Fish Diversity along the Mekong River and Delta Inferred by Environmental-DNA in a Period of Dam Building and Downstream Salinization
Abstract
:1. Introduction
2. Materials and Methods
2.1. Study Area
2.2. Water Sampling for Environmental-DNA (eDNA) Analysis of Fishes
2.3. Statistical Analyses
2.4. Reconstruction of the Species Distribution Range
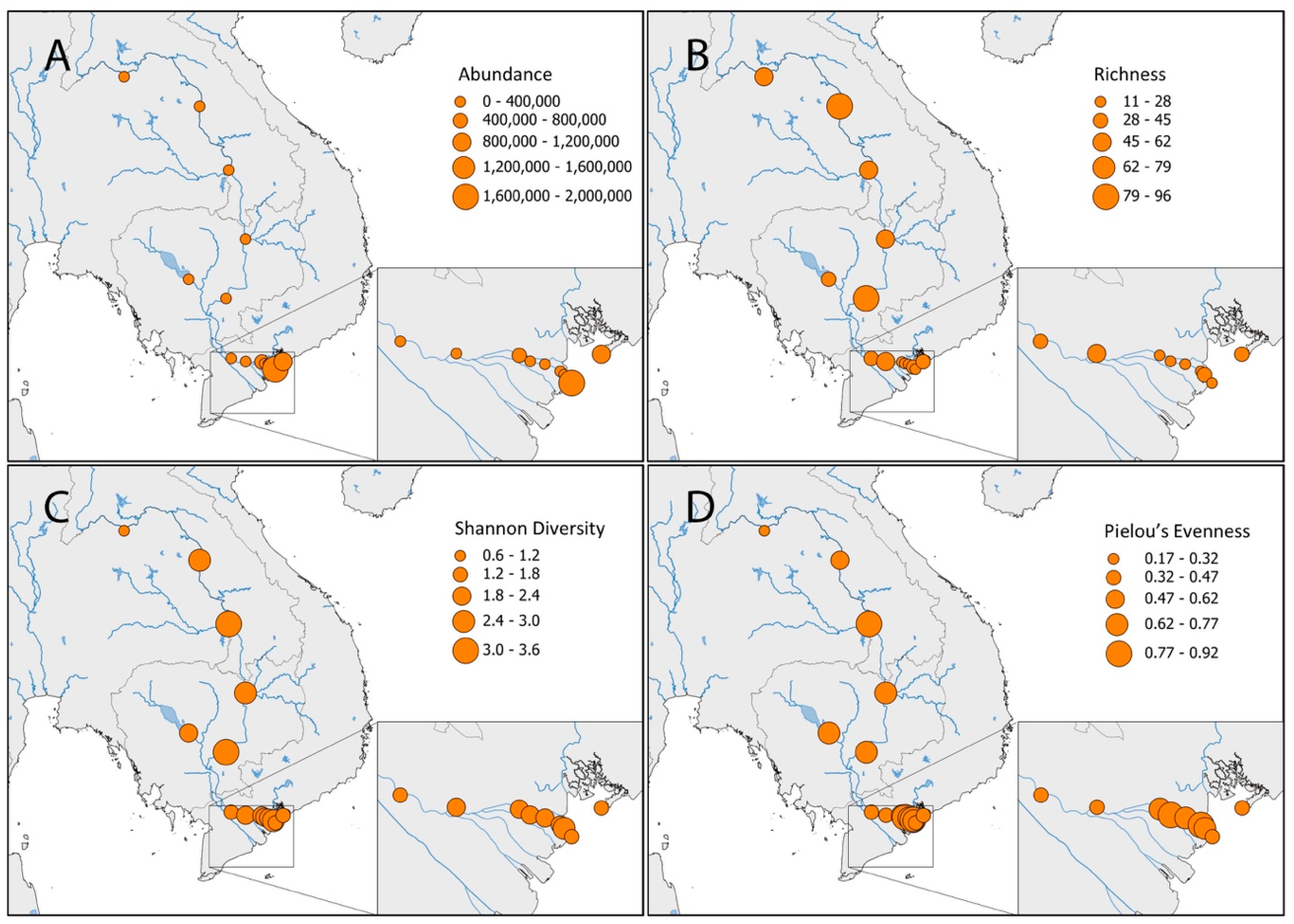
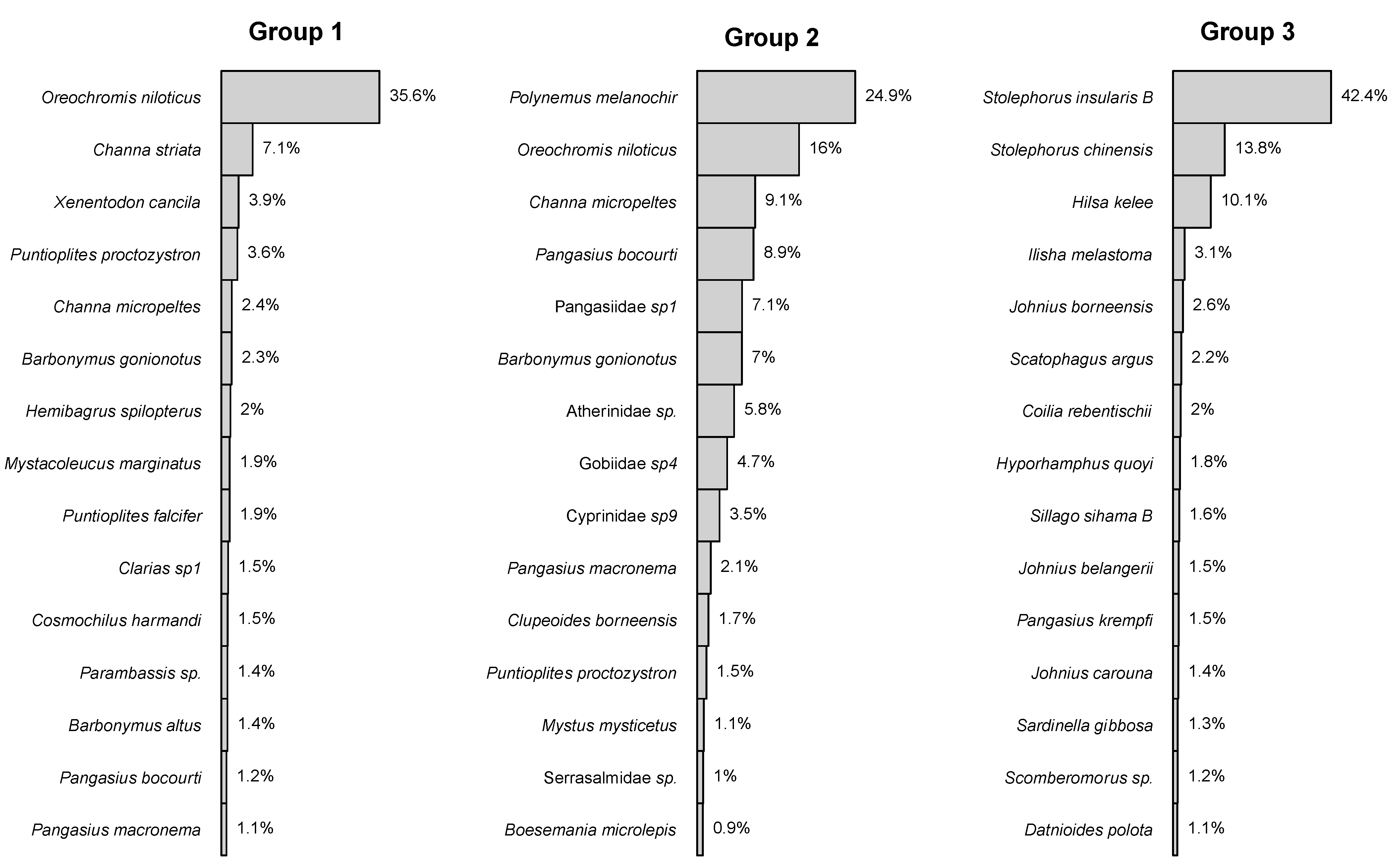
3. Results
4. Discussion
4.1. Limits and Advantages of the eDNA Approach in the Mekong River and Delta
4.2. Mekong Aquatic Environment and Fish Distributions
4.3. Migratory Fishes in the Mekong and Their Conservation
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Pörtner, H.O.; Peck, M.A. Climate change effects on fishes and fisheries: Towards a cause-and-effect understanding. J. Fish Biol. 2010, 77, 1745–1779. [Google Scholar] [CrossRef] [PubMed]
- Cheung, W.W.L.; Lam, V.W.Y.; Sarmiento, J.L.; Kearney, K.; Watson, R.; Pauly, D. Projecting global marine biodiversity impacts under climate change scenarios. Fish Fish 2009, 10, 235–251. [Google Scholar] [CrossRef]
- Daufresne, M.; Boet, P. Climate change impacts on structure and diversity of fish communities in rivers. Glob. Change Biol. 2007, 13, 2467–2478. [Google Scholar] [CrossRef]
- Lejeusne, C.; Chevaldonné, P.; Pergent-Martini, C.; Boudouresque, C.F.; Pérez, T. Climate change effects on a miniature ocean: The highly diverse, highly impacted Mediterranean Sea. Trends Ecol. Evol. 2010, 25, 250–260. [Google Scholar] [CrossRef] [PubMed]
- Pörtner, H.O.; Farrell, A.P. Physiology and climate change. Science 2008, 322, 690–692. [Google Scholar] [CrossRef]
- Liu, S.; Lu, P.; Liu, D.; Jin, P. Pinpointing source of Mekong and measuring its length through analysis of satellite imagery and field investigations. Geo-Spat. Inf. Sci. 2007, 10, 51–56. [Google Scholar] [CrossRef]
- Baran, E.; Baird, I.G.; Cans, G. Fisheries Bioecology at the Khone Falls (Mekong River, Southern Laos); WorldFish Center: Penang, Malaysia, 2005. [Google Scholar]
- Coates, D.; Poeu, O.; Suntornratana, U.; Tung, N.T.; Viravong, S. Biodiversity and fisheries in the Lower Mekong Basin; Phnom Penh: Mekong development series, 2, 30; Mekong River Commission: Phnom Penh, Cambodia, 2003. [Google Scholar]
- Kang, B.; Huang, X. Mekong fishes: Biogeography, migration, resources, threats, and conservation. Rev. Fish. Sci. Aquac. 2022, 30, 170–194. [Google Scholar] [CrossRef]
- Hortle, K.G. Chapter 9—Fisheries of the Mekong River Basin. In The Mekong; Campbell, I.C., Ed.; Elsevier Publishers: Amsterdam, The Netherlands, 2009; pp. 197–249. [Google Scholar]
- Kano, Y.; Dudgeon, D.; Nam, S.; Samejima, H.; Watanabe, K.; Grudpan, C.; Grudpan, J.; Magtoon, W.; Musikasinthorn, P.; Nguyen, P.T.; et al. Impacts of dams and global warming on fish biodiversity in the Indo-Burma hotspot. PLoS ONE 2016, 11, e0160151. [Google Scholar] [CrossRef]
- Stone, R. Dam-building threatens Mekong fisheries. Science 2016, 354, 1084–1085. [Google Scholar] [CrossRef]
- Ficke, A.D.; Myrick, C.A.; Hansen, L.J. Potential impacts of global climate change on freshwater fisheries. Rev. Fish Biol. Fish. 2007, 17, 581–613. [Google Scholar] [CrossRef]
- Lauri, H.; de Moel, H.; Ward, P.J.; Räsänen, T.A.; Keskinen, M.; Kummu, M. Future changes in Mekong River hydrology: Impact of climate change and reservoir operation on discharge. Hydrol. Earth Syst. Sci. 2012, 16, 4603–4619. [Google Scholar] [CrossRef]
- Fukushima, M.; Jutagate, T.; Grudpan, C.; Phomikong, P.; Nohara, S. Potential effects of hydroelectric dam development in the Mekong River basin on the migration of Siamese Mud Carp (Henicorhynchus siamensis and H. lobatus) elucidated by otolith microchemistry. PLoS ONE 2014, 9, e103722. [Google Scholar] [CrossRef] [PubMed]
- Ferguson, J.W.; Healey, M.; Dugan, P.; Barlow, C. Potential effects of dams on migratory fish in the Mekong River: Lessons from salmon in the Fraser and Columbia Rivers. Environ. Manag. 2011, 47, 141–159. [Google Scholar] [CrossRef]
- Le, S.T.; Vo, C.D. The livelihood adaptability of households under the impact of climate change in the Mekong Delta. J. Agribus. Dev. Emerg. Econ. 2020, 11, 7–26. [Google Scholar] [CrossRef]
- Sebastian, L.S.; Sander, B.O.; Simelton, E.; Ngo, D.M. The Drought and Salinity Intrusion in the Mekong River Delta of Vietnam; Assessment Report; CGIAR Research Centers in Southeast Asia: Wageningen, The Netherlands, 2016. [Google Scholar]
- Nguyen, A.L.; Dang, V.H.; Bosma, R.H.; Verreth, J.A.J.; Leemans, R.; De Silva, S.S. Simulated impacts of climate change on current farming locations of striped catfish (Pangasianodon hypophthalmus; Sauvage) in the Mekong Delta, Vietnam. AMBIO 2014, 43, 1059–1068. [Google Scholar] [CrossRef] [PubMed]
- Ziv, G.; Baran, E.; Nam, S.; Rodríguez-Iturbe, I.; Levin, S.A. Trading-off fish biodiversity, food security, and hydropower in the Mekong River Basin. Proc. Natl. Acad. Sci. USA 2012, 109, 5609–5614. [Google Scholar] [CrossRef]
- Valbo-Jørgensen, J.; Coates, D.; Hortle, K. Chapter 8—Fish Diversity in the Mekong River Basin. In The Mekong; Campbell, I.C., Ed.; Elsevier Publishers: Amsterdam, The Netherlands, 2009; pp. 161–196. [Google Scholar]
- Kottelat, M. Diagnoses of a new genus and 64 new species of fishes from Laos (Teleostei: Cyprinidae, Balitoridae, Bagridae, Syngnathidae, Chaudhuriidae and Tetraodontidae). J. South Asian Nat. Hist. 2000, 5, 37–82. [Google Scholar]
- Campbell, I.C. The Mekong: Biophysical Environment of an International River Basin; Elsevier Publishers: Amsterdam, The Netherlands, 2009. [Google Scholar]
- Herder, J.E.; Valentini, A.; Bellemain, E.; Dejean, T.; van Delft, J.J.C.W.; Thomsen, P.F.; Taberlet, P. Environmental DNA—A Review of the Possible Applications for the Detection of (Invasive) Species; Stichting RAVON: Nijmegen, The Netherlands, 2014. [Google Scholar]
- Gillet, B.; Cottet, M.; Destanque, T.; Kue, K.; Descloux, S.; Chanudet, V.; Hughes, S. Direct fishing and eDNA metabarcoding for biomonitoring during a 3-year survey significantly improves number of fish detected around a South East Asian reservoir. PLoS ONE 2018, 13, e0208592. [Google Scholar] [CrossRef]
- Bellemain, E.; Patricio, H.; Gray, T.; Guegan, F.; Valentini, A.; Miaud, C.; Dejean, T. Trails of river monsters: Detecting critically endangered Mekong giant catfish Pangasianodon gigas using environmental DNA. Glob. Ecol. Conserv. 2016, 7, 148–156. [Google Scholar] [CrossRef]
- Osathanunkul, M.; Minamoto, T. A molecular survey based on eDNA to assess the presence of a clown featherback (Chitala ornata) in a confined environment. PeerJ 2020, 8, e10338. [Google Scholar] [CrossRef]
- Jerde, C.L.; Mahon, A.R.; Campbell, T.; McElroy, M.E.; Pin, K.; Childress, J.N.; Armstrong, M.N.; Zehnpfennig, J.R.; Kelson, S.J.; Koning, A.A.; et al. Are genetic reference libraries sufficient for environmental DNA metabarcoding of Mekong River basin fish? Water 2021, 13, 1767. [Google Scholar] [CrossRef]
- Marques, V.; Milhau, T.; Albouy, C.; Dejean, T.; Manel, S.; Mouillot, D.; Juhel, J.-B. GAPeDNA: Assessing and mapping global species gaps in genetic databases for eDNA metabarcoding. Divers. Distrib. 2021, 27, 1880–1892. [Google Scholar] [CrossRef]
- Miya, M.; Sato, Y.; Fukunaga, T.; Sado, T.; Poulsen, J.Y.; Sato, K.; Minamoto, T.; Yamamoto, S.; Yamanaka, H.; Araki, H.; et al. MiFish, a set of universal PCR primers for metabarcoding environmental DNA from fishes: Detection of more than 230 subtropical marine species. R. Soc. Open Sci. 2015, 2, 150088. [Google Scholar] [CrossRef] [PubMed]
- Riaz, T.; Shehzad, W.; Viari, A.; Pompanon, F.; Taberlet, P.; Coissac, E. ecoPrimers: Inference of new DNA barcode markers from whole genome sequence analysis. Nucleic Acids Res. 2011, 39, e145. [Google Scholar] [CrossRef] [PubMed]
- Froese, R.; Pauly, D. FishBase. World Wide Web Electronic Publication. 2022. Available online: www.fishbase.org (accessed on 15 February 2022).
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2021; Available online: https://www.r-project.org (accessed on 15 June 2022).
- Rourke, M.L.; Fowler, A.M.; Hughes, J.M.; Broadhurst, M.K.; DiBattista, J.D.; Fielder, S.; Wilkes Walburn, J.; Furlan, E.M. Environmental DNA (eDNA) as a tool for assessing fish biomass: A review of approaches and future considerations for resource surveys. Environ. DNA 2022, 4, 9–33. [Google Scholar] [CrossRef]
- Yates, M.C.; Fraser, D.J.; Derry, A.M. Meta-analysis supports further refinement of eDNA for monitoring aquatic species-specific abundance in nature. Environ. DNA 2019, 1, 5–13. [Google Scholar] [CrossRef]
- Murtagh, F.; Legendre, P. Ward’s hierarchical agglomerative clustering method: Which algorithms implement Ward’s criterion? J. Classif. 2014, 31, 274–295. [Google Scholar] [CrossRef]
- Thioulouse, J.; Dray, S.; Dufour, A.; Siberchicot, A.; Jombart, T.; Pavoine, S. Multivariate Analysis of Ecological Data with ade4; Springer: New York, NY, USA, 2018. [Google Scholar] [CrossRef]
- De Cáceres, M.; Legendre, P. Associations between species and groups of sites: Indices and statistical inference. Ecology 2009, 90, 3566–3574. [Google Scholar] [CrossRef]
- De Cáceres, M.; Legendre, P.; Moretti, M. Improving indicator species analysis by combining groups of sites. Oikos 2010, 119, 1674–1684. [Google Scholar] [CrossRef]
- Dufrêne, M.; Legendre, P. Species assemblages and indicator species: The need for a flexible asymmetrical approach. Ecol. Monogr. 1997, 67, 345–366. [Google Scholar] [CrossRef]
- Osathanunkul, M. An eDNA detection of captive-bred Mekong Giant Catfish in the Chao Phraya River basin for further environmental impacts assessment. Aquaculture 2022, 546, 737328. [Google Scholar] [CrossRef]
- Rainboth, W.J.; Vidthayanon, C.; Yen, M.D. Fishes of the Greater Mekong Ecosystem with Species List and Photographic Atlas; Museum of Zoology, University of Michigan: Ann Arbor, MI, USA, 2010. [Google Scholar]
- Boyd, D.A.; Nithirojpakdee, P.; Deein, G.; Vidthayanon, C.; Grudpan, C.; Tangjitjaroen, W.; Pfeiffer, J.M.; Randall, Z.S.; Srisombat, T.; Page, L.M. Revision of the horseface loaches (Cobitidae, Acantopsis), with descriptions of three new species from Southeast Asia. Zootaxa 2017, 4341, 151–192. [Google Scholar] [CrossRef] [PubMed]
- Kimura, S.; Katahira, K.; Kuriiwa, K. The red-fin Decapterus group (Perciformes: Carangidae) with the description of a new species, Decapterus smithvanizi. Ichthyol. Res. 2013, 60, 363–379. [Google Scholar] [CrossRef]
- Fricke, R.; Eschmeyer, W.N.; van der Laan, R. (Eds.) Eschmeyer’s Catalog of Fishes: Genera, Species, References. 2022. Available online: http://researcharchive.calacademy.org/research/ichthyology/catalog/fishcatmain.asp (accessed on 15 April 2022).
- Pham, M.H.; Hoang, D.H.; Panfili, J.; Ponton, D.; Durand, J.-D. Diversity of fishes collected with light traps in the oldest marine protected area in Vietnam revealed by DNA barcoding. Mar. Biodivers. 2022, 52, 30. [Google Scholar] [CrossRef]
- Ratnasingham, S.; Hebert, P.D.N. A DNA-based registry for all animal species: The barcode index number (BIN) system. PLoS ONE 2013, 8, e66213. [Google Scholar] [CrossRef]
- Durand, J.-D.; Borsa, P. Mitochondrial phylogeny of grey mullets (Acanthopterygii: Mugilidae) suggests high proportion of cryptic species. Comptes Rendus Biol. 2015, 338, 266–277. [Google Scholar] [CrossRef]
- Hubert, N.; Meyer, C.P.; Bruggemann, H.J.; Guérin, F.; Komeno, R.J.L.; Espiau, B.; Causse, R.; Williams, J.T.; Planes, S. Cryptic diversity in Indo-Pacific coral-reef fishes revealed by DNA-barcoding provides new support to the centre-of-overlap hypothesis. PLoS ONE 2012, 7, e28987. [Google Scholar] [CrossRef]
- Zemlak, T.S.; Ward, R.D.; Connell, A.D.; Holmes, B.H.; Hebert, P.D.N. DNA barcoding reveals overlooked marine fishes. Mol. Ecol. Resour. 2009, 9, 237–242. [Google Scholar] [CrossRef]
- Whitfield, A.K. Why are there so few freshwater fish species in most estuaries? J. Fish Biol. 2015, 86, 1227–1250. [Google Scholar] [CrossRef]
- Nukazawa, K.; Hamasuna, Y.; Suzuki, Y. Simulating the advection and degradation of the environmental DNA of common carp along a river. Environ. Sci. Technol. 2018, 52, 10562–10570. [Google Scholar] [CrossRef]
- Barnes, M.A.; Turner, C.R.; Jerde, C.L.; Renshaw, M.A.; Chadderton, W.L.; Lodge, D.M. Environmental conditions influence eDNA persistence in aquatic systems. Environ. Sci. Technol. 2014, 48, 1819–1827. [Google Scholar] [CrossRef] [PubMed]
- Jo, T.; Murakami, H.; Yamamoto, S.; Masuda, R.; Minamoto, T. Effect of water temperature and fish biomass on environmental DNA shedding, degradation, and size distribution. Ecol. Evol. 2019, 9, 1135–1146. [Google Scholar] [CrossRef] [PubMed]
- Lance, R.F.; Klymus, K.E.; Richter, C.A.; Guan, X.; Farrington, H.L.; Carr, M.R.; Thompson, N.; Chapman, D.C.; Baerwaldt, K.L. Experimental observations on the decay of environmental DNA from bighead and silver carps. Manag. Biol. Invasions 2017, 8, 343. [Google Scholar] [CrossRef]
- Stoeckle, B.C.; Beggel, S.; Cerwenka, A.F.; Motivans, E.; Kuehn, R.; Geist, J. A systematic approach to evaluate the influence of environmental conditions on eDNA detection success in aquatic ecosystems. PLoS ONE 2017, 12, e0189119. [Google Scholar] [CrossRef]
- Strickler, K.M.; Fremier, A.K.; Goldberg, C.S. Quantifying effects of UV-B, temperature, and pH on eDNA degradation in aquatic microcosms. Biol. Conserv. 2015, 183, 85–92. [Google Scholar] [CrossRef]
- Jo, T.; Minamoto, T. Complex interactions between environmental DNA (eDNA) state and water chemistries on eDNA persistence suggested by meta-analyses. Mol. Ecol. Resour. 2021, 21, 1490–1503. [Google Scholar] [CrossRef]
- Abell, R.; Thie0me, M.L.; Revenga, C.; Bryer, M.; Kottelat, M.; Bogutskaya, N.; Coad, B.; Mandrak, N.; Balderas, S.C.; Bussing, W.; et al. Freshwater ecoregions of the world: A new map of biogeographic units for freshwater biodiversity conservation. BioScience 2008, 58, 403–414. [Google Scholar] [CrossRef]
- Grill, G.; Ouellet Dallaire, C.; Fluet Chouinard, E.; Sindorf, N.; Lehner, B. Development of new indicators to evaluate river fragmentation and flow regulation at large scales: A case study for the Mekong River Basin. Ecol. Indic. 2014, 45, 148–159. [Google Scholar] [CrossRef]
- Sokheng, C.; Chhea, C.K.; Viravong, S.; Bouakhamvongsa, K.; Suntornratana, U.; Yoorong, N.; Tung, N.T.; Bao, T.Q.; Poulsen, A.F.; Jorgensen, J.V. Fish migrations and spawning habits in the Mekong mainstream: A survey using local knowledge (basin-wide). In Assessment of Mekong Fisheries: Fish Migrations and Spawning and the Impact of Water Management Project (AMFC); AMFP Report, 2, 99; Mekong River Commission: Vientiane, Laos, 1999. [Google Scholar]
- Tran, N.T.; Labonne, M.; Chung, M.-T.; Wang, C.-W.; Huang, K.-F.; Durand, J.-D.; Chaiwut, G.; Chan, B.; Hoang, H.D.; Panfili, J. Natal origin and migration pathways of Mekong catfish (Pangasius krempfi) using strontium isotops and trace element concentrations in environmental water and otoliths. PLoS ONE 2021, 16, e0252769. [Google Scholar] [CrossRef]
- Poulsen, A.F.; Poeu, O.; Viravong, S.; Suntornratana, U.; Nguyen, T.T. Fish Migrations of the Lower Mekong River Basin: Implications for Development, Planning and Environmental Management; MRC Technical Paper No. 8; Mekong River Commission: Phnom Penh, Cambodia, 2002. [Google Scholar]
- Nuon, V.; Lek, S.; Ngor, P.B.; So, N.; Grenouillet, G. Fish community responses to human-induced stresses in the Lower Mekong Basin. Water 2020, 12, 3522. [Google Scholar] [CrossRef]
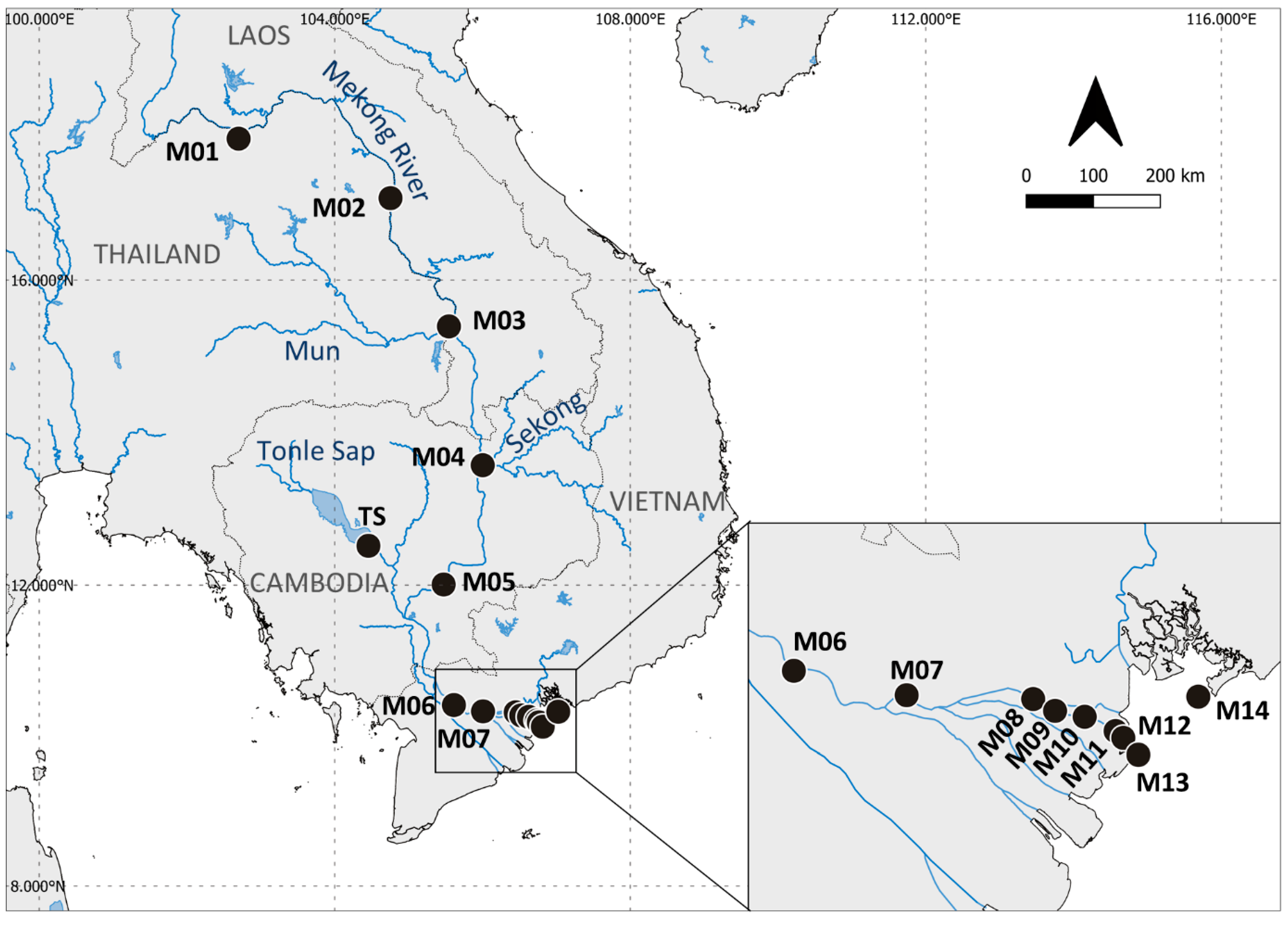



| Site Code | Location | Coordinates | Sampling Date | Salinity | Distance from the Sea (km) | |
|---|---|---|---|---|---|---|
| Latitude | Longitude | |||||
| M01 | Nong Khai, Thailand | 17°49.740′ N | 102°41.878′ E | Sept. 2019 | 0.0 | 1529 |
| M02 | Nakhon Phanom, Thailand | 17°3.735′ N | 104°45.268′ E | Sept. 2019 | 0.0 | 1158 |
| M03 | Ubon Ratchathani, Thailand | 15°23.823′ N | 105°32.737′ E | Sept. 2019 | 0.0 | 903 |
| M04 | Khone Falls, Stung Treng, Cambodia | 13°34.815′ N | 106°0.178′ E | Aug. 2019 | 0.0 | 646 |
| TS | Tonle Sap, Kampong Thom, Cambodia | 12°31.099′ N | 104°27.334′ E | June 2019 | 0.0 | 443 |
| M05 | Kampong Cham, Cambodia | 12°0.442′ N | 105°28.461′ E | June 2019 | 0.0 | 417 |
| M06 | Dong Thap, Vietnam | 10°24.514′ N | 105°36.764′ E | May 2019 | 0.0 | 143 |
| M07 | Tien Giang, Vietnam | 10°19.488′ N | 106°0.396′ E | May 2018 | 0.1 | 90 |
| M08 | Chau Thanh, Ben Tre, Vietnam | 10°18.699′ N | 106°26.909′ E | Apr. 2018 | 1.0 | 39 |
| M09 | Binh Dai, Ben Tre, Vietnam | 10°16.266′ N | 106°31.494′ E | Apr. 2018 | 5.9 | 29 |
| M10 | Binh Dai, Ben Tre, Vietnam | 10°15.048′ N | 106°37.698′ E | Apr. 2018 | 10.7 | 18 |
| M11 | Binh Dai, Ben Tre, Vietnam | 10°12.114′ N | 106°44.166′ E | Apr. 2018 | 16.6 | 5 |
| M12 | Binh Dai, Ben Tre, Vietnam | 10°10.703′ N | 106°45.793′ E | Apr. 2018 | 21.3 | 2 |
| M13 | Binh Dai, Ben Tre, Vietnam | 10°7.254′ N | 106°48.948′ E | Apr. 2018 | 26.7 | 0 |
| M14 | Vung Tau, Vietnam | 10°19.188′ N | 107°1.494′ E | Apr. 2018 | 30.0 | 0 |
| Order | Group | p |
|---|---|---|
| Familly | ||
| Species | ||
| Osteoglossiformes | ||
| Notopteridae | ||
| Chitala ornata | 1 | 0.0286 * |
| Clupeiformes | ||
| Engraulidae | ||
| Stolephorus chinensis | 3 | 0.0005 *** |
| Stolephorus insularis B | 3 | 0.0012 ** |
| Coilia rebentischii | 3 | 0.0245 * |
| Clupeidae | ||
| Clupeichthys aesarnensis | 1 | 0.0308 * |
| Clupeoides borneensis | 2 | 0.0108 * |
| Hilsa kelee | 3 | 0.0045 ** |
| Gonorynchiformes | ||
| Chanidae | ||
| Channa striata | 1 | 0.0170 * |
| Cypriniformes | ||
| Botiidae | ||
| Syncrossus helodes | 1 | 0.0304 * |
| Cyprinidae | ||
| Amblyrhynchichthys micracanthus | 1 | 0.0304 * |
| Bangana sp. | 1 | 0.0041 ** |
| Barbonymus altus | 1 | 0.0006 *** |
| Barbonymus gonionotus | 1 + 2 | 0.0005 *** |
| Barbonymus sp1 | 1 | 0.0049 ** |
| Barbonymus sp2 | 1 | 0.0006 *** |
| Cosmochilus harmandi | 1 | 0.0320 * |
| Cosmochilus sp2 | 1 | 0.0320 * |
| Cyprinidae sp12 | 1 | 0.0308 * |
| Cyprinidae sp13 | 1 | 0.0308 * |
| Cyprinidae sp16 | 1 | 0.0055 ** |
| Cyprinidae sp20 | 1 | 0.0294 * |
| Cyprinidae sp9 | 1 + 2 | 0.0399 * |
| Hypsibarbus malcolmi | 1 | 0. 0055 * |
| Hypsibarbus wetmorei | 1 | 0. 0049 ** |
| Labeo chrysophekadion | 1 | 0.0163 * |
| Mystacoleucus marginatus | 1 | 0.0311 * |
| Paralaubuca sp2 | 1 | 0.0165 * |
| Paralaubuca sp3 | 1 | 0.0311 * |
| Probarbus jullieni | 1 | 0.0311 * |
| Puntioplites falcifer | 1 | 0.0014 ** |
| Puntioplites proctozystron | 1 + 2 | 0.0088 ** |
| Puntioplites sp. | 2 | 0.0328 * |
| Sikukia gudgeri C | 1 | 0. 0047 ** |
| Sikukia sp2 | 1 | 0.0294 * |
| Siluriformes | ||
| Siluriformes sp1 | 1 | 0.0308 * |
| Bagridae | ||
| Hemibagrus spilopterus | 1 | 0.0048 ** |
| Mystus bocourti | 1 | 0.0308 * |
| Pangasiidae | ||
| Pangasius bocourti | 1 + 2 | 0.0005 *** |
| Pangasius larnaudii | 1 + 2 | 0.0440 * |
| Pangasius macronema | 1 + 2 | 0.0111 * |
| Siluridae | ||
| Belodontichthys truncatus | 1 | 0.0304 * |
| Gobiiformes | ||
| Gobiidae | ||
| Gobiidae sp4 | 2 | 0.0077 ** |
| Synbranchiformes | ||
| Mastacembelidae | ||
| Mastacembelus sp. | 1 | 0.0047 ** |
| Cichliformes | ||
| Cichlidae | ||
| Oreochromis niloticus | 1 + 2 | 0.0408 * |
| Beloniformes | ||
| Belonidae | ||
| Xenentodon cancila | 1 | 0.0006 *** |
| Perciformes | ||
| Polynemidae | ||
| Polynemus melanochir | 2 | 0.0081 ** |
| Acanthuriformes | ||
| Sciaenidae | ||
| Johnius carouna | 3 | 0.0042 ** |
| Johnius borneensis | 3 | 0.0246 * |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Durand, J.-D.; Simier, M.; Tran, N.T.; Grudpan, C.; Chan, B.; Nguyen, B.N.L.; Hoang, H.D.; Panfili, J. Fish Diversity along the Mekong River and Delta Inferred by Environmental-DNA in a Period of Dam Building and Downstream Salinization. Diversity 2022, 14, 634. https://doi.org/10.3390/d14080634
Durand J-D, Simier M, Tran NT, Grudpan C, Chan B, Nguyen BNL, Hoang HD, Panfili J. Fish Diversity along the Mekong River and Delta Inferred by Environmental-DNA in a Period of Dam Building and Downstream Salinization. Diversity. 2022; 14(8):634. https://doi.org/10.3390/d14080634
Chicago/Turabian StyleDurand, Jean-Dominique, Monique Simier, Ngan Trong Tran, Chaiwut Grudpan, Bunyeth Chan, Bao Ngoc Le Nguyen, Huy Duc Hoang, and Jacques Panfili. 2022. "Fish Diversity along the Mekong River and Delta Inferred by Environmental-DNA in a Period of Dam Building and Downstream Salinization" Diversity 14, no. 8: 634. https://doi.org/10.3390/d14080634
APA StyleDurand, J.-D., Simier, M., Tran, N. T., Grudpan, C., Chan, B., Nguyen, B. N. L., Hoang, H. D., & Panfili, J. (2022). Fish Diversity along the Mekong River and Delta Inferred by Environmental-DNA in a Period of Dam Building and Downstream Salinization. Diversity, 14(8), 634. https://doi.org/10.3390/d14080634







