; Berruyer, R.; Hamon, B.; Bataille-Simoneau, N.; Benichou, S.; Kihal, M.; Henni, D.E.; Simoneau, P. Alternaria species associated with early blight epidemics on tomato and other Solanaceae crops in northwestern Algeria. Eur. J. Plant Pathol. 2017, 148, 181–197. [Google Scholar] [CrossRef]
- El-Ganainy, S.M.; El-Abeid, S.E.; Ahmed, Y.; Iqbal, Z. Morphological and molecular characterization of large-spored Alternaria species associated with potato and tomato early blight in Egypt. Int. J. Agric. Biol. 2021, 25, 1101–1110. [Google Scholar] [CrossRef]
- Ding, S.; Meinholz, K.; Cleveland, K.; Jordan, S.A.; Gevens, A.J. Diversity and Virulence of Alternaria spp. Causing Potato Early Blight and Brown Spot in Wisconsin. Phytopathology 2019, 109, 436–445. [Google Scholar] [CrossRef]
- Einspanier, S.; Susanto, T.; Metz, N.; Wolters, P.J.; Vleeshouwers, V.G.; Lankinen, Å.; Liljeroth, E.; Landschoot, S.; Ivanović, Ž.; Hückelhoven, R.; et al. Whole genome sequencing elucidates the species-wide diversity and evolution of fungicide resistance in the early blight pathogen Alternaria solani. Evol. Appl. 2022, 01, 1–16. [Google Scholar] [CrossRef]
- Adhikari, T.B.; Muzhinji, N.; Halterman, D.; Louws, F.J. Genetic diversity and population structure of Alternaria species from tomato and potato in North Carolina and Wisconsin. Sci. Rep. 2021, 11, 17024. [Google Scholar] [CrossRef]
- Kudryavtseva, N.N.; Pobedinskaya, M.A.; Balabko, P.N.; Kokaeva, L.Y.; Zaichik, B.T.; Kutuzova, N.A.; Azarkovich, M.I.; Elansky, S.N.; Valueva, T.A. The proteolytic activity and virulence of Alternaria alternata strains, isolated from tomato. Mikol. I Fitopatol. 2017, 51, 110–116. [Google Scholar]
- Lourenço, V.; Moya, A.; González-Candelas, F.; Carbone, I.; Maffia, L.A.; Mizubuti, E.S.G. Population Biology Molecular Diversity and Evolutionary Processes of Alternaria solani in Brazil Inferred Using Genealogical and Coalescent Approaches. Phytopathology 2009, 99, 765–774. [Google Scholar] [CrossRef]
- Peixoto, C.C.; Cabral, C.S.; Fonseca, M.E.N.; Boiteux, L.S.; Reis, A. Species diversity, novel interactions and absence of well-supported host-guided phylogenetic groupings of Neotropical Alternaria isolates causing foliar lesions in Solanaceae. J. Appl. Microbiol. 2021, 131, 2466–2487. [Google Scholar] [CrossRef] [PubMed]
- Zhao, D.; Fan, S.; Zhang, D.; Pan, Y.; Gu, Q.; Wang, J.; Yang, Z.; Zhu, J. Parasexual reproduction in Alternaria solani: Simple sequence repeat molecular evidence for haploidization. Mycologia 2021, 113, 949–955. [Google Scholar] [CrossRef] [PubMed]
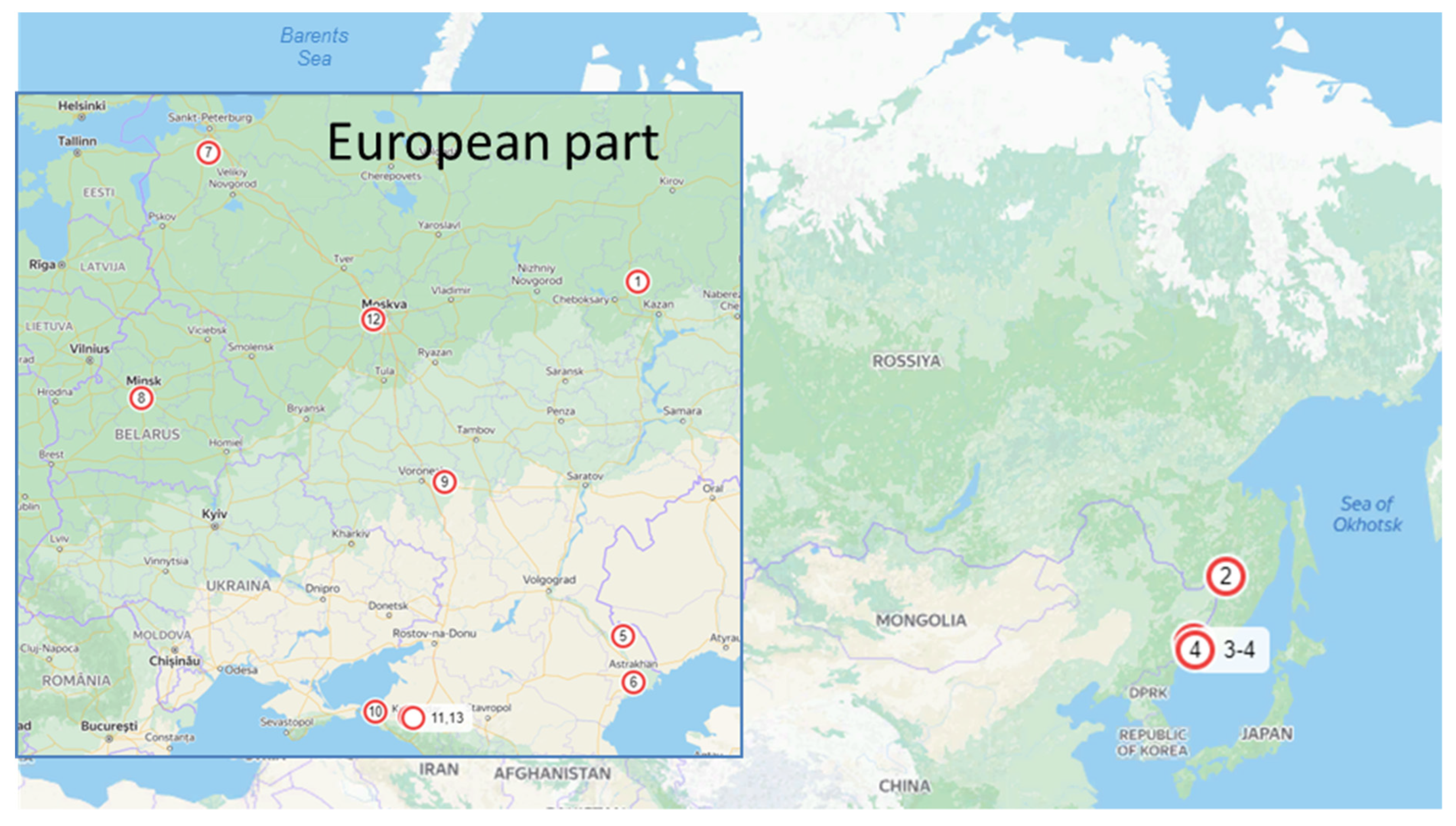
| Name | Strain Number | Year of Isolation | Host/Substrate | Locality of Collection Site (See Figure 1) | GenBank Accession Numbers | ||||
|---|---|---|---|---|---|---|---|---|---|
| ITS | TEF | RPB2 | GADPH | Alt a 1 | |||||
| Alternaria grandis | A16UsPL21 | 2016 | S. t., leaf | 3 | OM640142 | MN580515 | MN580526 | MN544407 | MN562255 |
| A16PrPL22 | 2016 | S. t., leaf | 3 | OM640143 | ON098332 | ON098335 | ON098326 | ON098329 | |
| A17VlPL41a | 2017 | S. t., leaf | 4 | OM640144 | ON098333 | ON098336 | ON098327 | ON098330 | |
| A16KhPL41 | 2016 | S. t., leaf | 2 | OM640145 | ON098334 | ON098337 | ON098328 | ON098331 | |
| Alternaria solani | A17SpbPL10 | 2017 | S. t., leaf | 7 | OM640146 | MN580516 | MN580527 | MN544406 | MN562256 |
| A21KrTL8 | 2021 | S.l., leaf | 11 | OM640147 | ON098290 | ON098292 | ON098286 | ON098288 | |
| A21MTSt3 | 2021 | S.l., stem | 12 | OM640148 | ON098291 | ON098293 | ON098287 | ON098289 | |
| Alternaria alternariacida | A16PrPL21 | 2016 | S. t., leaf | 3 | OM348531 | MN580518 | MN580529 | MN544404 | MN562258 |
| Alternaria protenta | A16PrPL11 | 2016 | S. t., leaf | 3 | OM640149 | MN580517 | MN580528 | MN544405 | MN562257 |
| A17VlPL41 | 2017 | S. t., leaf | 4 | OM640150 | ON098306 | ON098312 | ON098294 | ON098300 | |
| A17KhPL51 | 2017 | S. t., leaf | 2 | OM640151 | ON098307 | ON098313 | ON098295 | ON098301 | |
| A16PrPL45 | 2016 | S. t., leaf | 3 | OM640152 | MN580523 | MN580534 | MN593316 | MN593309 | |
| A17VlPL31 | 2017 | S. t., leaf | 4 | OM640153 | MN580524 | MN580535 | MN593317 | MN593310 | |
| A16UsPL31 | 2016 | S. t., leaf | 3 | OM640154 | ON098308 | ON098314 | ON098296 | ON098302 | |
| A17VlPL51 | 2017 | S. t., leaf | 9 | OM640155 | ON098309 | ON098315 | ON098297 | ON098303 | |
| A16KhPL11 | 2016 | S. t., leaf | 2 | OM640156 | ON098310 | ON098316 | ON098298 | ON098304 | |
| A17VlPL51a | 2017 | S. t., leaf | 9 | OM640157 | ON098311 | ON098317 | ON098299 | ON098305 | |
| Alternaria linariae | 7AHTF 11a | 2017 | S.l., fruit | 5 | KY496637 | MN580520 | MN580531 | MN593313 | MN562260 |
| A17AHTL 14e/2 | 2017 | S.l., leaf | 5 | OM640158 | ON135533 | ON135537 | ON135525 | ON135529 | |
| A18MYKTL7 | 2018 | S.l., leaf | 1 | OM640159 | ON135534 | ON135538 | ON135526 | ON135530 | |
| A18MYKTL18/1 | 2018 | S.l., leaf | 1 | OM640160 | ON135535 | ON135539 | ON135527 | ON135531 | |
| A17AHTL3a* | 2017 | S.l., leaf | 5 | OM640161 | ON135536 | ON135540 | ON135528 | ON135532 | |
| A17VlPL31a | 2017 | S. t., leaf | 4 | OM640162 | MN580519 | MN580530 | MN593312 | MN562259 | |
| A17MYKTL10/1 | 2017 | S.l., leaf | 1 | OM640163 | MN580521 | MN580532 | MN593314 | MN562261 | |
| A18MYKTL25/2(1) | 2018 | S.l., leaf | 1 | OM640164 | ON098322 | ON098324 | ON098318 | ON098320 | |
| A17MYKTL11/2 | 2017 | S.l., leaf | 1 | OM640165 | ON098323 | ON098325 | ON098319 | ON098321 | |
| A18BlTF1 | 2018 | S.l., fruit | 8 | OM640166 | MN580522 | MN580533 | MN593315 | MN562262 | |
| A18VTL10/2 | 2018 | S.l., leaf | 9 | OM640167 | ON149482 | ON149496 | ON149454 | ON149468 | |
| A16KhTL21 | 2016 | S.l., leaf | 2 | OM640168 | ON149483 | ON149497 | ON149455 | ON149469 | |
| A18AKTL117/7 | 2018 | S.l., leaf | 6 | OM640169 | ON149484 | ON149498 | ON149456 | ON149470 | |
| A20KrTL14 | 2020 | S.l., leaf | 10 | OM640170 | ON149485 | ON149499 | ON149457 | ON149471 | |
| A20KrTL16 | 2020 | S.l., leaf | 10 | OM640171 | ON149486 | ON149500 | ON149458 | ON149472 | |
| A21KrTS1.2 | 2021 | S.l., seed | 10 | OM640172 | ON149487 | ON149501 | ON149459 | ON149473 | |
| A21KrTL3 | 2021 | S.l., leaf | 11 | OM640173 | ON149488 | ON149502 | ON149460 | ON149474 | |
| A21KrTL5 | 2021 | S.l., leaf | 11 | OM640174 | ON149489 | ON149503 | ON149461 | ON149475 | |
| A21KrTL6 | 2021 | S.l., leaf | 11 | OM640175 | ON149490 | ON149504 | ON149462 | ON149476 | |
| A21KrTL10 | 2021 | S.l., leaf | 11 | OM640176 | ON149491 | ON149505 | ON149463 | ON149477 | |
| A21KrTS22 | 2021 | S.l., seed | 10 | OM640177 | ON149492 | ON149506 | ON149464 | ON149478 | |
| A21MTSt2 | 2021 | S.l., stem | 12 | OM640178 | ON149493 | ON149507 | ON149465 | ON149479 | |
| A21MTSt6 | 2021 | S.l., stem | 13 | OM640179 | ON149494 | ON149508 | ON149466 | ON149480 | |
| A21MTSt7 | 2021 | S.l., stem | 13 | OM640180 | ON149495 | ON149509 | ON149467 | ON149481 | |
| Current Species Name | Old Species Name | Strain | Host/Substrate | Locality | GenBank Accession Numbers | ||||
|---|---|---|---|---|---|---|---|---|---|
| ITS | GAPDH | Alt a1 | TEF1 | RPB2 | |||||
| A. alternariacida | A. solani | CBS 105.51 | S. lyc., fruit | UK | KJ718105 | KJ717959 | KJ718625 | KJ718454 | KJ718279 |
| A. grandis | CBS 109158 | S. tuber., leaf | USA | KJ718239 | JQ646341 | JQ646425 | EU130547 | KJ718414 | |
| CBS 116695 | S. tuber., leaf | USA | KJ718241 | KJ718070 | KJ718748 | KJ718587 | KJ718416 | ||
| A. linariae | A. solani | CBS 108.53 | – | – | KJ718181 | KJ718025 | KJ718693 | KJ718529 | KJ718354 |
| A. solani | CBS 107.61 | – | Belgium | KJ718182 | KJ718026 | KJ718694 | KJ718530 | KJ718355 | |
| A. tomatop-hila | CBS 109156 | S. lyc., leaf | USA | KJ718183 | JQ646347 | GQ180101 | KJ718531 | KJ718356 | |
| A. subcylin-drica | CBS 109161 | S. lyc., leaf | USA | KJ718184 | JQ646345 | JQ646429 | KJ718532 | KJ718357 | |
| A. cretica | CBS 109164 | S. lyc., leaf | Greece | KJ718185 | JQ646342 | JQ646426 | EU130545 | KJ718358 | |
| A. tomatophila | CBS 116704 | S. lyc., leaf | USA | KJ718188 | KJ718029 | KJ718697 | KJ718535 | KJ718361 | |
| CPC 21620 | S. lyc., leaf | Thailand | KJ718189 | KJ718030 | KJ718698 | KJ718536 | KJ718362 | ||
| A. nitrimali | CBS 109163 | S. viarum leaf | Puerto Rico | KJ718201 | JQ646358 | KJ718710 | KJ718547 | KJ71837 | |
| A. protenta | A. solani | CBS 347.79 | S. lyc., fruit | New Zealand | KJ718219 | KJ718054 | KJ718728 | KJ718565 | KJ718392 |
| A. solani | CBS 116651 | S. tuber., tuber | USA | KC584217 | KC584139 | GQ180097 | KC584688 | KC584430 | |
| A. solani | CBS 135189 | S. tuber., | New Zealand | KJ718224 | GQ180082 | GQ180098 | KJ718570 | KJ718397 | |
| A. solani | CBS 106.21 | – | – | KJ718236 | KJ718066 | KJ718743 | KJ718582 | KJ718410 | |
| CBS 111.41 | S. aviculare, leaf | – | KJ718237 | KJ718067 | KJ718744 | KJ718583 | KJ718411 | ||
| A. danida | CBS 109157 | S. tuber., leaf | USA | KJ718238 | GQ180080 | KJ718746 | KJ718585 | KJ718413 | |
| A. solaninigri | A. cyphoman-drae | CBS 113403 | S. nigrum, leaf | New Zealand | KJ718243 | KJ718071 | KJ718749 | KJ718589 | KJ718418 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kokaeva, L.Y.; Yarmeeva, M.M.; Kokaeva, Z.G.; Chudinova, E.M.; Balabko, P.N.; Elansky, S.N. Phylogenetic Study of Alternaria Potato and Tomato Pathogens in Russia. Diversity 2022, 14, 685. https://doi.org/10.3390/d14080685
Kokaeva LY, Yarmeeva MM, Kokaeva ZG, Chudinova EM, Balabko PN, Elansky SN. Phylogenetic Study of Alternaria Potato and Tomato Pathogens in Russia. Diversity. 2022; 14(8):685. https://doi.org/10.3390/d14080685
Chicago/Turabian StyleKokaeva, Lyudmila Yu., Maria M. Yarmeeva, Zarema G. Kokaeva, Elena M. Chudinova, Petr N. Balabko, and Sergey N. Elansky. 2022. "Phylogenetic Study of Alternaria Potato and Tomato Pathogens in Russia" Diversity 14, no. 8: 685. https://doi.org/10.3390/d14080685
APA StyleKokaeva, L. Y., Yarmeeva, M. M., Kokaeva, Z. G., Chudinova, E. M., Balabko, P. N., & Elansky, S. N. (2022). Phylogenetic Study of Alternaria Potato and Tomato Pathogens in Russia. Diversity, 14(8), 685. https://doi.org/10.3390/d14080685






