Abstract
The Italian peninsula represented one of the main glacial refugia during climatic oscillations of the Pleistocene, currently being a biodiversity hotspot. In this work, we analysed for the first time the genetic diversity of harvest mouse populations in Italy, and we compared them with those of the rest of Eurasia. Mitochondrial cytochrome-b gene was amplified from 12 samples from throughout the Italian range. We recorded a very low genetic diversity, in line with the rest of the harvest mouse range. In the comparative phylogenetic tree, Northern Italy samples clustered together as a sister group of the rest of Europe, whereas those from Central Italy clustered with Central Europe samples. Harvest mice have recently conquered Southern Europe, i.e., possibly at the start of the Holocene. The global genetic homogeneity might be due to accidental human-mediated introductions or to the sharp decline of the habitat of the harvest mouse, which may in turn have caused severe bottlenecks in the populations of this small rodent.
1. Introduction
Mediterranean peninsulas (Iberia, Balkan peninsula, and Italy) acted as glacial refugia for many European taxa and still represent interesting concentrations of genetic diversity and unicity, with the highest percentage of endemic taxa in Europe [1,2,3,4]. Divergence of genetic lineages in Mediterranean peninsulas is apparent even for large sized species (e.g., the roe deer Capreolus capreolus, the brown bear Ursus arctos, and the grey wolf Canis lupus [5,6,7]), thus being much stronger and more definite in small mammals (Rodentia and Eulipotyphla), which remained isolated for long times from other European conspecific populations [2]. Accordingly, the majority of endemic mammal species in Italy (i.e., 8 out of 10 endemic species [2,3]) belongs to the group of “small mammals” [8]. Furthermore, Italian small mammal species include several other divergent lineages, which may represent new species or at least well-supported subspecies or Evolutionary Divergent Units, e.g., the pygmy shrew Sorex minutus [9], the blind mole Talpa caeca [10], the European hedgehog Erinaceus europaeus in Sicily [11], the bank vole Chlethrionomys glareolus in Southern Italy [12,13], the edible dormouse Glis glis, and the hazel dormouse Muscardinus avellanarius in Central-Southern Italy [14,15]. The Italian peninsula hosts specific genotypes amongst small mammals, i.e., the most differentiated lineage of the Eurasian red squirrel Sciurus vulgaris clade from Spain to Korea, currently identified as a separated species endemic to Southern Italy, Sciurus meridionalis [7]. No genetic differentiation occurs for those species which, despite being widely distributed, may be accidentally transported by humans throughout their range (e.g., the Etruscan shrew Suncus etruscus [16]).
Research efforts and conservation of these species are therefore paramount, given also that most rodents represent reliable bioindicators of good environmental management practices [8]. Besides their remarkable taxonomic, evolutionary, and conservation importance, small mammals (and rodents in particular) are often neglected in conservation plans as this group is often connected to disease spread and damages to agriculture, forestry, and ecosystems [8]. Despite this, the detection of localized, endemic taxa is indeed pivotal to preserve them. The ongoing habitat loss and human environmental exploitation represents the main cause of the global biodiversity loss [17], and several rodent species are strictly linked to vulnerable habitats [8,18,19,20]. Amongst those, the harvest mouse Micromys minutus, the smallest European rodent, shows a wide distribution from Northern Spain to Japan [21]. Despite this, populations are becoming more and more localized, and the status of the global population is “declining” according to the International Union for the Conservation of Nature [22,23]. The harvest mouse is a typical species of open areas and wetlands characterized by the presence of reed beds with Phragmites australis (Cav.) Trin. ex Steud., which are imperiled habitats, threatened by the climatic change—with consequent droughts and wildfires—and by human actions, e.g., management interventions by reclamation consortia [23,24,25]. Thus, the harvest mouse is a reliable bioindicator of environmental health and of land-use change [8,21]. The importance of the harvest mouse as a prey for many mammalian carnivores and diurnal/nocturnal raptor birds implies its conservation interest [26]. Phylogeographic investigations on this species have been conducted through the cytochrome-b gene of the mitochondrial DNA, showing a very low nucleotide divergence between Europe and Eastern Asia [21,27]. Northern Vietnam hosts a different species, the Far East harvest mouse Micromys erythrotis [28]. Fossil evidence shows that this small rodent underwent several cycles of extinction and recolonization in Europe through the Quaternary Ice Ages [21]. However, previous studies did not include any sample of harvest mouse from Italy and the detected genetic homogeneity does not necessarily imply that Italian individuals would also show a similar genetic diversity (e.g., [29], for the Eurasian red squirrel). The harvest mouse is an understudied, often overlooked species in Italy (Appendix A), with a very low number of studies published in the last 45 years.
In this work, we aimed at investigating the sequence variation of the cytochrome-b mitochondrial gene of harvest mice from Italian populations to determine the phylogenetic relationships with the rest of the extent of occurrence of this species. Given the importance of the Italian peninsula as a glacial refugium, we predicted that isolated Italian populations of the harvest mouse would differ from those of the rest of Eurasia.
2. Materials and Methods
Given the objective difficulty to trap this species [30], we asked for harvest mouse samples from all museum collections from Northern Italy covering the distribution of this species. However, we were able to collect only 19 harvest mouse samples from the remnant extent of occurrence of the harvest mouse in Italy (Refs. [31,32,33], Figure 1). Amongst those, we were able to extract well-preserved DNA only from 12 (Figure 1). Two harvest mouse samples were obtained from Central Italy from individuals killed by domestic cats [34].
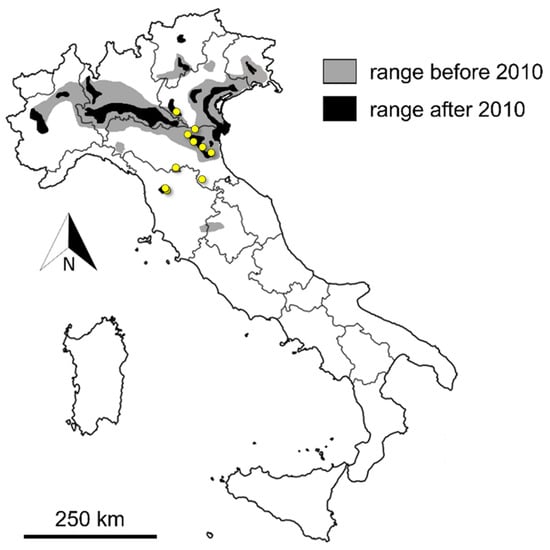
Figure 1.
Past (2000–2010) and present (2010–2022) distribution of Micromys minutus in Italy [31,32,33]. Yellow dots represent sampling sites. As to Central Tuscany (Padule di Fucecchio), first records date back to 1990s and have been recently confirmed [34].
DNA was extracted from 30 mg tissue samples previously preserved in absolute ethanol or from frozen samples using the QIAGEN Blood and Tissue kit (Qiagen®, Hilden, Germany), following manufacturer’s instructions. We conducted Sanger sequencing of cytochrome-b PCR amplifications (1140 base pairs, hereafter cytb) using primers already tested for the harvest mouse: L14115 (5′-GACATGAAAAATCATCGTTG-3′) and H15300 (5′-GTTTACAAGACCAGAGTAAT-3′) [21]. Each PCR mix contained 1 mM of MgCl2, 200 μM of dNTPs, 8–10 pmol of each primer, 1 unit of Taq DNA Polymerase recombinant (Thermo Fisher Scientific®, Dreieich, Germany), and 35 ng/μL of each DNA template. PCR reactions were run through a 2720 Thermal Cycler (Applied Biosystems®, Foster City, California, USA), following this profile: 10 min at 94 °C, 40 cycles of 30 s at 94 °C, 60 s at 50 °C, and 120 s at 72 °C, followed by 10 min at 72 °C as final extension. The electrophoresis was run for 45 min (100 V) on a GSTAR-stained 2.0% agarose gel. PCR products were purified using ExoSAP-IT PCR clean-up Kit (GE Healthcare®, Piscataway, New Jersey, USA) and sequenced with a sequencing kit (ABI Big Dye Terminator Cycle Sequencing v. 2.0- ABI PRISM, Applied Biosystems®, Foster City, California, USA). GenBank accession numbers are reported in Table 1. Electropherograms were visualized with CHROMAS 1.45 (http://www.technelysium.com.au; accessed on 12 July 2022). Sequences, once manually corrected, were aligned using CLUSTALX 1.81 (https://www.igbmc.u-strasbg.fr/BioInfo/Clustal; accessed on 12 July 2022). In order to determine if sequences were nuclear (NUMTs [35,36]) or mitochondrial copies, we followed three steps. First, sequence chromatograms were checked for double signals. Then, coding sequence alignments were inspected for frameshift mutations and/or stop codons. Afterwards, the corrected sequences were compared to those in GenBank: we compared our sequences to the ones deposited in the NCBI database using BLASTx and BLASTn (Table 1).

Table 1.
Origins of harvest mouse samples analysed in our work: Label, Location and Region of origin, Year of collection, Museum specimen code, GenBank Accession Numbers. *: sequences from other authors. NA, not applicable.
The phylogenetic reconstruction was performed by Neighbour-Joining (NJ), Bayesian (BI), and Maximum Likelihood (ML) analyses. Analyses were carried out using genetic models selected by jModelTest 2 [37], with the Akaike Information Criterion (AIC). The TN93 (TAMURA-NEI) nucleotide substitution model was selected and corrected for rate heterogeneity among sites with a Gamma (G) distribution. The NJ was performed by MEGA software and 1000 bootstrap replicates. The BI analysis was performed with MrBayes v.3.12 [38], using the best model selected. Four chains of Markov Chain Monte Carlo (MCMC) were run simultaneously and sampled every 1000 generations for 4 million generations. The first 1000 sampled trees from each run were discarded as burn-in. The ML phylogenetic analysis was conducted through the SeaView software [39]. We selected the optimized choices, and we obtained the tree-searching operations by Nearest-Neighbour Interchange (NNI) and Subtree Pruning–Regrafting (SPR).
A haplotype network of all sequences was built through TCS v.1.21 using median-joining algorithm [40,41]. The network was plotted with tcsBU [42]. Convergence of runs was estimated by the average standard deviation of split frequencies and the potential scale reduction factor. In addition, stationarity was verified by examining posterior probability, log likelihood, and parameters by the effective sample sizes (ESSs) in the program Tracer v1.7 [43]. As the runs converge onto the stationary distribution, we expect the average standard deviation of split frequencies to approach zero, reflecting the fact that the tree samples become increasingly similar each other. The proportion of trees that contained the clade was given as the posterior probability (PP) to estimate the robustness of each clade. Branch supports were assessed by 100 non-parametric bootstrap replicates.
To conclude, the software BEAST version 2.2.1 was used to estimate divergence time between main lineages [44]. We used two substitution rates for cytb of rodents (1.6% by [45] and 9.75% as an average by [46]) to determine divergence times between the major clades. To check the validity of a strict molecular clock for the entire dataset, the likelihood ratio method implemented in the Mega version 6 [47] was used with the TN model. The null hypothesis was rejected (p < 0.05) and, therefore, the uncorrelated log-normal relaxed molecular clock prior was used. For all analyses, chain length was set to 50 million iterations with data logged every 1000th iteration. Tree annotator version 1.7.4 [48] was used to summarize the posterior sample of trees, after removal of the first 10% of trees as burn-in, as a maximum clade credibility tree. Results obtained with BEAST were checked in Tracer to determine adequate burn-in and MCMC chain mixing through ESS values. Burn-in was set to 2 × 10−6, corresponding to 10% of the total samples in each run. Finally, the phylogenetic trees were visualized in Figtree version 1.4.2 [49].
3. Results
We were able to obtain cytb sequences from all analysed samples. All sequences generated in the present study were deposited in GenBank (Table 1). The alignment of the cytb gene consists of 1100 nucleotides for 13 taxa, 42 of which are variable and 15 parsimony-informative. The average ratio of TS/TV is 1.56. Nucleotide genetic diversity was 0.00898 (± 0.0017) and the average number of divergences was found 7.773. An ML tree is presented in Figure 2. Topologies were very similar for ML, BI, and NJ trees; support values at each node for the three methods were shown (Figure 2). The monophyly of M. minutus, including all European samples, was strongly supported (Figure 2). The Italian clade (showing a lower support) was split in two groups, where individuals from Padule di Fucecchio (Central Italy) showed more phylogenetic relationship with the other European samples (United Kingdom, France, Germany, and Spain), rather than with the other Italian ones. The Asian samples (South Korea + Japan, and Russia + China + Taiwan) appeared as sister groups of European samples (Figure 2).
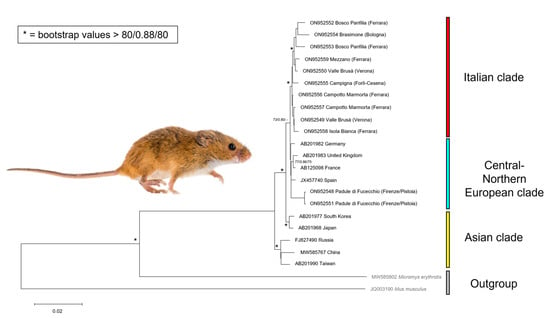
Figure 2.
ML tree obtained from the analysis of cytb for 1150 positions. For each node, support values (NJ Bootstrap support/Bayesian probabilities/ML Bootstrap support) above 80/0.88/80 were shown as asterisks. We used Micromys erythrotis and Mus musculus as outgroups.
In the phylogeographic network, two haplogroups could be identified: (A) including all Italian sequences, except for Padule di Fucecchio samples; (B) including all the other Eurasian samples (Figure 3).
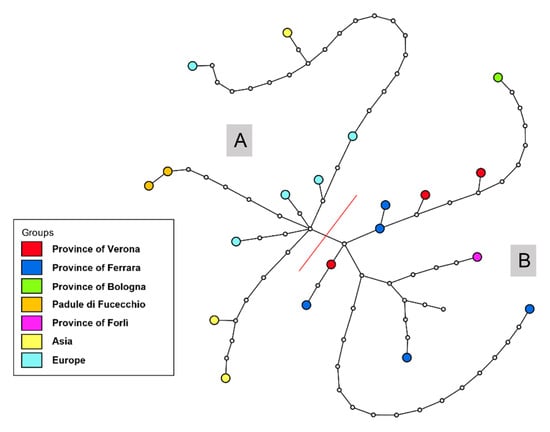
Figure 3.
Median-Joining haplotype network topology obtained for the cytb data of Micromys minutus in Italy, Europe, and Asia. Haplotypes are depicted according to sampling sites. Uncoloured circles represent mutation steps. The red line separates (A) Italian haplotypes from (B) Asian, European, and two Italian (Padule di Fucecchio) haplotypes (average: 12.31 nucleotide differences among populations).
Estimates for the time to the most recent common ancestor (TMRCA) of each clade, as well as their 95% credibility intervals, were obtained for two substitution rates reported in the scientific literature for rodents [45,46] separately (Figure 4A,B).
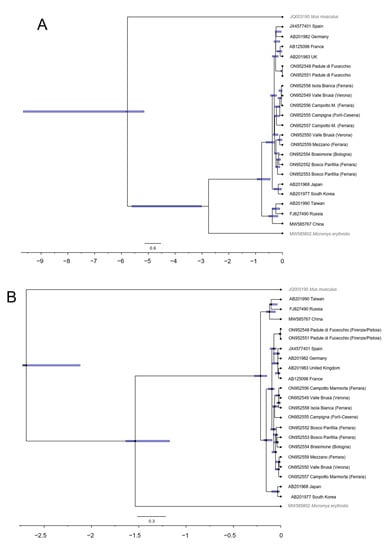
Figure 4.
Divergence time chronograms of Micromys minutus using an uncorrelated relaxed clock method by BEAST and two rates estimated for rodents ((A): 1.60%; (B): 9.75%). Branches are proportional to time in millions of years. Blue bars indicate 95% confidence intervals for the ages of basal branches in the tree and major lineages.
4. Discussion
Our work described for the first time the genetic diversity of the harvest mouse in Italy. We confirmed that, despite the presence of the Alps [12,14], also including Italian populations, this small rodent shows in general low levels of genetic variation (i.e., max 1.3% for cytb: net nucleotide divergence = 0.01: [21]) across its distribution range.
Italian populations showed levels of genetic diversity (i.e., nucleotide divergence = 0.0089; k = 7.773) larger than other European populations (π = 0.0003; k = 4.364) and smaller compared to Asiatic ones of Micromys minutus (π = 0.012; k = 12.54 [21]). However, genetic diversity values were comparable with (e.g., Gerbillus campestris, [50]; Apodemus agrarius [51]; other species [52]), or lower [53,54,55] than those found in other rodents.
Although traditional taxonomy identifies 20 subspecies [31], only four clades can be confirmed: the one including most Italian samples, the one including the rest of European (also comprising Padule di Fucecchio samples) specimens (commonly reported as Micromys minutus minutus), the one including South Korea + Japan samples (Micromys minutus hondonis), and the one including Russia + China + Taiwan samples (Micromys minutus takasagoensis) [31,56]. Apart from Central Italy individuals, the rest of the Italian group seems to diverge from the rest of Europe; however, this clade relies only on a few parsimony-informative nucleotides.
It has been suggested that the isolated populations of the harvest mouse in Central Italy may occur in this area as a result of progressive rarefactions of previously widespread populations following habitat loss [31]. However, genetic data here reported re-emerge the hypothesis of presence following passive transport (e.g., crop trade [31]). Accordingly, the Padule di Fucecchio population, currently the southernmost one in Italy [30], is genetically similar to other European populations, although the small number of variable and parsimony-informative sites of the cytb and our small sample size require caution in conclusions.
Our results provide support to Yasuda et al. [21], who reported that the harvest mouse may have undergone recent extreme population fluctuations (e.g., bottlenecks), which might have reduced the genetic variability of this species [57]. Furthermore, the most ancient fossil documentation of this species in Europe dates back to late Pliocene, and no fossil record is known to occur before Holocene in Italy (i.e., I century BC—I century AC), suggesting a recent colonization of Western Europe including the Italian peninsula by this species [31,58,59]. However, Italy may have acted as a glacial refugium for the harvest mouse. Particularly, following [46] who corrected previous literature [45], the time of the divergence of the harvest mouse Italian population may be placed at 120,000 years ago. Therefore, it may represent the result of natural distribution of this small rodent, rather than an outgrowth linked to human activities during the Holocene.
In several other cases amongst rodents, the Alps have separated Italian populations from those in the rest of Europe, thus promoting the genetic differentiation of endemic lineages [7,12,13,14]. Despite showing limited dispersal abilities and movement patterns, crop trade routes and the pet trade may have helped harvest mice to conquer several parts of their current range [21,31,60]. Differently from what was previously reported [21], accidental introductions of harvest mice did not only occur on islands, but also on the mainland, particularly at the borders of the natural distribution range, e.g., in Northern Europe [27]. These human-mediated animal movements might have promoted the genetic homogenization of the harvest mouse throughout its range [61]. The low genetic diversity of the harvest mouse and the strong evidence for genetic population structuring have been suggested to be due to the susceptibility of this species to climatic change, which in turn reduces the coverage of the suitable habitat (as also observed in Italy: cf. Figure 1), i.e., moist grasslands [21,23,24,62].
5. Conclusions
Conservation of rodent species is often neglected by the fact that, despite being key prey species for most carnivores, helping pollination, and representing good bioindicators of habitat management and seed dispersers, they are often treated as pests [8]. However, in Mediterranean habitats, most endemic species amongst mammals are represented by rodents [2,10,12,29], thus highlighting the importance of an extensive knowledge on their species diversity, including genetic analyses. Despite this, in most cases, lack of funds prevent researchers from conducting detailed analyses on these taxa (e.g., on cryptic species and on taxa requiring expertise to be correctly classified), which are also poorly recorded in museums and private collections [63]. At a first sight, harvest mice can be easily confused with juvenile individuals of common field mice Apodemus spp. Or with invasive domestic mice Mus musculus and Mus domesticus. The negligible research effort so far exerted on this species in Italy (cf. Appendix A), together with the lack of records and samples in museums and private collections, may bring researchers to overlook the population decline of this locally imperiled species. The mitochondrial lineage divergence—and thus clustering—of this species in Italy, observed for the first time in our work, may imply the designation of a different subspecies or Evolutionary Significant Unit, which have been separated from the rest of European populations by over 100,000 years.
Author Contributions
Conceptualization, E.M., A.V. and S.M.; sampling, S.M. and D.S.; methodology, M.B. and D.S.; software, M.B.; formal analysis, E.M. and M.B.; resources, M.B.; data curation, A.V. and A.B.; writing—original draft preparation, E.M., A.V. and M.B. All authors have read and agreed to the published version of the manuscript.
Funding
E.M. and A.V. were supported by Mur-FOE-Project Capitale Naturale-Task “Biodiversità”.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
All sequences are deposited on GenBank.
Acknowledgments
Authors would like to thank Alessio Bartolini, Enrico Zarri, Fabio Ussi, and Fabio Bona for the help they provided in data collection. D.S. is grateful to the Ghislieri Foundation for supporting his research activity through an accommodation scholarship. Two anonymous reviewers kindly took the time to improve our MS with their comments. E. Basset revised our MS and improved our English language.
Conflicts of Interest
The authors declare no conflict of interest.
Appendix A

Table A1.
List of published works (numbered) in 1976–2022 dealing with harvest mice in Italy.
Table A1.
List of published works (numbered) in 1976–2022 dealing with harvest mice in Italy.
| Year | Full Reference | Method | Topic |
|---|---|---|---|
| 1976 | (1) Barbieri, F.; Fogliani, G.; Fasola, M. Aspetti della predazione di barbagianni (T. alba). Atti V Simp. Cons. Nat. Bari 1976, 5, 293–302. | Owl pellet analysis | Distribution |
| 1978 | (2) Bertazzini, M.; Sala, B. Prime indagini sulle associazioni a micromammiferi del Basso Ferrarese. Studi Trentini Scienze Nat. 1978, 55, 35–46. | Owl pellet analysis | Distribution |
| 1980 | (3) Gerdol, R.; Mantovani, E. Dati preliminari sulla predazione di Tyto alba (Scopoli) nel ferrarese. Avocetta 1980, 4, 83–86. | Owl pellet analysis | Distribution |
| 1982 | (4) Boldreghini, P.; Casini, L.; Santolini, R. Dati sulla predazione di Tyto alba (Scop.) su micromammiferi nelle Valli Bertuzzi (Delta del Po). Boll. Zool. 1982, 49, 23–24. | Owl pellet analysis | Distribution |
| 1988 | (5) Boldreghini, P.; Casini, L.; Santolini, R. Variazioni stagionali della dieta di Tyto alba nel Bosco della Mesola (Delta del Po). Nat. Sicil. 1988, 4, 151–153. | Owl pellet analysis | Distribution |
| 1992 | (6) Canova, L. Distribution and habitat preference of small mammals in a biotope of the north Italian plain. Ital. J. Zool. 1992, 59, 417–420. | Captures | Ecology |
| 1992 | (7) Bon, M.; Roccaforte, P.; Sirna, G. Primi dati sui micromammiferi della gronda lagunare di Venezia tramite borre di Tyto alba (Scopoli, 1796). Boll. Mus. Civ. Sto. Nat. Venezia 1992, 41, 256–273. | Owl pellet analysis | Distribution |
| 1993 | (8) De Marinis, A.M.; Agnelli, P. Guide to the microscope analysis of Italian mammals hairs: Insectivora, Rodentia and Lagomorpha. Ital. J. Zool. 1993, 60, 225–232. | Hair structure analysis | Morphology |
| 1995 | (9) Agnelli, P.; Lazzaretti, A. On the distribution of Micromys minutus in Italy. Ital. J. Zool. 1995, 62, 395–399. | Review | Distribution |
| 1997 | (10) Gotta, A.; Pigozzi, G. Trophic niche of the barn owl and little owl in a rice field habitat in northern Italy. Ital. J. Zool. 1997, 64, 55–59. | Owl pellet analysis | Distribution |
| 1998 | (11) Castioni, C.; Debernardi, P.; Patriarca, E. L’alimentazione invernale del Gufo comune (Asio otus) nel Parco del Ticino (Italia nord-occidentale). Riv. Piem. Sto. Nat. 1998, 19, 299–312. | Owl pellet analysis | Distribution |
| 1998 | (12) Mazzotti, S.; Caramori, G. Analysis of small mammal communities in South-Eastern Po valley. Gortania 1998, 20, 253–262. | Owl pellet analysis | Distribution |
| 1999 | (13) Paci, A.; Romano, C. Micromammalia umbra: aggiornamento allo status 1983. Atti Conv. Naz. Biol. Selv. 1999, 4, 142. | Owl pellet analysis | Distribution |
| 2000 | (14) Ragni, B.; Chiappini, M.M. Micromys minutus (Mammalia, Rodentia) nel Lago Trasimeno (Italia, Umbria). Riv. Idrobiol. 2000, 89, 215–220. | Owl pellet analysis | Distribution |
| 2000 | (15) Pirovano, A.; Rubolini, D.; Brambilla, S.; Ferrari, N. Winter diet of urban roosting Long-eared Owls Asio otus in northern Italy: the importance of the Brown Rat Rattus norvegicus. Bird Study 2000, 47, 242–244. | Owl pellet analysis | Distribution |
| 2001 | (16) Manganelli, G.; Pezzo, F.; Piazzini, S. Micromys minutus (Mammalia, Rodentia, Muridae) nel comprensorio dei Laghi di Chiusi e Montepulciano (Toscana—Umbria). Atti Soc. Tosc. Sci. Nat. 2001, 108, 109–111. | Owl pellet analysis | Distribution |
| 2004 | (17) Agnelli, P.; Nappi, A.; Maio, N. Conclusive remarks about the synonymy of Mus meridionalis OG Costa, 1844 (Mammalia, Rodentia, Muridae). Ital. J. Zool. 2004, 71, 353–357. | Taxonomic assessment | Taxonomy |
| 2005 | (18) Mazzotti, S.; Lunardi, S. Struttura e fenologia delle comunità della microteriofauna di Valle Brusà. Quad. Staz. Ecol. Civ. Mus. Sto. Nat. Ferrara 2005, 15, 113–124. | Captures | Ecology |
| 2009 | (19) Lapini, L. Small mammals of the Natural Reserve” Lake of Cornino” (Forgaria nel Friuli, Udine, North-Eastern Italy). Gortania 2009, 31, 143–170. | Captures | Distribution |
| 2009 | (20) Gaggi, A.; Paci, A.M. Note sull’orientamento trofico del barbagianni Tyto alba in Umbria. Uccelli d’Italia 2009, 34, 19–34. | Owl pellet analysis | Distribution |
| 2013 | (21) Mazzotti, S.; Tiozzi, E. Impatto dei cambiamenti climatici sulle comunità di micromammiferi (Mammalia: Soricomorpha, Rodentia) del Delta del Po. Quad. Mus. Sto. Nat. Ferrara 2013, 1, 111–117. | Owl pellet analysis | Distribution |
| 2014 | (22) Gaggi, A.; Paci, A.M. Atlante degli Erinaceomorfi dei Soricomorfi e dei Piccoli Roditori dell’Umbria. Dimensione Grafica Snc, Spello (Perugia), Italy, 2014. | Owl pellet analysis | Distribution |
| 2014 | (23) Ferri, V.; Soccini, C.; Battisti, C. Check-List dei Mammiferi della Riserva Naturale di Monticchie (Lodi; Italia Settentrionale)—1985–2012. Nat. Hist. Sci. 2014, 1, 49–54. | Owl pellet analysis | Distribution |
| 2020 | (24) Bini, A.; Cecere, F.; Rossetti, G.; Leandri, F.; Mori, E. Il ruolo dei micromammiferi nella dieta del gufo comune nella Pianura Padana Cremonese. Quad Mus Civ. Sto Nat. Ferrara 2020, 8, 93–99. | Owl pellet analysis | Distribution |
| 2022 | (25) Mori, E.; Bartolini, A.; Zarri, E.; Bini, A.; Bona, F.; Ussi, F.; Viviano, A. On the safe side of the rock: harvest mice still occur in “Padule di Fucecchio” wetland (Tuscany, Central Italy). Atti del XII Congresso Italiano di Teriologia, Cogne (AO), 08-11/06/2022 2022, 1. | Owl pellet analysis | Distribution |
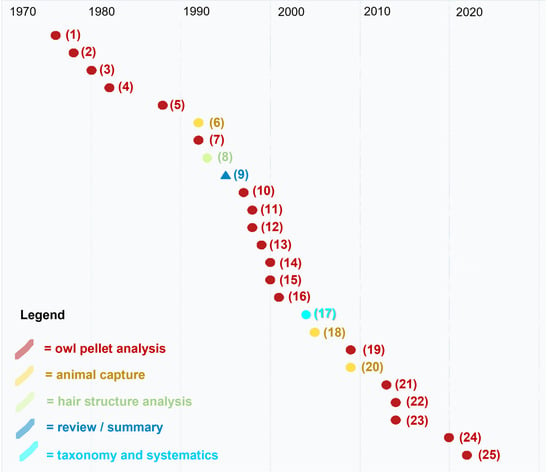
Figure A1.
Published works in 1976–2022 dealing with harvest mice in Italy. Different topics are labeled with different shapes and colours. Numbers refer to publication number shown in Table A1.
References
- Hewitt, G.M. Mediterranean peninsulas: The evolution of hotspots. In Biodiversity Hotspots; Zachos, F.E., Habel, J.C., Eds.; Springer: Amsterdam, The Netherlands, 2011; pp. 123–147. [Google Scholar]
- Amori, G.; Castiglia, R. Mammal endemism in Italy: A review. Biogeographia 2018, 33, 19–31. [Google Scholar] [CrossRef] [Green Version]
- Loy, A.; Aloise, G.; Ancillotto, L.; Angelici, F.M.; Bertolino, S.; Capizzi, D.; Castiglia, R.; Colangelo, P.; Contoli, L.; Cozzi, B.; et al. Mammals of Italy: An annotated checklist. Hystrix 2019, 30, 87–106. [Google Scholar]
- Canestrelli, D.; Cimmaruta, R.; Costantini, V.; Nascetti, G. Genetic diversity and phylogeography of the Apennine yellow-bellied toad Bombina pachypus, with implications for conservation. Molec. Ecol. 2005, 15, 3741–3754. [Google Scholar] [CrossRef]
- Randi, E.; Alves, P.C.; Carranza, J.; Milošević-Zlatanović, S.; Sfougaris, A.; Mucci, N. Phylogeography of roe deer (Capreolus capreolus) populations: The effects of historical genetic subdivisions and recent nonequilibrium dynamics. Mol. Ecol. 2004, 13, 3071–3083. [Google Scholar] [CrossRef]
- Randi, E. Genetics and conservation of wolves Canis lupus in Europe. Mammal Rev. 2011, 41, 99–111. [Google Scholar] [CrossRef]
- Loy, A.; Genov, P.; Galfo, M.; Jacobone, M.G.; Vigna Taglianti, A. Cranial morphometrics of the Apennine brown bear (Ursus arctos marsicanus) and preliminary notes on the relationships with other southern European populations. Ital. J. Zool. 2008, 75, 67–75. [Google Scholar] [CrossRef] [Green Version]
- Bertolino, S.; Colangelo, P.; Mori, E.; Capizzi, D. Good for management, not for conservation: An overview of research, conservation and management of Italian small mammals. Hystrix 2015, 26, 25–35. [Google Scholar]
- Vega, R.; Amori, G.; Aloise, G.; Cellini, S.; Loy, A.; Searle, J.B. Genetic and morphological variation in a Mediterranean glacial refugium: Evidence from Italian pygmy shrews, Sorex minutus (Mammalia: Soricomorpha). Biol. J. Linn. Soc. 2010, 100, 774–787. [Google Scholar] [CrossRef] [Green Version]
- Bannikova, A.A.; Zemlemerova, E.D.; Colangelo, P.; Sözen, M.; Sevindik, M.; Kidov, A.A.; Dzuev, R.I.; Kryštufek, B.; Lebedev, V.S. An underground burst of diversity—A new look at the phylogeny and taxonomy of the genus Talpa Linnaeus, 1758 (Mammalia: Talpidae) as revealed by nuclear and mitochondrial genes. Zool. J. Linn. Soc. 2015, 175, 930–948. [Google Scholar] [CrossRef] [Green Version]
- Santucci, F.; Emerson, B.C.; Hewitt, G.M. Mitochondrial DNA phylogeography of European hedgehogs. Molec. Ecol. 1998, 7, 1163–1172. [Google Scholar] [CrossRef]
- Colangelo, P.; Aloise, G.; Franchini, P.; Annesi, F.; Amori, G. Mitochondrial DNA reveals hidden diversity and an ancestral lineage of the bank vole in the Italian peninsula. J. Zool. 2012, 287, 41–52. [Google Scholar] [CrossRef]
- Chiocchio, A.; Colangelo, P.; Aloise, G.; Amori, G.; Bertolino, S.; Bisconti, R.; Castiglia, R.; Canestrelli, D. Population genetic structure of the bank vole Myodes glareolus within its glacial refugium in peninsular Italy. J. Zool. System. Evol. Res. 2019, 57, 959–969. [Google Scholar] [CrossRef]
- Lo Brutto, S.; Sarà, M.; Arculeo, M. Italian Peninsula preserves an evolutionary lineage of the fat dormouse Glis glis L. (Rodentia: Gliridae). Biol. J. Linn. Soc. 2011, 102, 11–21. [Google Scholar] [CrossRef] [Green Version]
- Mouton, A.; Mortelliti, A.; Grill, A.; Sarà, M.; Kryštufek, B.; Juškaitis, R.; Latinne, A.; Amori, G.; Randi, E.; Büchner, S.; et al. Evolutionary history and species delimitations: A case study of the hazel dormouse, Muscardinus avellanarius. Conserv. Gen. 2017, 18, 181–196. [Google Scholar] [CrossRef]
- Rotondi, C.; Annesi, F.; Amori, G.; Aloise, G.; Mori, E.; Castiglia, R. The “mistery of the Etruscan”: Preliminary data on the phylogeographic relationships of Italian populations of the Etruscan shrew, Suncus etruscus (Savi, 1822). In Proceedings of the Atti del III Convegno Nazionale sui Piccoli Mammiferi, Colle Val d’Elsa, Italy, 8–9 November 2017; Volume 3, p. 37. [Google Scholar]
- Sandor, M.E.; Elphick, C.S.; Tingley, M.W. Extinction of biotic interactions due to habitat loss could accelerate the current biodiversity crisis. Ecol. Appl. 2022, 1, e2608. [Google Scholar] [CrossRef]
- Mortelliti, A.; Amori, G.; Capizzi, D.; Cervone, C.; Fagiani, S.; Pollini, B.; Boitani, L. Independent effects of habitat loss, habitat fragmentation and structural connectivity on the distribution of two arboreal rodents. J. Appl. Ecol. 2011, 48, 153–162. [Google Scholar] [CrossRef]
- Bertolino, S. Distribution and status of the declining garden dormouse Eliomys quercinus. Mammal Rev. 2017, 47, 133–147. [Google Scholar] [CrossRef]
- Chevret, P.; Renaud, S.; Helvaci, Z.; Ulrich, R.G.; Quéré, J.P.; Michaux, J.R. Genetic structure, ecological versatility, and skull shape differentiation in Arvicola water voles (Rodentia, Cricetidae). J. Zool. System. Evol. Res. 2020, 58, 1323–1334. [Google Scholar] [CrossRef]
- Yasuda, S.P.; Vogel, P.; Tsuchiya, K.; Han, S.H.; Lin, L.K.; Suzuki, H. Phylogeographic patterning of mtDNA in the widely distributed harvest mouse (Micromys minutus) suggests dramatic cycles of range contraction and expansion dujring the mid- to late Pleistocene. Can. J. Zool. 2005, 83, 1411–1420. [Google Scholar] [CrossRef] [Green Version]
- Kryštufek, B.; Lunde, D.P.; Meinig, H.; Aplin, K.; Batsaikhan, N.; Henttonen, H. Micromys minutus. The IUCN Red List of Threatened Species 2019. E.T13373A119151882. Available online: https://dx.doi.org/10.2305/IUCN.UK.2019-1.RLTS.T13373A119151882.en (accessed on 26 May 2022).
- Harris, S. History, distribution, status and habitat requirements of the harvest mouse in Britain. Mammal Rev. 1979, 4, 159–171. [Google Scholar] [CrossRef]
- Canova, L. Distribution and habitat preference of small mammals in a biotope of the North Italian plain. Ital. J. Zool. 1992, 59, 417–420. [Google Scholar] [CrossRef] [Green Version]
- Marks, M.; Lapin, B.; Randall, J. Phragmites australis (P. communis): Threats, management, and monitoring. Nat. Areas J. 1994, 14, 285–294. [Google Scholar]
- Darinot, F. The harvest mouse (Micromys minutus Pallas, 1771) as prey: A literature review. J. Vert. Biol. 2016, 65, 117–137. [Google Scholar]
- Råberg, L.; Loman, J.; Hellgren, O.; van der Kooij, J.; Isaksen, K.; Solheim, R. The origin of Swedish and Norwegian populations of the Eurasian harvest mouse (Micromys minutus). Acta Theriol. 2013, 58, 101–104. [Google Scholar] [CrossRef]
- Abramov, A.V.; Meschersky, I.G.; Rozhnov, V.V. On the taxonomic status of the harvest mouse Micromys minutus (Rodentia: Muridae) from Vietnam. Zootaxa 2009, 2199, 58–68. [Google Scholar] [CrossRef] [Green Version]
- Wauters, L.A.; Amori, G.; Aloise, G.; Gippoliti, S.; Agnelli, P.; Galimberti, A.; Casiraghi, M.; Preatoni, D.; Martinoli, A. New endemic mammal species for Europe: Sciurus meridionalis (Rodentia, Sciuridae). Hystrix 2017, 28, 1–8. [Google Scholar]
- Vogel, P.; Gander, A. Live trapping design for the harvest mouse (Micromys minutus) in its summer habitat. Rev. Suisse Zool. 2020, 122, 143–148. [Google Scholar]
- Amori, G.; Contoli, L.; Nappi, A. Mammalia II: Erinaceomorpha, Soricomorpha, Lagomorpha, Rodentia; Calderini Editions: Bologna, Italy, 2008. [Google Scholar]
- Bon, M. Nuovo Atlante dei Mammiferi del Veneto; WBA Monographs Editions: Verona, Italy, 2017. [Google Scholar]
- Deflorian, M.C.; Caldonazzi, M.; Zanghellini, S.; Pedrini, P. Atlante dei Mammiferi della provincia di Trento; Monografie del Museo delle Scienze 6; Soc. Veneziana di Scienze Naturali: Trento, Italy, 2019. [Google Scholar]
- Mori, E.; Bartolini, A.; Zarri, E.; Bini, A.; Bona, F.; Ussi, F.; Viviano, A. On the safe side of the rock: Harvest mice still occur in “Padule di Fucecchio” wetland (Tuscany, Central Italy). In Proceedings of the Atti del XII Congresso Italiano di Teriologia, Cogne, Italy, 8–11 June 2022. [Google Scholar]
- Buhay, J.E. COI-like sequences are becoming problematic in molecular systematic and DNA barcoding studies. J. Crustacean Biol. 2009, 29, 96–110. [Google Scholar] [CrossRef]
- Song, H.; Buhay, J.E.; Whiting, M.F.; Crandall, K.A. Many species in one: DNA barcoding overestimates the number of species when nuclear mitochondrial pseudogenes are coamplified. Proc. Nat. Acad. Sci. USA 2008, 105, 13486–13491. [Google Scholar] [CrossRef] [Green Version]
- Darriba, D.; Taboada, G.L.; Doallo, R.; Posada, D. jModelTest 2: More models, new heuristics and parallel computing. Nat. Methods 2012, 30, 772. [Google Scholar] [CrossRef] [Green Version]
- Ronquist, F.; Huelsenbeck, J.P. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 2003, 19, 1572–1574. [Google Scholar] [CrossRef] [Green Version]
- Gouy, M.; Guindon, S.; Gascuel, O. SeaView version 4: A multiplatform graphical user interface for sequence alignment and phylogenetic tree building. Mol. Biol. Evol. 2010, 27, 221–224. [Google Scholar] [CrossRef] [Green Version]
- Clement, M.; Posada, D.; Crandall, K.A. TCS: A computer program to estimate gene genealogies. Mol. Ecol. 2000, 9, 1657–1660. [Google Scholar] [CrossRef] [Green Version]
- Bandelt, H.J.; Forster, P.; Röhl, A. Median-joining networks for inferring intraspecific phylogenies. Mol. Biol. Evol. 1999, 16, 37–48. [Google Scholar] [CrossRef]
- Santos, A.M.; Cabezas, M.P.; Tavares, A.I.; Xavier, R.; Branco, M. tcsBU: A tool to extend TCS network layout and visualization. Bioinformatics 2016, 32, 627–628. [Google Scholar]
- Rambaut, A.; Drummond, A.J.; Xie, D.; Baele, G.; Suchard, M.A. Posterior summarization in Bayesian phylogenetics using Tracer 1.7. System. Biol. 2018, 67, 901–904. [Google Scholar] [CrossRef] [Green Version]
- Bouckaert, R.; Heled, J.; Kühnert, D.; Vaughan, T.; Wu, C.H.; Xie, D.; Suchard, M.A.; Rambaut, A.; Drummond, A.J. BEAST 2: A software platform for Bayesian evolutionary analysis. PLoS Comp. Biol. 2014, 10, e1003537. [Google Scholar] [CrossRef] [Green Version]
- Martin, Y.; Gerlach, G.; Schlötterer, C.; Meyer, A. Molecular phylogeny of European muroid rodents based on complete cytochrome b sequences. Mol. Phyl. Evol. 2000, 16, 37–47. [Google Scholar]
- Arbogast, B.S.; Browne, R.A.; Weigel, P.D. Evolutionary genetics and Pleistocene biogeography of North American tree squirrels (Tamiasciurus). J. Mammal. 2001, 82, 302–319. [Google Scholar]
- Tamura, K.; Battistuzzi, F.U.; Billing-Ross, P.; Murillo, O.; Filipski, A.; Kumar, S. Estimating divergence times in large molecular phylogenies. Proc. Natl. Acad. Sci. USA 2012, 109, 19333–19338. [Google Scholar] [CrossRef] [Green Version]
- Drummond, A.J.; Suchard, M.A.; Xie, D.; Rambaut, A. Bayesian Phylogenetics with BEAUti and the BEAST 1.7. Mol. Biol. Evol. 2012, 29, 1969–1973. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rambaut, A. FigTree. Ver.1.4. 2; University of Edinburgh Press: Edinburgh, UK, 2012. [Google Scholar]
- Nicolas, V.; Ndiaye, A.; Benazzou, T.; Souttou, K.; Delapre, A.; Denys, C. Phylogeography of the North African dipodil (Rodentia: Muridae) based on cytochrome-b sequences. J. Mammal. 2014, 95, 241–253. [Google Scholar] [CrossRef] [Green Version]
- Kim, H.R.; Park, Y.C. Genetic diversity and genetic structure of the striped field mouse Apodemus agrarius coreae (Muridae, Rodentia) in Korea. Gene 2015, 572, 292–297. [Google Scholar] [CrossRef] [PubMed]
- Barbosa, S.; Pauperio, J.; Searle, J.B.; Alves, P.C. Genetic identification of I berian rodent species using both mitochondrial and nuclear loci: Application to noninvasive sampling. Mol. Ecol. Res. 2013, 13, 43–56. [Google Scholar] [CrossRef] [PubMed]
- Bryja, J.; Mikula, O.; Patzenhauerová, H.; Oguge, N.O.; Šumbera, R.; Verheyen, E. The role of dispersal and vicariance in the pleistocene history of an e ast a frican mountain rodent, p raomys delectorum. J. Biogeogr. 2014, 41, 196–208. [Google Scholar] [CrossRef]
- McDonough, M.M.; Šumbera, R.; Mazoch, V.; Ferguson, A.W.; Phillips, C.D.; Bryja, J. Multilocus phylogeography of a widespread savanna–woodland-adapted rodent reveals the influence of Pleistocene geomorphology and climate change in Africa’s Zambezi region. Mol. Ecol. 2015, 24, 5248–5266. [Google Scholar] [CrossRef]
- Ghawar, W.; Chaouch, M.; Ben Abderrazak, S.; Snoussi, M.A.; Salem, S.; Chouchen, S.; Bouaoun, A.; Salah, A.B.; Bettaieb, J. Evaluation of the Taxonomic Status of Lesser Egyptian Jerboa, Jaculus jaculus: First Description of New Phylogroups in Tunisia. Animals 2022, 12, 758. [Google Scholar] [CrossRef]
- Ellerman, J.R.; Morrison-Scott, T.C.S. Checklist of Palaearctic and Indian Mammals 1758 to 1946; Natural History Museum: London, UK, 1951. [Google Scholar]
- Hewitt, G.M. Some genetic consequences of ice ages, and their role in divergence and speciation. Biol. J. Linn. Soc. 1996, 58, 247–276. [Google Scholar] [CrossRef]
- Bon, M.; Trabucco, R.; Delfino, M. La fauna del Pozzo Romano di Lova (Laguna di Venezia, I sec. a.C.—I sec- d. C.). Boll. Mus. Civ. Sto. Nat. Venezia 2001, 51, 159–185. [Google Scholar]
- Spitzenberger, F. Micromys minutus (Pallas, 1771). In The Atlas of European Mammals; Mitchell-Jones, A.J., Amori, G., Bogdanowicz, W., Krystufek, B., Reijnders, P.J.H., Spitzenberger, F., Stubbe, M., Thissen, J.B.M., Vohralik, V., et al., Eds.; Academic Press: London, UK, 1999; pp. 264–265. [Google Scholar]
- Musser, G.G.; Carleton, M.D. Family Muridae. In Mammal Species of the World. A Taxonomic and Geographic Reference; Wilson, D.E., Reeder, D.H., Eds.; Smithsonian Institute Press: Washington, DC, USA, 1993; pp. 501–755. [Google Scholar]
- Mamuris, Z.; Sfougaris, A.I.; Stamatis, C. Genetic structure of Greek brown hare (Lepus europaeus) populations as revealed by mtDBNA RFLP-PCR analysis: Implications for conserving genetic diversity. Biol. Conserv. 2001, 101, 187–196. [Google Scholar] [CrossRef]
- Darinot, F.; Le Petitcorps, Q.; Arnal, V.; Coulon, A.; Montgelard, C. Effects of landscape features and flooding on the genetic structure of a small wetland rodent, the harvest mouse (Micromys minutus). Landsc. Ecol. 2021, 36, 1755–1771. [Google Scholar] [CrossRef]
- Cagnacci, F.; Cardini, A.; Ciucci, P.; Ferrari, N.; Mortelliti, A.; Preatoni, D.G.; Russo, D.; Scandura, M.; Wauters, L.A.; Amori, G. Less is more: Researcher survival guide in times of economic crisis. Hystrix 2012, 23, 1–7. [Google Scholar]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).